Abstract
Extracellular vesicle (EV)-mediated intercellular communication acts as a critical culprit in cancer development. The selective packaging of oncogenic molecules renders tumor-derived EVs capable of altering the tumor microenvironment and thereby modulating cancer developments that may contribute to drug resistance and cancer recurrence. Moreover, the molecular and functional characteristics of cancer through its development and posttreatment evolve over time. Tumor-derived EVs are profoundly involved in this process and can, therefore, provide valuable real-time information to reflect dynamic changes occurring within the body. Because they bear unique molecular profiles or signatures, tumor-derived EVs have been highlighted as valuable diagnostic and predictive biomarkers as well as novel therapeutic targets. In addition, the use of an advanced EV-based drug delivery system for cancer therapeutics has recently been emphasized in both basic and clinical studies. In this review, we highlight comprehensive aspects of tumor-derived EVs in oncogenic processes and their potential clinical applications.
Keywords: Extracellular vesicles, exosome, tumor-derived EVs, homeostasis, tumor microenvironment, EV isolation, biomarker, drug delivery
Background
Basic characteristics of EVs
EVs are transportable vesicles that participate in the exchange of biological molecules between cells. They are pivotal in maintaining cellular and body homeostasis [1, 2]. The transfer of EVs serves as an efficient and specific delivery system that carries different types of cellular cargo, such as nucleic acids, lipids, proteins, and metabolites, to their target destinations [3]. Exosomes are defined as a specific subset of EVs that range from 30 to 150 nm in size. They are originally endosomal-derived intraluminal vesicles (ILVs) that are subsequently released into the extracellular milieu through the fusion of multivesicular endosomes or multivesicular bodies (MVBs) with the plasma membrane [4–6]. Exosomes retain a conserved series of proteins that are shared with the secreting cell during their biogenesis. However, their uniqueness stems from the variety of molecular constituents and lipid contents derived from their cell of origin and status. Given their intricate characteristics, exosomes display potent influence on recipient cells and show promises in revealing cell-to-cell communication.
In 1877, serum particles were investigated and described as “motes floating in the sunlight” by Edmunds [7]. Later, Peter Barland et al. investigated the structure of cellular vesicles under an electron microscope [8]. However, the function of these cellular vesicles remained unclear until 1967, when Peter Wolf identified lipid-rich particles that displayed coagulant properties that he suggested had originated from the granules of platelets [9]. In 1981, the term “exosome” was first coined to describe extracellular vesicles with an average diameter of 500 to 1000 nm [10]. Later, the Johnstone group and the Stahl team independently reported that bioactive molecules shed from reticulocytes, such as transferrin receptors, were incorporated within vesicles (approximately 50 nm in diameter) and released by exocytosis in MVBs [11–13]. In 1987, Rose M. Johnstone et al. further described the functional link between exosomes and reticulocyte maturation [6]. Although exosomes are secreted by a wide range of mammalian cell types [4, 5], exosomes enclose limited cytosol from their parent cells with their lipid bilayers and are devoid of cellular organelles. The compositions of exosomes reflect the physiological and/or pathological states of their parent cells and are associated with their environmental conditions and/or stimuli [14]. Moreover, the distinct contents of exosomes heavily depend on their parent cell types and functions, which suggests that exosomes have the unique property of cargo selectivity [15]. Currently, exosomes can be isolated from almost all types of cells and various physiological and pathological fluids, such as blood, saliva, milk, urine, cerebrospinal fluid, ascites, tears, and pleural effusions [16–18].
Indeed, exosomes, the predominant form of microvesicles, are stable and abundant in bodily fluids (> 109 vesicles/mL of blood) [19]. Cancer cells especially secrete more exosomes than healthy cells [20], suggesting that exosomes function as critical mediators of cancer development.
EVs in homeostasis
EVs maintain cellular homeostasis by transporting bioactive and/or regulatory molecules between cells and tissues. For instance, exosome secretion ablates the harmful cytoplasmic accumulation of nuclear deoxyribonucleic acid (DNA) in cells by preventing the aberrant innate immune response [21]. Consistently, the inhibition of exosome secretion, such as the depletion of alpha-1,3/1,6-mannosyltransferase (ALG2)-interacting protein X (Alix) or Rab27a, induced the cytoplasmic accumulation of nuclear DNA and subsequently activated a stimulator of the interferon genes, a cytoplasmic double stranded DNA (dsDNA) sensor, contributing to the reactive oxygen species-dependent DNA damage response [21]. In neural EVs, cysteine string protein α, which regulates refolding pathways at the synapse, is involved in the EV-mediated cellular export of disease-associated proteins such as polyglutamine expanded protein 72Q huntingtinex°n1 or superoxide dismutase-1G93A [22]. In contrast, a loss-of-function mutation of cysteine string protein α ablated the EV-mediated cellular export of disease-associated proteins [22], suggesting the critical role of the EV-mediated removal of toxic proteins in neurons.
The interaction between receptor activator of nuclear factor-κB-ligand (RANKL)/ receptor activator of nuclear factor -κB (RANK) induces osteoclast differentiation and function in bone homeostasis. Osteoblast-derived EVs have been shown to participate in this process by transferring RANKL to osteoclast precursors to promote osteoclast formation [23]. In contrast, RANK-enriched EVs regulated bone homeostasis by competing with RANK to interact with RANKL on the surfaces of osteoclasts [24]. Recently, maturing osteoclasts-derived small EVs that contain RANK have been shown to induce RANKL reverse signaling in osteoblasts via the activation of Runt-related transcription factor 2 to facilitate bone formation [25]. Together, these studies indicate the importance of EVs in the maintenance of the homeostatic cellular balance.
EVs in cancer communication
EVs regulate the dynamic and functional communication between cancer stem cells and cancer cells/the tumor microenvironment during cancer development [26]. Indeed, exosomes secreted by C-X-C chemokine receptor type 4 (CXCR4)-overexpressing breast cancer cells exhibited high levels of stemness-related markers and metastatic-related messenger ribonucleic acids (mRNAs) [27]. Moreover, recipient cells treated with exosomes derived from CXCR4-overexpressing cells also demonstrated high expression levels of stemness-related markers and an increase in the invasive ability and metastatic potential of cancer cells [27]. Moreover, adipose-derived mesenchymal stem cells secrete exosomes to facilitate cancer migration and proliferation in a wingless/integrated (Wnt)/β-catenin signal-dependent manner [28]. In colorectal cancer, exosome derived from cancer-associated fibroblasts have been shown to prime cancer stem cells and to contribute to drug resistance and chemoresistance through Wnt signaling [29]. Additionally, the chemotherapeutic agent gemcitabine induced the upregulation and secretion of miR-146a and Snail in cancer-associated fibroblast-derived exosomes, facilitating proliferation and drug resistance in recipient pancreatic cancer cells [30]. During gemcitabine treatment, the inhibition of exosome generation by the inactivation of neutral sphingomyelinase significantly reduced the survival of cocultured pancreatic cancer cells [30]. Fibroblast-derived exosomes that contain Wnt have been shown to contribute to chemotherapy resistance by restoring cancer stem cell characteristics in colorectal cancer cells in a Wnt/β-catenin signal-dependent manner [31], suggesting an important effect of EVs derived from the tumor microenvironment on drug resistance in cancer. Moreover, chemotherapeutic drugs, such as paclitaxel or doxorubicin, elicited EVs enriched in annexin A6 that were shown to active endothelial cells, induce pulmonary C-C motif chemokine ligand 2 expression, and increase C-C chemokine receptor type 2-positive monocyte expansion, leading to mammary tumor metastasis [32]. These studies indicated the critical role of EVs in cell-to-cell communication during cancer development.
Association between tumor-derived EVs and cancer development
Tumor-derived EVs with protumorigenic activity regulate cancer development by promoting cancer aggressiveness, cancer invasiveness, the remodeling of the extracellular matrix, angiogenesis, drug resistance, and immunosuppression [14, 33], suggesting the important effects of tumor-derived EVs on cancer development and cancer therapy.
The transfer of metastatic components (i.e., oncogenic proteins or oncogenic microRNAs, oncomiRs) can trigger and reprogram signaling cascades, phenotypes, and the functions of recipient cells [34, 35]. Tumor-derived EVs, especially exosomes, with prometastatic effects can also govern the pathogenesis of cancer invasion and metastasis. For example, exosomal cluster of differentiation 44 (CD44) is transferred from ovarian cancer cells to peritoneal mesothelial cells, which subsequently results in the mesenchymal and spindle morphology of peritoneal mesothelial cells and contributes to cancer invasion [36]. Indeed, numerous studies have shown exosomes to be involved in epithelial–mesenchymal-transition (EMT) during the development of malignant cancer [37]. The uptake of pancreatic cancer-derived exosomes by Kupffer cells elicited premetastatic niche formation through increasing transforming growth factor β (TGF-β) secretion and fibronectin expression by hepatic stellate cells and subsequently promoted liver metastasis [38, 39]. Moreover, tumor-derived exosomal miR-1247-3p has been shown to activate the β1-integrin/nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling axis, resulting in the activation of cancer-associated fibroblasts to foster the metastasis of liver cancer to the lung [34]. The transfer of EV microRNAs, such as miR-200, from highly metastatic breast cancer cells to poorly metastatic cells altered gene expression and facilitated mesenchymal-to-epithelial transition, which promoted metastasis within the weaker metastatic cells [40]. Moreover, highly metastatic melanoma-derived exosomes exhibited a prometastatic phenotype caused by an increase in MET expression in educating bone marrow progenitor cells [41]. Indeed, exosomes derived from highly metastatic melanomas altered their metastatic activity to produce poorly metastatic melanomas [42], implicating the strong connection between exosomes and cancer malignancy. Furthermore, tumor-derived exosomes promoted hypoxia-driven pro-angiogenic tumor responses [43] during cancer development, indicating the importance of tumor-derived exosome in the regulation of sustained angiogenesis. The transfer of miR-130a from gastric cancer cells to vascular endothelial cells through exosomes facilitated angiogenesis and cancer growth by targeting c-MYB [44]. It is believed that EVs function as critical mediators of cancer development and malignancy. The following describes in detail the contents of EVs, their isolation and the analytical approaches used in tumor-derived EV studies.
