Abstract
DNA replication mechanisms are conserved across all organisms. The proteins required to initiate, coordinate, and complete the replication process are best characterized in model organisms such as Escherichia coli. These include nucleotide triphosphate‐driven nanomachines such as the DNA‐unwinding helicase DnaB and the clamp loader complex that loads DNA‐clamps onto primer–template junctions. DNA‐clamps are required for the processivity of the DNA polymerase III core, a heterotrimer of α, ε, and θ, required for leading‐ and lagging‐strand synthesis. DnaB binds the DnaG primase that synthesizes RNA primers on both strands. Representative structures are available for most classes of DNA replication proteins, although there are gaps in our understanding of their interactions and the structural transitions that occur in nanomachines such as the helicase, clamp loader, and replicase core as they function. Reviewed here is the structural biology of these bacterial DNA replication proteins and prospects for future research.
Keywords: DNA replication, helicase, primase, DNA polymerase, DNA clamp, clamp loader complex, single‐stranded DNA binding protein, macromolecular structure, protein‐DNA interaction, antimicrobials
Introduction
The transmission of genetic instructions used in life processes is essential to all known living organisms and viruses. Bacterial cells can replicate DNA with remarkable speed and fidelity: in Escherichia coli, the in vitro rate is estimated at ~1000 bp/s (based on chromosome size and replication time) and the rate of spontaneous base‐pair substitutions has been estimated at 2 × 10−10 mutations per nucleotide per generation.1 Additionally, the replisome must overcome obstacles such as damaged DNA and active transcription machinery. Bacterial DNA replication serves not only as a model for understanding DNA replication processes, but is also a source of novel targets for antibacterial agents.2, 3 A detailed (but incomplete) understanding of bacterial DNA replication has resulted from decades of research, mostly using the model organism E. coli.
Replication begins with the assembly of a multiprotein complex at a predefined locus (multiple loci in Archaea and Eukaryota) on a (usually circular) chromosome, which is called the origin of chromosomal replication (oriC in bacteria). Two replication forks are assembled at the origin and advanced in opposite directions around the chromosome. The long‐established model of DNA replication is of a semi‐discontinuous process: the leading strand is synthesized continuously as a single chain and the lagging strand discontinuously in ~2 kb Okazaki fragments. This idealized view has given way to an appreciation that, even in the absence of DNA‐damaging agents, leading strand synthesis is discontinuous.4 Replication terminates in E. coli when the two replication forks meet at the Ter region opposite oriC on the circular chromosome.5, 6 When the replication forks converge and intervening DNA is unwound, remaining gaps are filled and ligated, and any catenanes are removed.7
Several proteins (“the replisome”; Fig. 1) coordinate and catalyze the enzymatic activities required for coupled DNA replication. The replisome has been described as consisting of a hierarchy of strong and weak functional interactions (K D values range from low pM to high μM) and irreversible steps involving nucleotide hydrolysis or incorporation.8 At the head of the replication fork is the primosome (NTP‐powered helicase DnaB and primase DnaG). Single‐stranded DNA is protected by forming a complex with single‐stranded DNA‐binding protein (SSB). The polymerase core comprises PolIIIαεθ, together with the (PolIIIβ)2 sliding clamp, efficiently duplicates DNA from single‐stranded templates. The polymerase core requires primers to commence DNA synthesis; short (~10 nt) RNA primers are synthesized by the DnaG primase. Sliding clamps act as mobile tethers on dsDNA and are required for the processivity of the polymerase core. Sliding clamps are loaded onto primed templates by the clamp loader complex (CLC), comprises PolIIIδ(γ/τ)3δ'ψχ subunits. The clamp loader binds to template DNA with bound RNA primer. Functions in the polymerase core are divided into synthesis (α) and proofreading (ε, a 3′ → 5′ exonuclease). The (non‐essential) θ subunit stabilizes ε.9
Figure 1.
