Abstract
Small cell lung cancer (SCLC) is an aggressive form of lung cancer for which therapeutic options are very limited. A new study (Augert et al. Sci. Signal. 2019;12: eaau2922) shows that inhibition of the LSD1 demethylase can induce derepression of NOTCH receptor genes and subsequent activation of the NOTCH pathway, potently suppressing the growth of SCLC.
The NOTCH signaling pathway is a cell-communication pathway that controls cell fate decisions during development and regeneration. In cancer, NOTCH signaling can be either oncogenic or tumor-suppressive, and a major issue in the field has been to develop strategies to activate or inhibit the NOTCH pathway depending on the context (reviewed in [1]). A recent study from Augert et al. in Science Signaling identifies a new way to activate NOTCH signaling in cancer cells [2].
Small cell lung cancer (SCLC) is a highly aggressive form of lung malignancy with dismal survival rates. Resection is used in the few cases detected before metastatic spread and when lungs are sufficiently healthy for the patient to undergo surgery. Chemotherapy is standard-of-care, in conjunction with radiation therapy, but is only transiently efficient. Emerging immunotherapies may be effective in a limited number of cases. Overall, there is a great need for novel targeted therapies (reviewed in [3]).
SCLC is a neuroendocrine (NE) cancer, and strong evidence indicates that NOTCH pathway activation and NE cell fate are antagonistic during development and regeneration, including in the lungs (reviewed in [4]). One mechanism by which NOTCH inhibits NE cell fate is by inhibition and downregulation of ASCL1 (Achaete-Scute family BHLH transcription factor 1), a transcription factor that controls NE gene programs and is also crucial for the long-term survival of SCLC cells [5]. Accordingly, genomic sequencing of human SCLC identified recurrent inactivating mutations in genes encoding NOTCH receptors, suggesting that NOTCH signaling is tumor-suppressive in SCLC [6]. Indeed, inhibition of NOTCH signaling promotes the development of SCLC-like tumors from normal human cells in culture [7]. Conversely, activation of NOTCH in SCLC cells results in decreased proliferation, increased cell death, and a switch to a non-NE state, accompanied by inhibition of ASCL1 and upregulation of REST (RE1-silencing transcription factor, also known as NRSF, neuron-restrictive silencer factor), a repressor of NE gene programs ([6,8] and references therein). These observations indicate that activation of NOTCH signaling in SCLC cells would provide a novel therapeutic option to inhibit this neuroendocrine cancer, but until now such a strategy had not been possible.
SCLC exhibits frequent mutations in chromatin-regulating genes, suggesting that an altered epigenetic landscape is one of the driving mechanisms behind its development [6]. In their recent work, Augert, Eastwood, and colleagues investigated the functional role of an epigenetic regulator, LSD1 (lysine-specific histone demethylase 1A), which controls gene expression by coordinating histone methylation and acetylation, and which is highly expressed in SCLC. Whereas inhibition of LSD1 has been shown to have some therapeutic efficacy in preclinical studies in acute myeloid leukemia (AML) and SCLC models, the underlying mechanisms have been unclear ([9] for a review on LSD1).
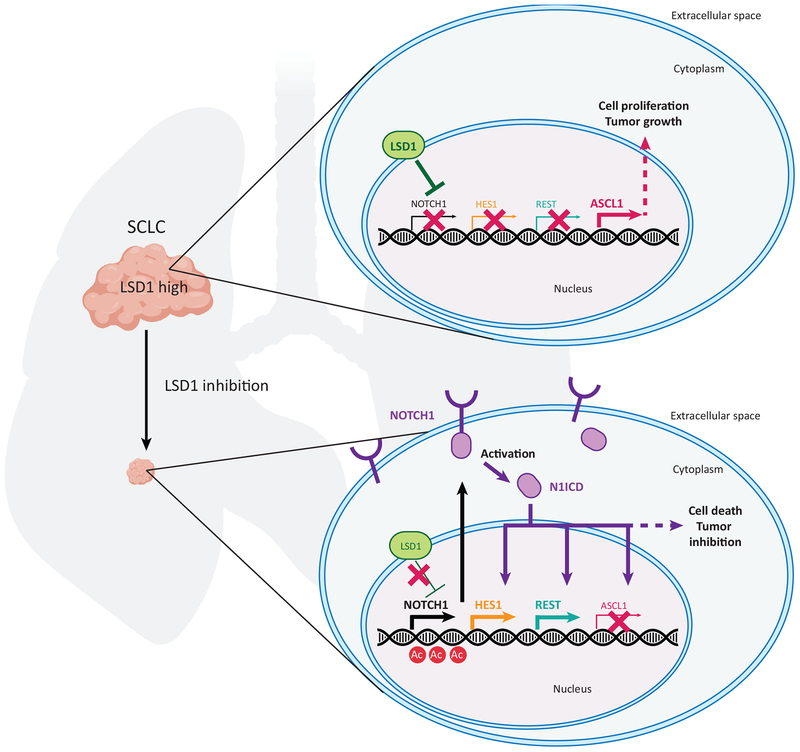
To address this question the authors analyzed transcriptional changes in a panel of SCLC cell lines and patient-derived xenograft (PDX) models treated with the LSD1 inhibitor ORY-1001 [2] (Figure 1). Several of these models were sensitive to the LSD1 inhibitor, with a potent inhibitory effect on viability. Gene ontology analyses revealed strong enrichment in NOTCH signaling pathways, with upregulation of the expression of multiple NOTCH receptor family member genes, especially NOTCH1, and NOTCH target genes including REST/NRSF, whereas ASCL1 expression levels were decreased. Association between LSD1 and the NOTCH pathway was further validated using genetic inhibition of LSD1, which increased the expression of NOTCH1 and its target gene HES1, as well as of REST, but decreased ASCL1 expression. Decreased LSD1, increased NOTCH1, and decreased ASCL1 all led to decreased SCLC expansion. Importantly, pharmacological inhibition of NOTCH signaling partially rescued cells from growth suppression induced by the LSD1 inhibitor. Thus, although it is likely that LSD1 inhibition has broad consequences for the chromatin structure of cells, activation of NOTCH downstream of LSD1 inhibition is a crucial contributor to the cell death observed in SCLC cells. Activation of the transcription of NOTCH receptor genes was not associated with changes in the deposition of histone H3 dimethylated on lysine 4 (H3K4me2), which would have been expected based on LSD1 intrinsic enzymatic activity [9], but instead was associated with changes in histone acetylation (i.e., H3K27Ac), which is likely due to functional interactions between LSD1 and protein complexes that include histone deacetylase activity.