EV contents
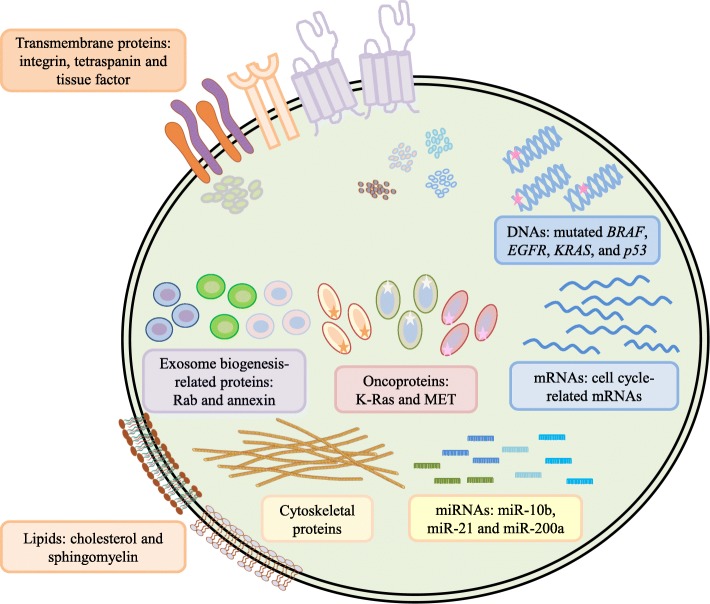
EVs containing varied molecular cargos (Figure 1), such as nucleic acids (i.e., DNAs, mRNAs, microRNAs, long noncoding RNAs and many noncoding RNAs), proteins, lipids, and metabolites, are directly internalized by recipient cells, leading to morphological and functional changes in the recipient cells [14, 33]. It has been highlighted in recent years that the transfer of oncogenic cargo through EVs and tumor-derived exosomes drives oncogenic signal transduction cascades in association with the development of cancer malignancies and tumor microenvironments [5, 35]. Table 1 lists the functional effects of EV cargos during cancer development.
Figure 1.
Summary of diverse bioactive molecules in tumor-derived EVs. Tumor-derived EVs are phospholipid bilayer-enclosed vesicles that contain diverse bioactive molecules. These bioactive molecules can be divided into general groups, such as nucleic acids, proteins, lipids, and metabolites. The nucleic acid group contains mutated oncogenes/ tumor suppressor genes, cell cycle-related mRNAs, and cancer-related miRNAs. The protein group can be divided into several subgroups: transmembrane proteins, growth factors, exosome biogenesis-related proteins, oncoproteins, and cytoskeletal proteins. Among members of the lipid group, cholesterol, sphingomyelin, prostaglandins, and leukotrienes can be detected in EVs. Some amino acids, pyruvate, lactate, and TCA-cycle intermediates are included in the metabolite group
Table 1.
Functional effects of EV cargos in cancers
| EV cargo | Type of cargo | Functional effects | Reference |
|---|---|---|---|
| Amino acids or TCA-cycle intermediates | Metabolites | Promote cancer growth | [74] |
| Annexin A6 | Protein | Pre-mPremetastatic niche formation | [31] |
| Integrins | Protein | Organotropic metastasis | [39] |
| Lethal-7 miRNA family | miRNA | Maintain the highly metastatic tumorigenic phenotype | [56] |
| MET | Protein | Pre-mPremetastatic niche formation | [41] |
| miR-223 | miRNA | Enhance cancer invasion | [57] |
| miR-10b and miR-21 | miRNA | Regulate cancer development | [59] |
| Mitochondrial DNA | Regulate escape from dormancy in therapy-resistant cancers | [47] | |
| Mutant K-Ras | Protein | Enhance three-dimensional growth of cells | [68] |
| TGF-β | Protein | Promote fibroblast-myofibroblast differentiation | [64, 65] |
| Tissue factor | Protein | Regulate pro-coagulant activity of endothelial cells | [66] |
DNAs
Oncogenes and tumor suppressor genes are key mediators during cancer progression and malignancy. Several studies have indicated that dsDNAs represent the largest proportion of exosomal DNAs (exoDNAs) in tumor-derived exosomes [45]. Indeed, double-stranded genomic DNA spanning all chromosomes has been detected in exosomes [46]. Moreover, fragments of mutated KRAS and p53, the most frequently mutated oncogenes or tumor suppressor genes, within exosomes derived from pancreatic cancer cells have been investigated in clinical studies [46]. Additionally, exoDNAs contain similar mutations, such as BRAF (V600E) and mutated epidermal growth factor receptor (EGFR), as the cancer cell lines from which they originated [45], suggesting the potential role of exoDNAs as alternative biomarkers in the detection and diagnosis of cancers. Additionally, the complete circular mitochondrial genome packaged within exosomes has been shown to regulate exit from therapy-induced metabolic dormancy in hormonal therapy-resistant breast cancer [47]. Nevertheless, how genomic DNA or mitochondrial DNA is packaged into exosomes and the regulatory mechanisms or functional consequences of exosomal DNA in recipient cells remain controversial. Indeed, studies reported that retrotransposon elements present in EVs potentially had a genetic influence on disease development with low efficiency [48, 49].
mRNAs
mRNAs within tumor-derived EVs influence the translational profiles of recipient cells during tumor progression [50, 51]. In 2008, Skog et al. showed that functional mRNAs incorporated into EVs were delivered to and translated by recipient cells [50]. Functionally, EVs containing mRNAs for oncogenic proteins were enriched in angiogenic proteins and induced tubule formation in recipient endothelial cells [50]. Additionally, the enrichment of cell cycle-related mRNAs leading to endothelial cell proliferation was investigated in the transcriptome of human colorectal cancer-derived EVs [52]. Of note, fewer than one copy of nonribosomal RNA was indicated per EV, suggesting that the uptake of abundant tumor-derived EVs as well as EV-RNAs by recipient cells might be essential for the functional effects of EV-RNAs on recipient cells [53]. Mutant mRNA variants and miRNAs signatures found in glioblastoma-derived EVs were clinically detected in only patients with glioblastoma [50]. These studies emphasize that tumor-derived EVs with the oncogenic characteristics of the host serve as effective biomarkers for cancer diagnosis and potential therapeutic targets.
MicroRNAs
MicroRNAs (miRNAs) are small noncoding RNAs that inhibit gene expression by binding to the 3’ untranslated regions (UTRs) of messenger RNAs, which subsequently leads to mRNA destabilization, translational inhibition, or mRNA degradation [54]. During physiological and pathological processes, the transfer of miRNAs (i.e., oncomiRs) through EVs mediates cell-to-cell communication. Indeed, the encapsulation of miRNAs within exosomes protects miRNAs from degradation, thereby increasing the diagnostic value of miRNAs contained in exosomes in cancer pathogenesis [55].
Several studies have investigated the enrichment of selective miRNAs in EVs in malignant cancers. For example, Ohshima et al. found that members of the enriched lethal-7 (let-7) miRNA family, tumor suppressors targeting oncogenic Ras, were selectively enriched in only highly metastatic gastric cancer-derived exosomes, leading to the maintenance of a highly metastatic tumorigenic phenotype by exosome-mediated clearance [56]. Additionally, tumor-associated macrophages (TAMs) release EVs containing miRNAs (i.e., miR-223) that enhance the invasiveness of breast cancer through the myocyte enhancer factor 2C/β-catenin pathway [57]. Since angiogenesis facilitates the process of cancer malignancy, angiogenesis regulated by EV miRNAs has been highlighted in recent studies [58]. EVs released from renal cancer stem cells stimulated angiogenesis through the upregulation of angiogenic factors (i.e., vascular endothelial growth factor) or extracellular matrix (ECM) degradation/remodeling enzymes (i.e., matrix metallopeptidase 2 (MMP2) and MMP9) in premetastatic lung niches, leading to lung metastasis [58]. Of note, the association between miRNAs (i.e., miR-10b and miR-21) and the RNA-induced silencing complex-loading complex in breast cancer-derived exosomes processes precursor miRNAs into mature miRNAs in a cell-independent manner, resulting in cancer development [59].
Interestingly, the opposite effect of miRNAs within EVs during cancer development was observed in glioblastoma. The pro-oncogenic effects of tumor-derived EVs were ablated by miR-1 which targets the abundant protein annexin A2 in glioblastoma-derived EVs, leading to tumor suppression of the glioblastoma microenvironment [60]. This finding provides an alternative strategy for miRNA-based targeted therapy for cancer treatment.
Proteins
The notion that several enlisted proteins are selectively packaged into EVs rather than packaged through a random process is commonly accepted today. Unlike membrane vesicles released by apoptotic cells with limited amounts of bioactive proteins, exosomes are enriched with varied bioactive proteins originating from the plasma membrane (i.e., growth factor receptors, integrins and tetraspanins), the cytosol (e.g., Rabs and annexins) and other intracellular compartments dependent on the endocytic pathway [61, 62].
Integrins are a major family of cell surface receptors that mediate cell adhesion to the ECM and modulate the bidirectional integration of signals between the inside and outside of a cell. Similarly, exosomal integrins exhibit an adhesive function by directing exosomes to recipient cells [62, 63], rendering specific and efficient intercellular communication. Of note, tumor-derived exosomal integrins have been reported to be highly associated with the metastasis of human breast cancer to the lung [39], resulting in organotropic metastasis. Targeting integrin within tumor-derived exosomes can interrupt exosome uptake by recipient cells and ablate cancer metastasis [39], implicating the potency of exosomal integrins in targeted cancer therapies.