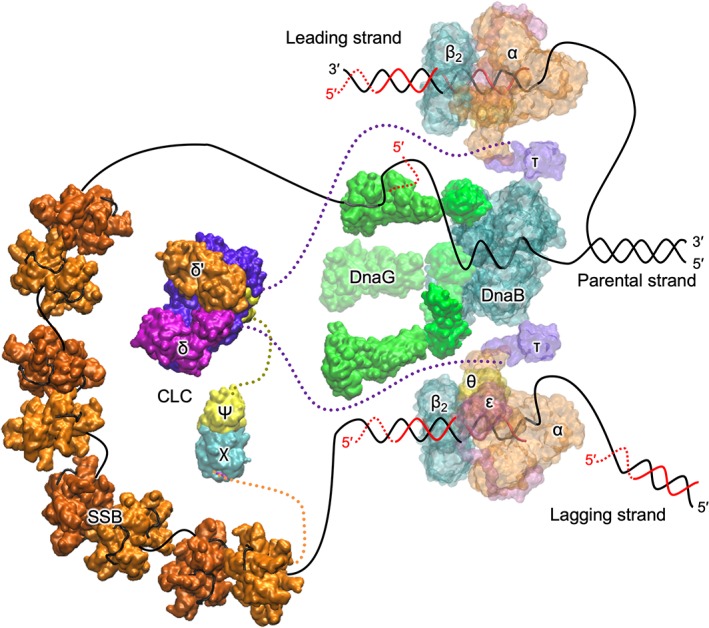
A schematic view of the replisome. Protein assemblies are modeled using representative structures from the PDB. Parent DNA strands are represented by continuous black and red lines for parental and nascent DNA strands, respectively. RNA primers are represented by dashed red lines. Disordered regions in proteins are indicated by dotted lines (lengths not to scale). On the lagging strand is DnaB helicase, which uses the energy of NTP hydrolysis to unwind dsDNA. DnaG primases bind to DnaB and synthesizes RNA primers, required by the polymerase on the lagging strand to initiate Okazaki fragment synthesis. The lagging strand ssDNA is protected by SSB. The clamp loader complex (CLC), with subunit composition δ(γ/τ)3δ'ψχ, uses ATP hydrolysis to load β2 clamps onto RNA‐primed templates DNA. The accessory ψ and χ subunits stimulate the CLC and bridge the CLC to SSB, respectively. The polymerase III core, commencing at primed‐templates, uses ssDNA as a template to synthesize new DNA on both the leading and lagging strands. The β2 sliding clamp acts as a mobile tether and is essential for the processivity of the polymerase. The C‐terminal domains of τ subunits are coupled to Pol III cores and (weakly) to DnaB.
Recent reviews (e.g., Refs. 8, 10) have tended to focus on functional aspects of bacterial DNA replication. Here, I focus on the structural aspects of the bacterial replisome. Structures of DNA‐replication proteins not covered include the Tus protein that binds to Ter sites on DNA regions opposite oriC to terminate DNA replication,5, 6 3′ → 5′ DNA helicases such as PriA, Rep, and UvrD that are involved in replication restart following fork collapse and/or removal of protein roadblocks, and proteins involved in maturation of Okazaki fragment (DNA polymerase I that removes the RNA primer of the downstream Okazaki fragment by its 5′ → 3′ exonuclease while extending the DNA, and DNA ligase that seals the remaining nick).
The Primosome
The primosome is the protein complex responsible for unwinding dsDNA and synthesizing RNA primers on single‐stranded DNA during DNA replication. In E. coli, the proteins considered as part of the primosome are DnaG (synthesizes RNA primers), DnaB (DNA‐unwinding helicase), and DnaC (helicase loader). Central to the replisome is the DnaB6:DnaG3 primase assembly. As the DnaB helicase unwinds dsDNA, the single‐stranded DNA extruded through the center of DnaB is utilized by DnaG to synthesize primers. It should be noted that PriA, PriB, PriC, and DnaT proteins facilitate loading of DnaB onto the lagging strand templates during replication restart.11
Primase
DNA polymerases require a template and primer. DnaG is the replicative primase that synthesizes RNA primers for extension by DNA polymerases. The E. coli primase transcribes roughly 2000 RNA primers per replication cycle.12 The process consists of five steps: template binding, NTP binding, initiation, extension of the primer, and transfer of the primer to DNA polymerase III. DnaG must bind to the DnaB helicase to synthesize primers near the replication fork.13 The E. coli DnaG primase is a monomer of three functional domains: a ~12 kDa N‐terminal zinc‐binding domain (ZBD) that binds specific sites on DNA, a ~37 kDa RNA polymerase domain (RPD), and C‐terminal domain (DnaGC) that interacts with the DnaB helicase and SSB [Fig. 2(a)]. No structure of full‐length DnaG has been determined, possibly reflecting the flexible nature of its interdomain linkers.
Figure 2.