Figure 1. The LSD1–NOTCH–ASCL1 Axis in Small Cell Lung Cancer (SCLC).
High levels of the LSD1 epigenetic regulator correlate with tumor growth in SCLC, a neuroendocrine cancer in which the NOTCH pathway is tumor-suppressive and the ASCL1 transcription factor is oncogenic. Upon inhibition of LSD1, the expression of genes coding for NOTCH receptors, especially NOTCH1, is derepressed by increased histone acetylation (Ac) in regulatory regions. Higher levels of NOTCH1 result in activation of the NOTCH signaling pathway after cleavage of the receptor to generate the intracellular form of NOTCH1 (N1ICD), which acts as a transcription factor. The subsequent induction of NOTCH target genes (e.g., HES1, REST) and repression of ASCL1 results in increased cell death and potent inhibition of SCLC growth.
These experiments raise several new questions. First, before LSD1 inhibition can be implemented as a new therapeutic strategy in SCLC patients, a major issue will be the identification of biomarkers of response to this inhibition. The absence of ASCL1 expression may be a first biomarker indicative of low/no response to LSD1 inhibition, as shown by the authors with one PDX model. It is also possible that different receptors, ligands, regulators, and mediators of the NOTCH pathway can help to predict how individual tumors will respond to LSD1 inhibition. Second, SCLC tumors are often heterogeneous and are composed of NE and non-NE populations, including non-NE cells generated via endogenous NOTCH pathway activation [8]. It would be interesting to know whether LSD1 inhibition will affect the ratio between these NE and non-NE populations of cancer cells within SCLC tumors. Because the NE/non-NE ratio in SCLC tumors may affect the response to chemotherapy and other stresses [8], combination therapies with LSD1 inhibitors will need to be carefully designed to avoid generating tumors that may become more resistant to standard-of-care treatment. Third, although SCLC and type I large cell neuroendocrine carcinoma (LCNEC) exhibit low NOTCH expression, type II LCNECs with high NOTCH expression have been characterized [10]. It is possible that treatment with LSD1 inhibitors and NOTCH activation may eventually transform SCLC tumors into type II LCNEC tumors. Finally, although the authors provide strong genetic evidence that NOTCH signaling is a major mediator of the antitumor effects of LSD1 inhibition, it will be important to identify NOTCH- independent effects of inhibiting LSD1, and this may help to design better ways to treat SCLC or block resistance mechanisms.
Together, these elegant experiments define a new LSD1–NOTCH–ASCL1 axis in SCLC cells. Inhibition of LSD1 can lead to the reactivation of NOTCH signaling and the subsequent inhibition of ASCL1, which normally serves as a driver of the expansion of these NE cancer cells. These studies represent a promising strategy to help SCLC patients.
References
- 1.Bray SJ (2016) Notch signalling in context. Nat. Rev. Mol. Cell Biol 17, 722–735 [DOI] [PubMed] [Google Scholar]
- 2.Augert A et al. (2019) Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci. Signal 12, eaau2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zimmerman S et al. (2019) 2017–2018 scientific advances in thoracic oncology: small cell lung cancer. J. Thorac. Oncol Published online February 11, 2019. 10.1016/j.jtho.2019.01.022 [DOI] [PubMed] [Google Scholar]
- 4.Collins BJ et al. (2004) Notch in lung development and lung cancer. Semin. Cancer Biol 14, 357–364 [DOI] [PubMed] [Google Scholar]
- 5.Augustyn A et al. (2014) ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proc. Natl. Acad. Sci. U. S. A 111, 14788–14793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.George J et al. (2015) Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HJ et al. (2019) Generation of pulmonary neuroendocrine cells and SCLC-like tumors from human embryonic stem cells. J. Exp. Med Published online February 8, 2019. 10.1084/jem.20181155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim JS et al. (2017) Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545, 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiques-Diaz A and Somervaille TC (2016) LSD1: biologic roles and therapeutic targeting. Epigenomics 8, 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George J et al. (2018) Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun 9, 1048. [DOI] [PMC free article] [PubMed] [Google Scholar]