Various studies have indicated that tumor-derived EV proteins shape the tumor microenvironment by remodeling the ECM, re-educating stromal cells, or activating angiogenesis, thereby facilitating cancer development. The transfer of growth factors (i.e., TGF-β) by tumor-derived exosomes influences the procancer stromal environment by elevating α-smooth muscle actin expression and promoting fibroblast-myofibroblast differentiation [64]. TGF-β-containing exosomes triggered fibroblast differentiation that supports angiogenesis and accelerates tumor progression [65]. Furthermore, mesenchymal-like cancer-derived EVs that exhibited an upregulated tissue factor, a transmembrane receptor for the coagulation factor VII/VIIa, affected the procoagulant activity of endothelial cells and led to cancer malignancy [66].
Importantly, tumor-derived exosomes carrying oncoproteins (i.e., mutant K-Ras) have been investigated in several types of cancers, such as human colorectal cancer [67, 68]. The transfer of mutant K-Ras from mutant K-Ras-expressing cells into nontransformed recipient cells by tumor-derived exosomes enhanced the three-dimensional growth of the nontransformed recipient cells [68]. Additionally, metastatic melanoma-derived exosomes transferred the MET oncoprotein to bone marrow-derived cells, enabling the promotion of premetastatic niche formation and indicating the importance of tumor-derived exosomes with a cancer metastasis-related protein signature [41].
Lipids and metabolites
Different types of lipids, such as cholesterol, diglycerides, sphingolipids, phospholipids, polyglycerophospholipids, and phosphatidylethanolamine, are predominantly expressed in EVs [69, 70]. Cholesterol, sphingomyelin, phosphatidylserine, and phosphatidylinositol especially promote EV membrane rigidity [69]. Some bioactive lipids (i.e., prostaglandins and leukotrienes) and lipid metabolism-related enzymes have also been detected in EVs [69, 71, 72], suggesting the potential role of EVs with cancer progression-related lipids in cancer development.
The intrinsic metabolic activity of EVs has demonstrated their ability to synthesize adenosine triphosphate by glycolysis as well as carry varied metabolites and metabolic enzymes, including pyruvate, lactate, and lactate dehydrogenase isoforms [73]. Thus, the metabolism of the recipient cells is altered by the uptake of EVs. Of note, a study by Zhao et al. [74] demonstrated that exosomes derived from cancer-associated fibroblasts provided diverse metabolites, such as amino acids or tricarboxylic acid (TCA)-cycle intermediates, to nutrient-deprived cancer cells to promote prostate or pancreatic cancer growth in a K-Ras independent manner.
Isolation of EVs
To understand how EVs participate in physiological and pathological processes, an efficient and reliable strategy for the isolation of EV with high purity — low contamination from other extracellular vesicles, soluble proteins, or broken cells — is challenging but necessary for basic experimental and clinical analyses. Based on the physical properties of exosomes, which have a specific buoyant density and different in flotation velocities, differential centrifugation is the most common method to isolate exosomes from cell culture conditioned media or physiological fluids [4, 75–77]. Accordingly, live or dead cells, cellular debris, and large particles in the cell culture conditioned media or physiological fluids are first separated by gradual centrifugal forces between 200 × g to 10,000 × g, followed by the application of ultracentrifugal force at 100,000 × g to isolate exosomes [75]. In the basic research setting, the most widely used sample for EV isolation is conditioned cell culture media [77]. Larger sample volumes are required for the isolation of EVs from cell culture conditioned media than the isolation of EVs from biofluids, such as plasma, serum, or urine [77]. Although some concerns, such as the time-consuming nature and high equipment cost for EV isolation, have been addressed, the ultracentrifugation method allows for EV isolation from large sample volumes and produces high yields of EVs [77, 78]. However, the differential centrifugation method often results in protein aggregates or contaminations with particulates with similar physical properties in the isolated EVs [75]. To resolve these impurities, modified EV isolation methods combining differential centrifugation and immunoadsorption techniques [79] or sucrose gradient ultracentrifugation [80] have been reported in several studies.
Due to increasingly high demand from the emerging field of EV-based therapeutics and diagnostics, several methods and commercially available kits for EV isolation based on size exclusive chromatography, microfluidics, immunoaffinity, or flow metrics provide easy and efficient methods for the enrichment of purified EVs from liquid samples, such as cell culture conditioned media or physiological fluids. In fact, the principle of these methods is separation according to the density, size, mass, surface charge and/or surface protein features of EVs. Sized-based isolation methods mainly depend on the size or molecular weight of EVs. For example, ultrafiltration combined with sequential filtration, which is a faster procedure than ultracentrifugation despite the observation of large vesicle deformation, is used for exosome isolation [81]. Ultrafiltration, which has no special equipment requirements, is more efficient than ultracentrifugation [82]. In clinical studies, a nanomembrane ultrafiltration concentrator was used to rapidly isolate exosomes from human urine samples [82]. Alternatively, size exclusive chromatography, which is easy and fast, to isolate EVs or exosomes from conditioned media or plasma samples depends on the hydrodynamic radius of the exosomes; nanoscale exosomes enter most of the porous beads and can be collected in the latter fractions, unlike larger particles, such as microvesicles or apoptotic bodies [83, 84].
Regarding precision in exosome isolation, immunoaffinity-based methods exhibit high specificity for exosome isolation. Accordingly, these methods rely on specific exosomal surface proteins, such as CD9, CD63, or CD81, that can be captured by their corresponding antibodies [85]. Although immunoaffinity-based methods are not suitable for EV isolation from large sample volume, high-purity EVs can be isolated from the conditioned media of cancer cells or plasma samples from patients with cancer [85–87]. Notably, exosomes purified by immunoaffinity-based methods only represent a subpopulation of the whole exosome population and exhibit various characteristics of the subpopulation corresponding to different antibodies, even when the antibody exhibits the same specificity against the same surface protein. Exosome precipitation involves altering the solubility or dispersibility of exosomes. The easy, rapid, and cost-effective method of EV isolation by polyethylene glycol is often employed to separate exosomes from the original soluble samples to discharge water molecules and force exosomes out of solution [81, 88]. In a longitudinal study, polyethylene glycol-based precipitation enriched EVs from human serum samples for subsequent EV miRNA analysis [89]. Inevitably, EVs purified with this method coprecipitate with contaminants, such as membrane-free macromolecular/protein aggregates or particulates, which results in impure isolated exosome populations.
In the clinical setting, it is very important to isolate EVs from small sample volumes within a fairly limited time period to improve the clinical potential of EVs in cancer diagnosis [90]. Numerous innovative methods of EV isolation, such as microfluidic- or flow cytometry-based methods, have been developed to enhance the sensitivity and specificity of EV isolation to fulfill clinical requirements [90]. Microfluidic-based EV purification methods, including sieving EVs from blood samples through nanoporous membranes [91], capturing EVs from clinical plasma samples by an immunoaffinity antibody [92–94], or trapping EVs on porous structures such as porous silicon nanowire-on-micropillar structures, are efficient and fast [95]. Notably, microfluidic-based methods require additional off-chip processes for sample precleaning/preparation and/or reagent mixing [93]. Due to the size limitation of detection by forward scattered light, magnetic beads coated with antibodies or commercial fluorescent-labeled antibodies against exosomal surface biomarkers allow exosomes to be detected and isolated by flow cytometry [96, 97]. Despite its low yields and high costs, a newly developed method based on the characterization, size-based separation, and quantification of exosomes by the asymmetrical flow field-flow fractionation (AF4) technique combined with multidetection systems, such as ultraviolet (UV) or multiangle light scattering (MALS) [98, 99], displays the powerful ability to produce high-purity EV subpopulations, such as large exosome vesicles, small exosome vesicles and exomeres, at a high resolution [100]. Table 2 summaries the pros and cons, such as yield, purity, processing time, cost, and standardization, of the current methods commonly utilized for EV isolation.
Table 2.
Summary of common EV isolation methods
| Method | Description | Example | Yield | Purity/quality | Equipment/Cost | Approximate processing time | Commercial products | Reference |
|---|---|---|---|---|---|---|---|---|
| Ultracentrifugation-based methods | Size- or density-dependent isolation | 1. Differential centrifugation/Ultracentrifugation 2. Sucrose gradient ultracentrifugation | High | Protein aggregates and particulates contaminations | High equipment cost | 2.5 – 48 h | No commercial available | [75, 77–80] |
| Size-based methods | Size- or molecular weight-dependent isolation | 1. Ultrafiltration 2. Size exclusive chromatography (SEC) 3. Asymmetrical-flow field-flow fractionation (AF4) | Low (AF4) | High purity (AF4, SEC); exosomes may be deformed (ultrafiltration) | Low/moderate equipment cost (ultrafiltration and SEC ); high equipment cost (AF4) | 1 - 1.5 h (Ultrafiltration); 0.5 - 1 h (SEC); 1h (AF4) | Available, .e.g.. EVSecond column (GL Sciences) or qEV column (Izon Science) | [82–84, 98–100] |
| Immunoaffinity-based methods | Antibody-antigen interaction-dependent isolation | 1. Immunocapture 2. Immunoadsorption | Low | High purity | High reagent cost | 4 - 5 h | Available, e.g. MagaCapture™ Exosome Isolation Kit (Wako) or Exosome-Human CD9 Isolation Reagent (ThermoFisher) | [85–87] |
| Precipitation-based methods | Solubility- or dispersibility-dependent isolation | Polyethylene glycol | Dependence | Contaminated precipitates | Low equipment cost | 1 h | Available, e.g. Total Exosome Isolation Kit (Invitrogen) or ExoQuick™ Exosome Precipitation (System Biosciences) | [81, 88, 89] |
| Microfluidic-based methods | Size-, density-, or antibody-antigen interaction-dependent isolation | Microfluidic device with nanoporous membrane, immuno-chip, or porous silicon nanowires-on-micropillar structure | Dependence | High purity (porous nanowires-on-micropillar structure); EVs may be damaged (nanoporous membrane-based filtration) | Low/moderate equipment cost | 2 h (porous membrane-based filtration); 1.5 h (immuno-chip) | No commercial available | [91–95] |
| Flow cytometry-based methods | Antibody-antigen interaction-dependent isolation | 1. Fluorescent-labeled antibody-based isolation 2. Immuno-magnetic-based isolation | Low | High purity | High equipment and reagent cost | 12 h | Avaialbe, e.g. Exosome Flow Cytometry Kit (Wako) | [96, 97] |
Characterization of EVs
In basic experimental and clinical studies, the characterization of isolated EVs by imaging, biochemical techniques, or physiochemical techniques is an essential step. The main methods used to characterize isolated EVs are electron microscopy, nanoparticle tracking analysis, Western blotting, and flow cytometry. Due to their nanometer size, the morphology of exosomes is subject to visualization and further determination by electron microscopy (EM). Typically, the rounded structure of exosomes is investigated by cryogenic electron microscopy [101]. Moreover, a modified method to characterize exosome, the immuno-EM method, which combines electron microscopy and antigen-specific immunolabeling, has been reported in many basic experimental studies [75]. Furthermore, nanoparticle tracking analysis (NTA, a NanoSightTM technology) based on the Brownian motion of particles allows for the determination of the size distribution and particle concentration of EVs [102]. Other commercial NTA systems with similar outputs are available on the market. According to the molecular composition of EVs, separation of EV proteins by SDS-PAGE, followed by immunoblotting for specific EV biomarkers, such as CD9, CD63, CD81, heat shock protein 70 (HSP70), HSP90, or Alix, enables the characterization of EVs [103]. Additionally, flow cytometry-based methods, such as AF4/UV-MALS, are used to determine the size and amount of the particles discussed earlier [100].