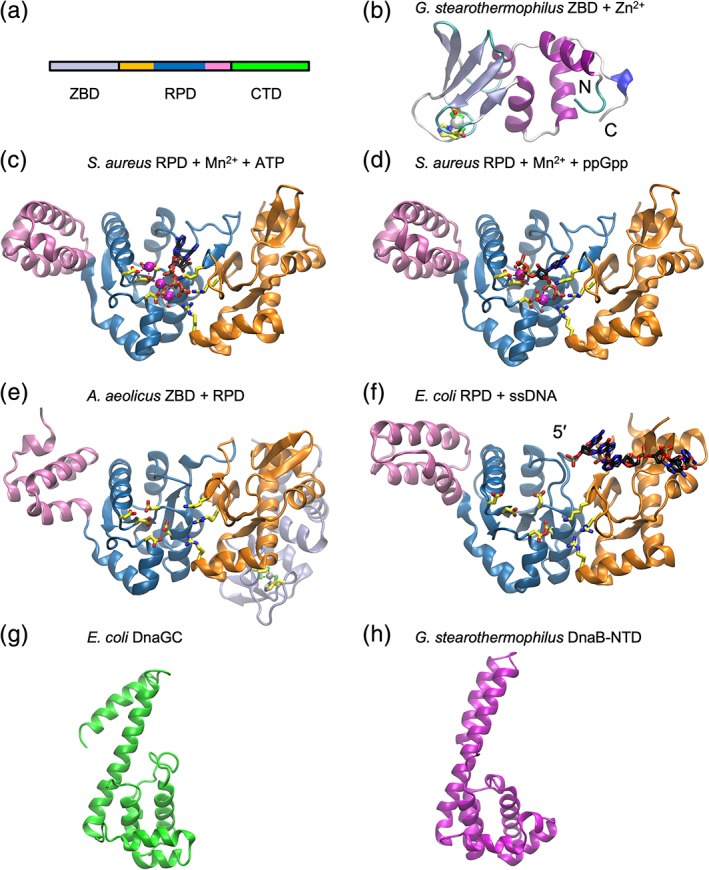
Cartoon representations of DnaG primase. (a) Arrangement of domains. The subdomains of RPD are indicated in orange (N‐terminal segment), blue (TOPRIM), and pink (C‐terminal segment). (b) ZBD from G. stearothermophilus (PDB ID 1D0Q) with Zn2+ (gray sphere) and zinc‐binding residues in stick form. (c) Staphylococcus aureus RPD domain with bound Mn2+ ions (magenta spheres) and ATP (PDB ID 4EDG). Bound ATP (black carbon atoms) and interacting side‐chains (carbon atoms yellow) are shown in stick form. (d) Staphylococcus aureus RPD domain with bound Mn2+ ions (magenta spheres) and alarmone ppGpp (PDB ID 4EDT). Bound ppGpp (black carbon atoms) and interacting residues (yellow carbon atoms) in stick form. (e) Aquifex aeolicus ZBD and RPD domains (PDB ID 2AU3). Zn2+ bound to ZBD is shown as a white sphere. Catalytic‐ and Zn2+‐binding residues shown in stick form. (f) Escherichia coli RPD domain with ssDNA (PDB ID 3B39). (g) DnaGC from E. coli (PDB ID 2HAJ). (h) DnaB‐NTD from G. stearothermophilus (PDB ID 2R6A).
The synthesis of RNA primers is initiated at specific triplet sequences [5′‐d(CTG) in E. coli]. The recognition mechanism of such initiation sites has remained elusive, but is known to involve the ZBD. The structure of a DnaG‐ZBD was first determined for the Geobacillus stearothermophilus homolog.14 Conserved across all viral, bacteriophage, prokaryotic, and eukaryotic DNA primases it contains the “zinc ribbon” topology decorated with helices [Fig. 2(b)]. The Zn2+ ion is coordinated by three cysteines and one histidine residue. The core of a zinc ribbon is composed of two structurally similar zinc‐chelating “knuckles” present as turns of β‐hairpins.15
The RPD is composed of three subdomains: the N‐terminal segment with a unique α/β fold, the central RNA polymerase subdomain belonging to the topoisomerase‐primase (TOPRIM) family, and the C‐terminal segment with an antiparallel, three‐helix bundle [Fig. 2(c)].16 The TOPRIM family is common to primase, topoisomerase, overcoming lysogenization defect nucleases, and RecR.17 It consists of a five‐stranded β sheet sandwiched by six α helices. The phosphotransferase activity in such enzymes is mediated by divalent metal ions (usually Mg2+), essential for binding DNA and NTPs. Directly observed in the Staphylococcus aureus RPD, a cluster of acidic residues in the polymerase subdomain coordinates catalytic metal ions, which in turn coordinate the phosphate groups of nucleotide triphosphate (NTP) substrates. The adjacent N‐terminal segment contains conserved basic residues that also contact the NTP moiety.18 The structure of RPD from S. aureus has been determined in complex with the alarmones ppGpp and pppGpp.18 These intracellular signaling molecules are produced in the stringent response to nutrient depletion and impede primer formation by directly binding to the NTP binding site [Fig. 2(d)], thus stalling DNA replication.