Notably, the International Society for Extracellular Vesicles (ISEV) has recently launched the “Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018)” guideline [104] by updating the MISEV2014 guideline to provide comprehensive mandatory considerations for EV characterization, including quantification, global characterization, and single EV characterization.
Roles and potential applications of tumor-derived EVs
The pathologic functions of tumor-derived EVs in cancer development and malignancy are highlighted by their bona fide effects; EVs facilitate transformation, survival, immunosuppression, epithelial-mesenchymal transition (EMT), invasion, angiogenesis, premetastatic niche formation, and metastasis. Understanding tumor-derived EVs renders a new avenue to monitor and treat cancer since cancer evolves over time during cancer development or treatment. The dynamic and comprehensive molecular information within tumor-derived EVs reflects de novo cancer evolution, which also sheds light on novel and valuable diagnostic and prognostic factors. Overall, the potential and potent applications of tumor-derived EVs will fulfill the fundamental needs of and strategies for precision medicine. The illustration of Figure 2 encompasses the potential roles and applications of tumor-derived EVs.
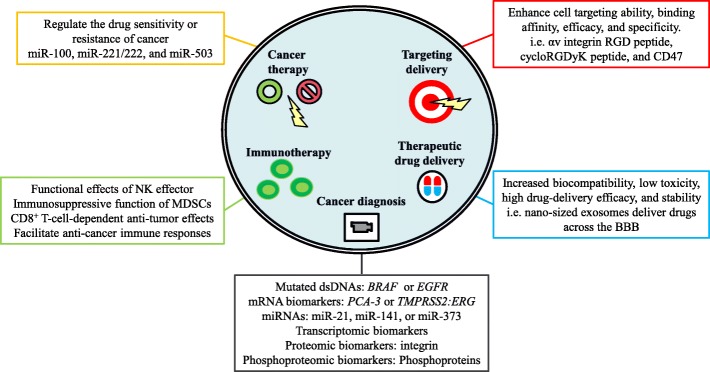
Figure 2.
Potential applications of tumor-derived EVs. Investigations of the specific expression patterns of mRNAs/miRNAs and transcriptomic, proteomic, and phosphoproteomic biomarkers in tumor-derived EVs, have indicated a powerful role for tumor-derived EVs in cancer diagnosis. The functional effects of tumor-derived EVs on the regulation of drug sensitivity or resistance in cancer suggest the role of tumor-derived EVs in cancer therapy. Regarding the role of EVs in immunotherapy, exosomes have been shown to modulate NK effector functional effects, reinforce the immunosuppressive function of MDSCs, and facilitate anticancer immune responses. A growing number of studies have indicated that the RGD peptide, cyclic RGDyK peptide, and CD47-modified exosomes promote the cell targeting, binding affinity, efficacy, and specificity of exosomes, suggesting the potential applications of EVs in targeted delivery. Moreover, the EV-based therapeutic drug delivery system exhibits increased biocompatibility, low toxicity, and stability
EVs in cancer diagnosis
The fundamental basis of precision medicine was recently highlighted by biomarker discovery through liquid biopsy, which allows for noninvasive, fast, dynamic, low-cost, and accurate diagnosis for the early and real-time detection of cancer. To gain comprehensive information on cancer development and progression, the use of several potential and meaningful biomarkers, such as circulating tumor cells (CTCs), cell free DNAs (cfDNAs), and EVs, in liquid biopsy aimed at the clinical detection of various types and stages of cancers has been intensively investigated over the past two decades [105]. Taking advantage of their highly dynamic and multispecies nature, abundance, and stability, cancer-related EVs have served as suitable and precise biomarkers in various clinical settings. Indeed, the exosomal protein tumor susceptibility gene 101 (TSG101) stored at -20 °C or -80 °C was stable for over 3 months [106]. Moreover, phosphoproteins from microvesicular and exosomal EVs from human plasma were stable for up to 5 years [107]. Notably, although CTCs and cfDNAs allow for the rapid and easily accessible diagnosis of malignant cancers, the limitations in detecting late-stage malignancies and the technical challenges of cfDNAs and CTCs have been indicated in several clinical utilities. Although cfDNAs enable the detection of tumor-specific mutations [108], the expeditious elimination of DNA by nucleases has been documented [109]. Although double-stranded DNA remains in the blood longer than single-stranded DNA, the rapid clearance of DNAs is indispensable regardless of its size or strandedness, which limits the application of cfDNA in cancer diagnosis [110]. Due to the rarity of CTCs in the bloodstream and the lack of meaningful information on the number of CTCs in patients’ circulating blood [111], improvements in detecting and capturing CTCs is required for the use of CTCs for clinical diagnosis.
EV miRNAs in cancer diagnosis
Versatile bioactive molecules enclosed within EVs are protected from degradation [112]. With this in mind, the content of EVs was profiled to decipher biomolecules with mechanistic and/or diagnostic specificity for varied types of cancers; because of their consistent and robust detection, these biomolecules are regarded as valuable biomarkers in liquid biopsy for cancer diagnosis and prognosis (Table 3). Consistently, in an RNA-based clinical analysis, exosomes protected miRNAs from degradation by ribonucleases in feces [121]. Indeed, miRNA expression profiling in circulating EVs, such as exosomes, has been performed to establish the predictive function of specific miRNA signatures in human peripheral blood [122]. Recent efforts have also been devoted to the discovery of early diagnostic biomarkers for cancer due to the significant improvement in the survival of cancer patients. For example, miR-21, miR-141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, and miR-214 were significantly elevated in exosomes from patients with ovarian cancer compared to those from patients with benign disease [116]. Additionally, exosomes derived from the serum of patients with hormone receptor-negative breast cancer (estrogen receptor-negative or progesterone receptor-negative) exhibited a higher level of miR-373 expression than those with the serum of patients with hormone receptor-positive breast cancer [115]. Moreover, upregulation of the exosome miR-21 in serum from patients with esophageal squamous cell carcinoma was highly correlated with advanced tumor classification, positive lymph node status, and metastasis [114], suggesting that EV miRNAs provide useful diagnostic information to evaluate the status of cancer development. Importantly, the development of EV-based liquid biopsy from saliva and urine provides an alternative, noninvasive and sensitive strategy for cancer detection. In this technique, exosomes derived from the saliva of mice with pancreatic cancer exhibited pancreatic cancer-specific salivary transcriptomic biomarkers [118]. The inhibition of exosome biogenesis altered this pancreatic cancer-specific transcriptomic biomarker profile in salivary exosomes [118]. Clinically, exosomes derived from the urine of patients with prostate cancer have also been documented to display specific prostate cancer mRNA biomarkers, such as prostate cancer antigen 3 and transmembrane protease serine 2:transforming protein ERG (TMPRSS2:ERG) [117].
Table 3.
EV cargos used as diagnostic biomarkers in cancers
| EV cargo | Type of cargo | Type of body fluid | Cancer type | Reference |
|---|---|---|---|---|
| CD63 and caveolin-1 | Protein | Plasma | Melanoma | [113] |
| Integrin | Protein | Plasma | Breast cancer | [39] |
| miR-21 | miRNA | Serum | Esophageal squamous cell carcinoma | [114] |
| miR-373 | miRNA | Serum | Breast cancer | [115] |
| miRNA signatures | miRNA | Serum | Ovarian cancer | [116] |
| Prostate cancer antigen 3 and TMPRSS2:ERG | RNA | Urine | Prostate cancer | [117] |
| Salivary transcriptomic biomarkers | RNA | Saliva | Pancreatic cancer | [118] |
| Specific phosphoproteins | Protein | Plasma | Breast cancer | [107] |
| Specific protein profile | Protein | Ascites | Colorectal cancer | [119] |
| Survivin | Protein | Plasma | Prostate cancer | [120] |
EV proteins in cancer diagnosis
Cancer-specific EVs carry specific and stabile protein cargo for intercellular signal exchange to regulate the tumor microenvironment. Likewise, the detection of unique EV proteins associated with cancer development and progression has been emphasized in various clinical utilities (Table 3). For instance, survivin expression was significantly higher in plasma exosomes from patients with prostate cancer compared to that in plasma exosomes from patients with pre-inflammatory benign prostatic hyperplasia or normal healthy controls [120]. Utilizing proteomic analysis, the specific protein expression profiles in diverse body fluids EVs, including ascites from colorectal cancer patients, were illustrated and determined [119]. Increased CD63 or caveolin-1 was detected in plasma exosomes from patients with melanoma compared to that in plasma exosomes from healthy donors [113]. Recently, our study indicated that tumor-derived exosomes with specific integrin expression profiles regulated organotropic metastasis [39]. This was the first study to show that bioactive molecules in exosomes could determine and predict the specific organ of cancer metastasis, further implicating exosome integrin profiles as biomarkers for organotropic metastasis [39]. Given that protein phosphorylation is essential in many cancer cell functions, the phosphoproteome analyses of tumor-derived EVs from human plasma provide valuable information for cancer diagnosis. Specific phosphoproteins in plasma EVs are significantly increased in patients with breast cancer compared to those in healthy controls [107]. Together, these studies suggest that the profiles of specific bioactive molecules in tumor-derived EVs function as novel and valuable biomarkers to diagnose or track the real-time status of cancer during cancer development and progression.
EV biology during cancer therapy
Given that neoadjuvant chemotherapy and chemoradiotherapy are effective anticancer therapeutic strategies in many types of cancers, understanding the detailed mechanisms of the pathologic changes in response to therapy is essential to optimize preoperative and postoperative treatments. Because they manage both bioactive molecules and cellular waste in cells, therapeutic treatment-induced EVs reflected the response of cancer cells upon encountering anticancer treatments [123]. Indeed, cisplatin-resistant cell-derived exosomes contained more platinum than those derived from cisplatin-sensitive cells [124]. Moreover, exosomes have been shown to regulate the cisplatin sensitivity of lung cancer [125]. Clinically, the secretion of annexin A3 is associated with exosomes released from patients with platinum-resistant ovarian cancer [123]. Together, these studies suggest the functional effects of EVs on the regulation of drug sensitivity and response.
The EV-mediated transfer of miRNAs has also been attributed to drug resistance. For instance, drug-resistant breast cancer-derived exosomes regulated the drug sensitivity of recipient drug-sensitive cells by modulating drug-induced apoptosis [126]. Mechanistically, specific miRNA profiles, including those of miR-100 and miR-222, in drug-resistant breast cancer-derived exosomes have been investigated [126]. In addition, the transfer of miR-221/222 from tamoxifen-resistant breast cancer to tamoxifen-sensitive breast cancer by exosomes led modified p27 and estrogen receptor alpha expression and resulted in a drug-resistant response in the recipient cells [127].
Moreover, the crosstalk between cancer and the tumor microenvironment by exosomal miRNAs also modulates the growth and response to drugs of cancers [128]. In a recent study, increased exosomal miRNAs, such as miR-503, in neoadjuvant chemotherapy-treated endothelial cells exhibited an anti-breast cancer response [129]. Increased plasma miR-503 has been detected in breast cancer patients with neoadjuvant chemotherapy [129], suggesting that stromal cells modulate cancer development by releasing EV miRNAs in response to anticancer therapies.
EVs in immunotherapy
Tumor-derived EVs in addition to tumor cells indeed participate in immunosuppression or immunostimulation in accordance with the development and progression of cancer [130–132]. Numerous studies have indicated that tumor-derived EVs mediate cancer development by inhibiting immune responses. For example, tumor-derived exosomes facilitated cancer immune evasion by triggering the downregulation the expression of natural killer group 2D, an activating receptor for natural killer (NK) cells, leading to NK effector functional defects [133]. Moreover, tumor-derived exosomes contained membrane-associated Hsp72, which interacts with myeloid-derived suppressor cells (MDSCs), reinforcing the signal transducer and activator of transcription 3-dependent immunosuppressive function of MDSCs [134]. Additionally, exosomes derived from Epstein-Barr virus-associated nasopharyngeal carcinoma exerted galectin-9, a ligand of T cell immunoglobulin and mucin domain-3 (TIM-3), to induce the apoptosis of mature T-helper type 1 lymphocytes [135]. In contrast, exosome-mediated apoptosis was blocked by both anti-Tim-3 and anti-galectin-9 antibodies [135]. Moreover, pancreatic cancer-derived exosomes also downregulated Toll-like receptor 4 and its downstream cytokines tumor necrosis factor-α and interleukin-12 in dendritic cells (DCs) via exosomal miR-203 [136]. Together, these comprehensive studies suggest that the ablation or inhibition of EV-mediated immune responses enhances the efficacy of immunotherapeutic anticancer therapies
Several studies indicated that exosomes secreted from antigen-presenting cells, such as B-cells, enable the induction of the immune response [137, 138]. Intriguingly, tumor-derived exosomes have been suggested to transfer tumor antigens to DCs, leading to potent CD8+ T-cell-dependent antitumor effects in vivo [139]. Clinical studies also suggested that tumor-derived exosomes isolated from ascites function as natural tumor rejection antigens [140]. Moreover, exosomes derived from NK cells expressed killing proteins, such as the Fas ligand and perforin molecules, suggesting anticancer activity [141]. An ex vivo study also indicated that healthy donor plasma-derived exosomes displayed NK markers with exosome-induced cytotoxicity [141]. Additionally, mast cell-derived exosomes were capable of facilitating the maturation of DCs and inducing immune responses [142]. Indeed, the concept of the exosome-elicited immune response has been under evaluation in a phase I clinical trial for the immunization of patients against melanoma by using autologous exosomes with MAGE 3 (melanoma-associated antigen 3) peptides [143]. In a phase I clinical trial, DC-derived exosomes loaded with cancer antigen induced anticancer immune responses [144]. Furthermore, exosomes from interferon-γ-maturated DCs were generated to facilitate anticancer immune responses in a phase II clinical trial [145]. Together, these studies highlight the feasibility of EV-based anticancer immunotherapy.
EV-based targeted delivery
Given their cell- and tissue-tropic features, EVs are thought to be ideal therapeutic carriers for anticancer targeted therapy. Typically, in addition to carrying bioactive luminal cargos, EVs contain cell-cell and cell-ECM adhesion receptors on their surfaces to recognize distinct receptors of their target cells and tissues. For example, tetraspanin-8-expressed exosomes preferentially target CD11b/CD54-positive cells [146]. Interestingly, exosomes expressing modified receptors, such as tetraspanins, fused with specific candidate proteins displayed enhanced cell targeting [147]. Likewise, the expression of a fusion protein containing the αv integrin arginyl-glycyl-aspartic acid (RGD) peptide and an exosomal transmembrane protein, such as lysosome-associated membrane glycoprotein 2b (Lamp2b), allowed dendritic cell-derived exosomes to target αv integrin-positive cancers [148]. Small interfering RNA (siRNA)-carrying exosomes containing a fusion protein between central nervous system–specific rabies viral glycoprotein peptide and Lamp2b specifically targeted the acetylcholine receptor of neurons, resulting in brain-specific gene knockdown in vivo [149]. Since cyclic (Arg-Gly-Asp-D-Tyr-Lys, RGDyK) peptide, c(RGDyK), has a high binding affinity with integrin αvβ3 on cerebral vascular endothelial cells, c(RGDyK)-conjugated exosomes loaded with curcumin were specifically directed toward the lesion-containing region of the ischemic brain, where they then ameliorated inflammatory responses and apoptosis [150].
In light of the EV-mediated transfer of biological molecules, magnet-conjugated transferrin bound to transferrin receptor-expressed blood exosomes has been shown to preferentially target magnets surrounding cancer cells, followed by the inhibition of cancer development [151]. Furthermore, engineered glycosylation prevented the proteolytic degradation of exosomal-targeting ligands, suggesting the high stability and efficient targeting of glycosylated exosomes [152]. Due to the CD47-mediated protection of exosomes from phagocytosis, cell-derived exosomes carrying siRNAs exhibited enhanced efficacy in targeting oncogenic KRAS in a CD47-dependent manner [153]. Taken together, these studies suggest that EV-based targeted delivery, particularly with some modifications, is a highly efficacious alternative to cancer therapies.
EVs as a therapeutic drug delivery system
Drug delivery is a critical determinant for the efficacy of clinical therapeutic treatment. As discussed above, EVs have emerged as a novel and promising drug delivery technology with the advantage of precise targeting, prolonged stability and controllable release. Despite the popularity of synthetic liposomes and polymeric nanoparticles in drug delivery [112, 154, 155], the instability and low biocompatibility of synthetic liposomes and polymeric nanoparticles, respectively, give rise to a degree of toxicity and lower efficacy in terms of clinical utility [156, 157]. In contrast, EVs exhibit biocompatibility, low toxicity, high drug delivery efficacy, specificity, and stability [158, 159]. More specifically, the lipid bilayer harbors many unique integral proteins with various posttranslational modifications that allow EVs to serve as a protective shelter for the sustainable release of anticancer drugs or cancer suppressors and to evade degradation and immune responses [160]. As a result, in human body fluids, EVs are proper nucleic acid drug (e.g., siRNAs or miRNAs) carriers. Consistently, anti-miR-9 delivered by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells reversed the expression of the multidrug transporter and sensitized the glioblastoma multiforme cells to chemotherapy drugs [161]. Additionally, exosomes can efficiently deliver microRNAs, such as let-7a, to breast cancer cells overexpressing EGFR, inhibiting cancer development in vivo [162]. Alternatively, the use of exosomes to deliver small molecules to treat cancers and other diseases has been demonstrated as well. For example, exosomes with encapsulated anti-inflammatory drugs, such as curcumin, exhibited increased solubility, stability, and drug bioavailability in vitro and in vivo [163].