The crystal structure of a DnaG fragment containing the ZBD and RPD from Aquifex aeolicus shows the ZBD is bound through hydrophobic and polar contacts at a face on the opposite side of the RPD from the active site [Fig. 2(e)]. SAXS experiments using E. coli DnaG show that, while its ZBD also docks against the RPD, its mode of engagement is distinct from that seen in A. aeolicus.19 A mechanism was proposed whereby the ZBD of one primase can undock from its RPD and in conjunction with the RPD domain of an adjacent subunit scan for an initiation site. Once bound, the ZBD works with the RPD domain in trans to commence primer synthesis.
The RPD is a relatively inefficient polymerase with weak affinity for DNA.20 Thus, complexes of RPD with ssDNA template have been difficult to observe. Crystal structures of RPD from E. coli 21 and Mycobacterium tuberculosis 22 in complex with DNA shed some light on enzyme‐template interactions. Corn and co‐workers used an innovative cysteine‐scanning/crosslinking approach to trap an RPD/ssDNA complex.21 The structure of the complex shows that the ssDNA template is loosely bound at a basic groove in the N‐terminal and TOPRIM segments [Fig. 2(f)].21 Interactions were observed between the phosphate backbone and protein, and congruent with the nonspecific nature of the RPD, no specific interactions were observed with base‐pair forming regions of the ssDNA. The 3′‐end of the template is oriented toward the catalytic site. There is currently no structure reported of a RPD with a DNA–RNA hybrid that would result from primer synthesis.
The structure of the helicase‐interacting domain DnaGC from E. coli has been determined in both crystal23 and solution states24 [e.g., Fig. 2(g)]. DnaGC has an N‐terminal helical bundle similar to the N‐terminal domain of DnaB with which it interacts [e.g., Fig. 2(h)] (vide infra), followed by a long helix and a C‐terminal helical hairpin. Structures of homologs from G. stearothermophilus 25 and Helicobacter pylori 26 have also been reported and show similar topology in spite of poor sequence conservation. Using bioinformatic and site‐directed mutagenesis, the region of the helical bundle that binds to the conserved C‐terminal residues of SSB was identified.27
Helicase
DNA helicases, powered by NTP hydrolysis, separate duplex DNA into single strands. The E. coli DNA unwinding helicase, DnaB, belongs to the SF4 family of hexameric helicases that include the replicative helicases of all bacteria as well as bacteriophages T4, T7 and others. DnaB is loaded onto ssDNA, a process requiring DnaA (at oriC) and DnaC (at oriC and during replication restart). Biochemical analysis identified two domains within the 52 kDa DnaB: a ~12 kDa N‐terminal domain (NTD) and a ~33 kDa C‐terminal domain (CTD) joined by a linker domain containing a helix [Fig. 3(a)].28 The NTD is important for both helicase activity and for binding specific partner proteins. The CTD of DnaB‐like helicases contains an AAA ATPase domain similar to the core fold first observed in RecA.29 Energy from nucleotide hydrolysis powers the helicase.30 DnaB can hydrolyze all NTPs with a preference for purine over pyrimidine nucleotides.31 Confusingly, the replicative helicase in some Gram‐positive species (e.g., in Bacillus subtilis) is designated DnaC, and the helicase loader is DnaI.
Figure 3.