EV-based therapy in brain disease
The blood-brain barrier (BBB) is a major obstacle for drug delivery to the central nervous system [164]. To mediate the delivery of misfolded proteins between neurons in neurodegenerative diseases [165], nanosized exosomes are presumably favorable for delivering agents/drugs across the BBB. Although nanoformulations are employed to improve the permeability of drugs across the BBB, toxicity and reticuloendothelial system-mediated or mononuclear phagocyte-mediated drug clearance both hinder the efficacy of man-made nanoformulations in treating diseases including cancers [166]. In contrast, EVs, which are naturally produced by cells, are guaranteed to exhibit biocompatibility and low antigenicity [158], which highlights the potential ability of EVs in treating brain disease, such as neurodegenerative diseases or brain cancers. Eventually, an exosome-based delivery system for antioxidants, such as catalase, that have a beneficial effect on patients with Parkinson’s disease will be approved [167]. Specific biological molecules must be used to cross the BBB or target brain tissue, and exosomes derived from brain endothelial cell displayed specific homing proteins that gave rise to increased transport across the BBB [168]. Thus, employing brain endothelial cells-derived exosomes to carry anticancer drugs, such as doxorubicin, is applicable for the suppression of brain cancer development [168].
Improvements in EV-based therapy
Nevertheless, some technical limitations in terms of the use of EVs as a drug delivery system, in particular, the efficiency of loading agents/drugs into EVs, are issues that remain to be resolved. Presumably, highly membrane-permeable small agents/drugs can easily be loaded into exosomes following incubation [169]. However, the loading of membrane-impermeable drugs, such as macromolecular drugs, siRNAs, and small DNAs, into exosomes using chemical approaches, such as temperature switching or detergents, is problematic. Recently, exosomes loaded with membrane-impermeable candidate nucleic acids or protein agents/drugs were produced by the pre-overexpression of these candidates in donor cells [161]. Alternatively, physical transfection methods, such as electroporation or liposome-mediated transfection, have been utilized to package membrane-impermeable agents/drugs into exosomes [170]. Unfortunately, the low efficiency of loading agents/drugs into exosomes through the use of membrane-permeable reagents (i.e., liposomes) remains unimproved [171]. Alternatively, several studies indicated that artificial exosome mimetics could be substitutes for exosome-based drug delivery [159]. Reportedly, exosome-mimetic nanovesicles loaded with chemotherapeutic drugs, such as doxorubicin, could target malignant cancers in vivo [172].
Conclusions
Cancer development is an evolving, dynamic and highly regulated process associated with the tumor microenvironment and even distant tissues. Given the participation of EVs in local and systemic intercellular communication, the essential roles of EVs in the regulation of cancer progression and malignancy have been highlighted in the past decade, although EVs were observed more than three decades ago. Until now, the biogenesis and heterogeneity of EVs, the regulatory mechanisms of diverse cargo packaging into EVs, and the in vivo functionality of tumor-derived EVs have remained largely unknown. Nonetheless, numerous studies have provided valuable information, such as detailed EV cargo profiles; unique EV biomarkers/signatures for the early detection, diagnosis and treatment of cancers; and powerful methods to isolate EVs from cell culture conditioned media or body fluids. These efforts provide the impetus for the promising applications of EVs in disease management and emphasize the importance of EV biology in precision medicine.
The molecular features of cancers change dynamically during cancer development; following anticancer therapy, tumor-derived EVs reflect the real-time state of cancer cells and allow the monitoring of disease progression. As a result, in addition to their diagnostic, predictive, and prognostic utilities, tumor-derived EVs serve as novel anticancer targets. However, optimal and/or standardized methods for EV isolation, storage, and characterization are required for basic research and clinical standardization in the diagnosis and treatment of cancer and other diseases. Notably, a guideline for EV studies suggested by the ISEV entitled“Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018)” [104] was discussed and published to provide comprehensive mandatory, mandatory if applicable and encouraged considerations. Nevertheless, any new guidelines will be updated in accordance with emerging studies and discoveries in EVs.
Multiple aspects of EVs in cancer biology have been indicated, which sheds a novel light in understanding the tumor microenvironment involved in cancer development and the potential uses of EVs in cancer management. As a result, EVs are an authentic key mediator in cancer biology.
Acknowledgements
Not applicable.
Funding
This work was supported by the Ministry of Science and Technology, Taiwan (105-2320-B-002-058-MY3) and Dragon-gate program, Ministry of Science and Technology, Taiwan (106-2911-I-002-569).
Availability of data and materials
Not applicable.
Abbreviations
- AF4
Asymmetrical-flow field-flow fractionation
- Alix
Alpha-1,3/1,6-mannosyltransferase (ALG2)-interacting protein X
- BBB
Blood-brain barrier
- CD
Cluster of differentiation
- cfDNAs
Cell free DNAs
- CTCs
Circulating tumor cells
- CXCR4
C-X-C chemokine receptor type 4
- DCs
Dendritic cells
- DNA
Deoxyribonucleic acid
- dsDNAs
Double-stranded DNAs
- ECMs
Extracellular matrixes
- EGFR
Epidermal growth factor receptor
- EM
Electron microscopy
- EMT
Epithelial-mesenchymal transition
- EV
Extracellular vesicle
- exoDNAs
Exosomal DNAs
- HSP
Heat shock protein
- ILVs
Intraluminal vesicles
- ISEV
International society for extracellular vesicles
- Lamp2b
Lysosome-associated membrane glycoprotein 2b
- let-7
Lethal-7
- MAGE 3
Melanoma-associated antigen 3
- MALS
Multi-angle light scattering
- MDSCs
Myeloid-derived suppressor cells
- miRNAs
microRNAs
- MISEV2018
Minimal information for studies of extracellular vesicles 2018
- MMP
Matrix metallopeptidase
- mRNAs
Messenger ribonucleic acids
- MVBs
Multivesicular bodies
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NK
Natural killer
- NTA
Nanoparticle tracking analysis
- oncomiRs
Oncogenic microRNAs
- RANK
Receptor activator of nuclear factor-κB
- RANKL
Receptor activator of nuclear factor κB-ligand
- RGD
Arginyl-glycyl-aspartic acid
- RGDyK
Arg-Gly-Asp-D-Tyr-Lys
- siRNA
Small interfering RNA
- TAMs
Tumor-associated macrophages
- TCA
Tricarboxylic acid
- TGF-β
Transforming growth factor β
- TIM-3
T cell immunoglobulin and mucin domain-3
- TMPRSS2:ERG
transmembrane protease serine 2:transforming protein ERG
- TSG101
tumor susceptibility gene 101
- UTRs
untranslated regions
- UV
ultraviolet
- Wnt
wingless/integrated
Authors' contributions
Y-L T, P-Y C, B-H L, K-C C, C-Y Y, W-H K, and T-L S wrote the manuscript. Y-L T and C-Y Y designed and illustrated table. Y-L T and P-Y C designed and illustrated figures. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yu-Ling Tai, Email: d98633002@ntu.edu.tw.
Pei-Yu Chu, Email: areteka@gmail.com.
Bao-Hong Lee, Email: f96b47117@ntu.edu.tw.
Ko-Chien Chen, Email: kc2388@gmail.com.
Chia-Yu Yang, Email: b03613031@ntu.edu.tw.
Wen-Hung Kuo, Email: brcancer@gmail.com.
Tang-Long Shen, Email: shentl@ntu.edu.tw.
References
- 1.Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27:172–188. doi: 10.1016/j.tcb.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuana Y, Sturk A, Nieuwland R. Extracellular vesicles in physiological and pathological conditions. Blood Rev. 2013;27:31–39. doi: 10.1016/j.blre.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Pathan M, Fonseka P, Chitti SV, Kang T, Sanwlani R, Van Deun J, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019;47:D516–D519. doi: 10.1093/nar/gky1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–9420. [PubMed] [Google Scholar]
- 7.Frazer AC, Stewart HC. Ultramicroscopic particles in normal human blood. J Physiol. 1937;90:18–30. doi: 10.1113/jphysiol.1937.sp003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barland P, Novikoff AB, Hamerman D. Electron microscopy of the human synovial membrane. J Cell Biol. 1962;14:207–220. doi: 10.1083/jcb.14.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 10.Trams EG, Lauter CJ, Salem N, Heine U. Exfoliation of Membrane Ecto-Enzymes in the Form of Micro-Vesicles. Biochim Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 11.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 12.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai YL, Chen KC, Hsieh JT, Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109:2364–2374. doi: 10.1111/cas.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witwer Kenneth W., Buzás Edit I., Bemis Lynne T., Bora Adriana, Lässer Cecilia, Lötvall Jan, Nolte-‘t Hoen Esther N., Piper Melissa G., Sivaraman Sarada, Skog Johan, Théry Clotilde, Wauben Marca H., Hochberg Fred. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles. 2013;2(1):20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigor'eva AE, Tamkovich SN, Eremina AV, Tupikin AE, Kabilov MR, Chernykh VV, et al. Characteristics of exosomes andmicroparticles discovered in human tears. Biomed Khim. 2016;62:99–106. doi: 10.18097/pbmc20166201099. [DOI] [PubMed] [Google Scholar]
- 18.Wolf T, Baier SR, Zempleni J. The Intestinal Transport of Bovine Milk Exosomes Is Mediated by Endocytosis in Human Colon Carcinoma Caco-2 Cells and Rat Small Intestinal IEC-6 Cells. J Nutr. 2015;145:2201–2206. doi: 10.3945/jn.115.218586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4:280–290. [Google Scholar]
- 20.Whiteside TL. Tumor-Derived Exosomes and Their Role in Cancer Progression. Adv Clin Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng J, Koutras C, Donnelier J, Alshehri M, Fotouhi M, Girard M, et al. Neurons Export Extracellular Vesicles Enriched in Cysteine String Protein and Misfolded Protein Cargo. Sci Rep. 2017;7:956. doi: 10.1038/s41598-017-01115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng L, Wang Y, Peng Y, Wu Y, Ding Y, Jiang Y, et al. Osteoblast-derived microvesicles: A novel mechanism for communication between osteoblasts and osteoclasts. Bone. 2015;79:37–42. doi: 10.1016/j.bone.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Huynh N, VonMoss L, Smith D, Rahman I, Felemban MF, Zuo J, et al. Characterization of Regulatory Extracellular Vesicles from Osteoclasts. J Dent Res. 2016;95:673–679. doi: 10.1177/0022034516633189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikebuchi Y, Aoki S, Honma M, Hayashi M, Sugamori Y, Khan M, et al. Coupling of bone resorption and formation by RANKL reverse signalling. Nature. 2018;561:195–200. doi: 10.1038/s41586-018-0482-7. [DOI] [PubMed] [Google Scholar]
- 26.Xu J, Liao K, Zhou W. Exosomes Regulate the Transformation of Cancer Cells in Cancer Stem Cell Homeostasis. Stem Cells Int. 2018;2018:4837370. doi: 10.1155/2018/4837370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez M, Silva J, Herrera A, Herrera M, Pena C, Martin P, et al. Exosomes enriched in stemness/metastatic-related mRNAS promote oncogenic potential in breast cancer. Oncotarget. 2015;6:40575–40587. doi: 10.18632/oncotarget.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin R, Wang S, Zhao RC. Exosomes from human adipose-derived mesenchymal stem cells promote migration through Wnt signaling pathway in a breast cancer cell model. Mol Cell Biochem. 2013;383:13–20. doi: 10.1007/s11010-013-1746-z. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, et al. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS One. 2015;10:e0125625. doi: 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36:1770–1778. doi: 10.1038/onc.2016.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu YB, Yan C, Mu L, Mi YL, Zhao H, Hu H, et al. Exosomal Wnt-induced dedifferentiation of colorectal cancer cells contributes to chemotherapy resistance. Oncogene. 2019;38:1951–1965. doi: 10.1038/s41388-018-0557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keklikoglou I, Cianciaruso C, Guc E, Squadrito ML, Spring LM, Tazzyman S, et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol. 2019;21:190–202. doi: 10.1038/s41556-018-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 34.Fang T, Lv H, Lv G, Li T, Wang C, Han Q, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura K, Sawada K, Kinose Y, Yoshimura A, Toda A, Nakatsuka E, et al. Exosomes Promote Ovarian Cancer Cell Invasion through Transfer of CD44 to Peritoneal Mesothelial Cells. Mol Cancer Res. 2017;15:78–92. doi: 10.1158/1541-7786.MCR-16-0191. [DOI] [PubMed] [Google Scholar]
- 37.Vella LJ. The emerging role of exosomes in epithelial-mesenchymal-transition in cancer. Front Oncol. 2014;4:361. doi: 10.3389/fonc.2014.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le MT, Hamar P, Guo C, Basar E, Perdigao-Henriques R, Balaj L, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao S, Ye Z, Li F, Meng Q, Qureshi M, Yang J, et al. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp Oncol. 2006;28:126–131. [PubMed] [Google Scholar]
- 43.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci U S A. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H, Zhang H, Ge S, Ning T, Bai M, Li J, et al. Exosome-Derived miR-130a Activates Angiogenesis in Gastric Cancer by Targeting C-MYB in Vascular Endothelial Cells. Mol Ther. 2018;26:2466–2475. doi: 10.1016/j.ymthe.2018.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch M, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A. 2017;114:E9066–E9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawamura Y, Yamamoto Y, Sato TA, Ochiya T. Extracellular vesicles as trans-genomic agents: Emerging roles in disease and evolution. Cancer Sci. 2017;108:824–830. doi: 10.1111/cas.13222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ekström Karin, Valadi Hadi, Sjöstrand Margareta, Malmhäll Carina, Bossios Apostolos, Eldh Maria, Lötvall Jan. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. Journal of Extracellular Vesicles. 2012;1(1):18389. doi: 10.3402/jev.v1i0.18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics. 2009;10:556. doi: 10.1186/1471-2164-10-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei Z, Batagov AO, Schinelli S, Wang J, Wang Y, El Fatimy R, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8:1145. doi: 10.1038/s41467-017-01196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 55.Cheng Lesley, Sharples Robyn A., Scicluna Benjamin J., Hill Andrew F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. Journal of Extracellular Vesicles. 2014;3(1):23743. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y, et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 59.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bronisz A, Wang Y, Nowicki MO, Peruzzi P, Ansari K, Ogawa D, et al. Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014;74:738–750. doi: 10.1158/0008-5472.CAN-13-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, et al. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 62.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 63.Rieu S, Geminard C, Rabesandratana H, Sainte-Marie J, Vidal M. Exosomes released during reticulocyte maturation bind to fibronectin via integrin alpha4beta1. Eur J Biochem. 2000;267:583–590. doi: 10.1046/j.1432-1327.2000.01036.x. [DOI] [PubMed] [Google Scholar]
- 64.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 65.Webber JP, Spary LK, Sanders AJ, Chowdhury R, Jiang WG, Steadman R, et al. Differentiation of tumour-promoting stromal myofibroblasts by cancer exosomes. Oncogene. 2015;34:290–302. doi: 10.1038/onc.2013.560. [DOI] [PubMed] [Google Scholar]
- 66.Garnier D, Magnus N, Lee TH, Bentley V, Meehan B, Milsom C, et al. Cancer cells induced to express mesenchymal phenotype release exosome-like extracellular vesicles carrying tissue factor. J Biol Chem. 2012;287:43565–43572. doi: 10.1074/jbc.M112.401760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ji H, Greening DW, Barnes TW, Lim JW, Tauro BJ, Rai A, et al. Proteome profiling of exosomes derived from human primary and metastatic colorectal cancer cells reveal differential expression of key metastatic factors and signal transduction components. Proteomics. 2013;13:1672–1686. doi: 10.1002/pmic.201200562. [DOI] [PubMed] [Google Scholar]
- 68.Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hannafon BN, Ding WQ. Intercellular communication by exosome-derived microRNAs in cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skotland T, Hessvik NP, Sandvig K, Llorente A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2019;60:9–18. doi: 10.1194/jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esser J, Gehrmann U, D'Alexandri FL, Hidalgo-Estevez AM, Wheelock CE, Scheynius A, et al. Exosomes from human macrophages and dendritic cells contain enzymes for leukotriene biosynthesis and promote granulocyte migration. J Allergy Clin Immun. 2010;126:1032–U213. doi: 10.1016/j.jaci.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 73.Fonseca P, Vardaki I, Occhionero A, Panaretakis T. Metabolic and Signaling Functions of Cancer Cell-Derived Extracellular Vesicles. Int Rev Cell Mol Biol. 2016;326:175–199. doi: 10.1016/bs.ircmb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 74.Zhao H, Yang L, Baddour J, Achreja A, Bernard V, Moss T, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006; Chapter 3:Unit 3 22. [DOI] [PubMed]
- 76.Xu R, Greening DW, Rai A, Ji H, Simpson RJ. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods. 2015;87:11–25. doi: 10.1016/j.ymeth.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 77.Gardiner Chris, Vizio Dolores Di, Sahoo Susmita, Théry Clotilde, Witwer Kenneth W., Wauben Marca, Hill Andrew F. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. Journal of Extracellular Vesicles. 2016;5(1):32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 80.Aalberts M, van Dissel-Emiliani FM, van Adrichem NP, van Wijnen M, Wauben MH, Stout TA, et al. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol Reprod. 2012;86:82. doi: 10.1095/biolreprod.111.095760. [DOI] [PubMed] [Google Scholar]
- 81.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PST, et al. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol-Renal. 2007;292:F1657–F1661. doi: 10.1152/ajprenal.00434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 84.Gamez-Valero A, Monguio-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borras FE. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles' characteristics compared to precipitating agents. Sci Rep. 2016;6:33641. doi: 10.1038/srep33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17:3558–3577. doi: 10.1039/C7LC00592J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 87.Sharma P, Ludwig S, Muller L, Hong CS, Kirkwood JM, Ferrone S, et al. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J Extracell Vesicles. 2018;7:1435138. doi: 10.1080/20013078.2018.1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lewis GD, Metcalf TG. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl Environ Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Andreu Z, Rivas E, Sanguino-Pascual A, Lamana A, Marazuela M, Gonzalez-Alvaro I, et al. Comparative analysis of EV isolation procedures for miRNAs detection in serum samples. J Extracell Vesicles. 2016;5:31655. doi: 10.3402/jev.v5.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li K, Rodosthenous RS, Kashanchi F, Gingeras T, Gould SJ, Kuo LS, et al. Advances, challenges, and opportunities in extracellular RNA biology: insights from the NIH exRNA Strategic Workshop. JCI Insight. 2018;3(7):e98942. 10.1172/jci.insight.98942. [DOI] [PMC free article] [PubMed]
- 91.Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip. 2012;12:5202–5210. doi: 10.1039/c2lc41006k. [DOI] [PubMed] [Google Scholar]
- 92.Whiteside TL. Extracellular vesicles isolation and their biomarker potential: are we ready for testing? Ann Transl Med. 2017;5:54. doi: 10.21037/atm.2017.01.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab Chip. 2015;15:2388–2394. doi: 10.1039/C5LC00240K. [DOI] [PubMed] [Google Scholar]
- 94.He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773–3780. doi: 10.1039/C4LC00662C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, et al. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip. 2013;13:2879–2882. doi: 10.1039/c3lc41343h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Oksvold MP, Neurauter A, Pedersen KW. Magnetic bead-based isolation of exosomes. Methods Mol Biol. 2015;1218:465–481. doi: 10.1007/978-1-4939-1538-5_27. [DOI] [PubMed] [Google Scholar]
- 97.Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530. doi: 10.3402/jev.v4.25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sitar S, Kejzar A, Pahovnik D, Kogej K, Tusek-Znidaric M, Lenassi M, et al. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal Chem. 2015;87:9225–9233. doi: 10.1021/acs.analchem.5b01636. [DOI] [PubMed] [Google Scholar]
- 99.Petersen KE, Manangon E, Hood JL, Wickline SA, Fernandez DP, Johnson WP, et al. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal Bioanal Chem. 2014;406:7855–7866. doi: 10.1007/s00216-014-8040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20:332–343. doi: 10.1038/s41556-018-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Conde-Vancells J, Rodriguez-Suarez E, Embade N, Gil D, Matthiesen R, Valle M, et al. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J Proteome Res. 2008;7:5157–5166. doi: 10.1021/pr8004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoshioka Yusuke, Konishi Yuki, Kosaka Nobuyoshi, Katsuda Takeshi, Kato Takashi, Ochiya Takahiro. Comparative marker analysis of extracellular vesicles in different human cancer types. Journal of Extracellular Vesicles. 2013;2(1):20424. doi: 10.3402/jev.v2i0.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017. pp. 755–768. [DOI] [PubMed] [Google Scholar]
- 106.Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, et al. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics. 2013;13:3354–3364. doi: 10.1002/pmic.201300282. [DOI] [PubMed] [Google Scholar]
- 107.Chen IH, Xue L, Hsu CC, Paez JS, Pan L, Andaluz H, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114:3175–3180. doi: 10.1073/pnas.1618088114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer. 2017;17:223–238. doi: 10.1038/nrc.2017.7. [DOI] [PubMed] [Google Scholar]
- 109.Emlen W, Mannik M. Kinetics and mechanisms for removal of circulating single-stranded DNA in mice. J Exp Med. 1978;147:684–699. doi: 10.1084/jem.147.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Emlen W, Mannik M. Effect of DNA size and strandedness on the in vivo clearance and organ localization of DNA. Clin Exp Immunol. 1984;56:185–192. [PMC free article] [PubMed] [Google Scholar]
- 111.Ferreira MM, Ramani VC, Jeffrey SS. Circulating tumor cell technologies. Mol Oncol. 2016;10:374–394. doi: 10.1016/j.molonc.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PloS one. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 115.Eichelser C, Stuckrath I, Muller V, Milde-Langosch K, Wikman H, Pantel K, et al. Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget. 2014;5:9650–9663. doi: 10.18632/oncotarget.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 117.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888–26897. doi: 10.1074/jbc.M113.452458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Choi DS, Park JO, Jang SC, Yoon YJ, Jung JW, Choi DY, et al. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics. 2011;11:2745–2751. doi: 10.1002/pmic.201100022. [DOI] [PubMed] [Google Scholar]
- 120.Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PloS one. 2012;7:e46737. doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Koga Y, Yasunaga M, Moriya Y, Akasu T, Fujita S, Yamamoto S, et al. Exosome can prevent RNase from degrading microRNA in feces. J Gastrointest Oncol. 2011;2:215–222. doi: 10.3978/j.issn.2078-6891.2011.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PloS one. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yin J, Yan X, Yao X, Zhang Y, Shan Y, Mao N, et al. Secretion of annexin A3 from ovarian cancer cells and its association with platinum resistance in ovarian cancer patients. J Cell Mol Med. 2012;16:337–348. doi: 10.1111/j.1582-4934.2011.01316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Safaei R, Larson BJ, Cheng TC, Gibson MA, Otani S, Naerdemann W, et al. Abnormal lysosomal trafficking and enhanced exosomal export of cisplatin in drug-resistant human ovarian carcinoma cells. Mol Cancer Ther. 2005;4:1595–1604. doi: 10.1158/1535-7163.MCT-05-0102. [DOI] [PubMed] [Google Scholar]
- 125.Xiao X, Yu S, Li S, Wu J, Ma R, Cao H, et al. Exosomes: decreased sensitivity of lung cancer A549 cells to cisplatin. PloS one. 2014;9:e89534. doi: 10.1371/journal.pone.0089534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PloS one. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wei Y, Lai X, Yu S, Chen S, Ma Y, Zhang Y, et al. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147:423–431. doi: 10.1007/s10549-014-3037-0. [DOI] [PubMed] [Google Scholar]
- 128.Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107(77):djv135. 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed]
- 129.Bovy N, Blomme B, Freres P, Dederen S, Nivelles O, Lion M, et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget. 2015;6:10253–10266. doi: 10.18632/oncotarget.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoshimura A, Sawada K, Kimura T. Is the exosome a potential target for cancer immunotherapy? Ann Transl Med. 2017;5:117. doi: 10.21037/atm.2017.01.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chaput N, Taieb J, Schartz N, Flament C, Novault S, Andre F, et al. The potential of exosomes in immunotherapy of cancer. Blood Cells Mol Dis. 2005;35:111–115. doi: 10.1016/j.bcmd.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 133.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180:7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 134.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120:457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klibi J, Niki T, Riedel A, Pioche-Durieu C, Souquere S, Rubinstein E, et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood. 2009;113:1957–1966. doi: 10.1182/blood-2008-02-142596. [DOI] [PubMed] [Google Scholar]
- 136.Zhou M, Chen J, Zhou L, Chen W, Ding G, Cao L. Pancreatic cancer derived exosomes regulate the expression of TLR4 in dendritic cells via miR-203. Cell Immunol. 2014;292:65–69. doi: 10.1016/j.cellimm.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 137.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shenoda BB, Ajit SK. Modulation of Immune Responses by Exosomes Derived from Antigen-Presenting Cells. Clin Med Insights Pathol. 2016;9:1–8. doi: 10.4137/CPath.S39925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wolfers J, Lozier A, Raposo G, Regnault A, Thery C, Masurier C, et al. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat Med. 2001;7:297–303. doi: 10.1038/85438. [DOI] [PubMed] [Google Scholar]
- 140.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, et al. Malignant effusions and immunogenic tumour-derived exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 141.Lugini L, Cecchetti S, Huber V, Luciani F, Macchia G, Spadaro F, et al. Immune surveillance properties of human NK cell-derived exosomes. J Immunol. 2012;189:2833–2842. doi: 10.4049/jimmunol.1101988. [DOI] [PubMed] [Google Scholar]
- 142.Skokos D, Botros HG, Demeure C, Morin J, Peronet R, Birkenmeier G, et al. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170:3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 143.Escudier B, Dorval T, Chaput N, Andre F, Caby MP, Novault S, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3:10. doi: 10.1186/1479-5876-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3:9. doi: 10.1186/1479-5876-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 2016;5:e1071008. doi: 10.1080/2162402X.2015.1071008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rana S, Yue S, Stadel D, Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol. 2012;44:1574–1584. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 147.Stickney Z, Losacco J, McDevitt S, Zhang Z, Lu B. Development of exosome surface display technology in living human cells. Biochem Biophys Res Commun. 2016;472:53–59. doi: 10.1016/j.bbrc.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 148.Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35:2383–2390. doi: 10.1016/j.biomaterials.2013.11.083. [DOI] [PubMed] [Google Scholar]
- 149.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 150.Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi C, et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. doi: 10.1016/j.biomaterials.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 151.Qi H, Liu C, Long L, Ren Y, Zhang S, Chang X, et al. Blood Exosomes Endowed with Magnetic and Targeting Properties for Cancer Therapy. ACS nano. 2016;10:3323–3333. doi: 10.1021/acsnano.5b06939. [DOI] [PubMed] [Google Scholar]
- 152.Hung ME, Leonard J. Stabilization of exosome-targeting peptides via engineered glycosylation. J Biol Chem. 2015;290:8166–8172. doi: 10.1074/jbc.M114.621383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S. Advances and Challenges of Liposome Assisted Drug Delivery. Front Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bolhassani A, Javanzad S, Saleh T, Hashemi M, Aghasadeghi MR, Sadat SM. Polymeric nanoparticles: potent vectors for vaccine delivery targeting cancer and infectious diseases. Hum Vaccin Immunother. 2014;10:321–332. doi: 10.4161/hv.26796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Raemdonck K, Braeckmans K, Demeester J, De Smedt SC. Merging the best of both worlds: hybrid lipid-enveloped matrix nanocomposites in drug delivery. Chem Soc Rev. 2014;43:444–472. doi: 10.1039/C3CS60299K. [DOI] [PubMed] [Google Scholar]
- 157.Li C, Zhang J, Zu YJ, Nie SF, Cao J, Wang Q, et al. Biocompatible and biodegradable nanoparticles for enhancement of anti-cancer activities of phytochemicals. Chin J Nat Med. 2015;13:641–652. doi: 10.1016/S1875-5364(15)30061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Physiol. 2014;306:C621–C633. doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 159.Aryani A, Denecke B. Exosomes as a Nanodelivery System: a Key to the Future of Neuromedicine? Mol Neurobiol. 2016;53:818–834. doi: 10.1007/s12035-014-9054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Camussi G, Quesenberry PJ. Perspectives on the Potential Therapeutic Uses of Vesicles. Exosomes Microvesicles. 2013;1(6):10.5772/57393. 10.5772/57393. [DOI] [PMC free article] [PubMed]
- 161.Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther. Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, et al. A novel nanoparticle drug delivery system: the anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18:1606–1614. doi: 10.1038/mt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. 2015;7:a020412. doi: 10.1101/cshperspect.a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guo JL, Lee VM. Cell-to-cell transmission of pathogenic proteins in neurodegenerative diseases. Nat Med. 2014;20:130–138. doi: 10.1038/nm.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haque S, Whittaker MR, McIntosh MP, Pouton CW, Kaminskas LM. Disposition and safety of inhaled biodegradable nanomedicines: Opportunities and challenges. Nanomedicine. 2016;12:1703–1724. doi: 10.1016/j.nano.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 167.Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, et al. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J Controlled Release. 2015;207:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32:2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hood JL. Post isolation modification of exosomes for nanomedicine applications. Nanomedicine (Lond). 2016;11:1745–1756. doi: 10.2217/nnm-2016-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hood JL, Scott MJ, Wickline SA. Maximizing exosome colloidal stability following electroporation. Anal Biochem. 2014;448:41–49. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shtam TA, Kovalev RA, Varfolomeeva EY, Makarov EM, Kil YV, Filatov M. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013;11:88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, et al. Bioinspired exosome-mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS nano. 2013;7:7698–7710. doi: 10.1021/nn402232g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.