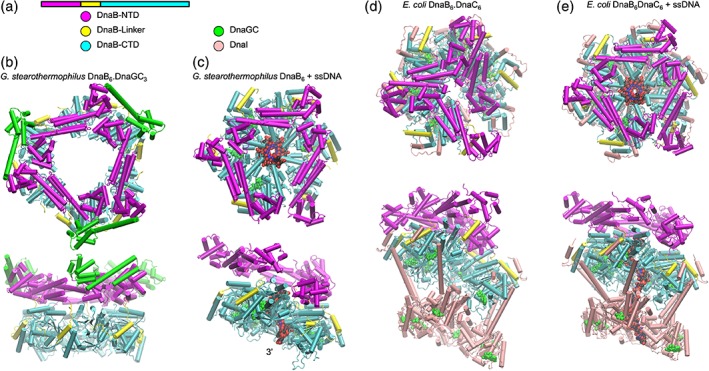
Cartoon representations of DnaB helicase. (a) Arrangement and color coding of DnaB domains and associated proteins. (b) Orthogonal views of G. stearothermophilus DnaB6.DnaGC3 complex (PDB ID 2R6A). (c) Orthogonal views of G. stearothermophilus DnaB6 in complex with ssDNA (VDW spheres) (PDB ID 4ESV). (d) Orthogonal views of the DnaB6.DnaC6 complex (E. coli) (PDB ID 6QEL). (e) Orthogonal views of the DnaB6.DnaC6 complex with ssDNA (E. coli) (PDB ID 6QEM). Bound nucleoside phosphates are represented as green VDW spheres.
Negative‐stain electron microscopy demonstrated symmetric arrangement of subunits into a ring with a pore diameter of 3–4 nm.32, 33 DnaB translocates along ssDNA (which passes through the central channel) in the 5′ → 3′ direction to provide the lagging strand template, whereas the leading strand is occluded. DnaB is associated with a loader protein, DnaC, which together with DnaA helps chaperone two DnaB hexamers onto ssDNA strands during initiation of replication.34 Furthermore, multiple quaternary states of DnaB were observed: in the presence of ATP, ATPγS, AMP‐PNP, or ADP, DnaB formed rings with C3 or C6 symmetry.33 Discerned from the first three‐dimensional structure of DnaB (by cryo‐EM) was the hexamer with two faces: one with C3 and the other with C6 symmetry.35 Published simultaneously, the solution Nuclear Magnetic Resonance (NMR)36 and crystal X‐ray structures37 of the NTD of E. coli DnaB revealed a helical bundle similar to the primary dimerization domain of E. coli gyrase A. The crystal structure of the G. stearothermophilus DnaB/DnaGC complex in multiple crystal forms38 revealed a flat, two‐tier configuration with the NTD “collar” forming a trimer of dimers, and the CTD ring with approximately sixfold symmetry [Fig. 3(b)]. No nucleotide analogue or DNA was bound. In this structure, the central channel is dilated: the diameter is ~50 Å, wide enough to accommodate dsDNA. Neighboring NTDs interact through helical hairpins to produce dumbbell‐shaped motifs. The NTD of M. tuberculosis DnaB has also been observed to form the trimer‐of‐dimers arrangement in isolation.39 DnaGC was observed to bind to interfaces between NTDs, giving a DnaB6.DnaGC3 stoichiometry. The crystal structure of hexameric G40P, a DnaB family helicase from B. subtilis bacteriophage SPP1 is similar to G. stearothermophilus DnaB,40 but with a narrower channel: NTD collar, ~42 Å; CTD ring, ~17 Å. Again, no NTP was observed bound in these structures. The helicase from A. aeolicus in complex with ADP shows a constricted arrangement of CTDs, and a highly constricted arrangement of the NTDs not observed previously.41 The co‐crystal structure of G. stearothermophilus DnaB with ssDNA and GDP‐AlF4 (mimicking the pentavalent transition state of GTP hydrolysis) [Fig. 3(c)] is highly informative, revealing a spiral arrangement of subunits around ssDNA, which in turn adopts a conformation observed in A‐form dsDNA.42 Eleven nucleotides of ssDNA were observed in the CTD ring, held by loops that each bind two phosphodiester bonds.
The available DnaB structures provide valuable but incomplete insight into the complex interplay between structural states and function. The significance of the dilated forms of DnaB is still unclear. In terms of mechanism, the spiral arrangement of G. stearothermophilus DnaB led Itsathitphaisarn and co‐workers to propose a hand‐over‐hand translocation mechanism in which sequential hydrolysis of NTP is coupled to two‐nucleotide translocation steps along ssDNA.42 The recent cryo‐EM structure of the bacteriophage T7 replisome suggests that a similar mechanism operates in the T7 primase/helicase gp4. In this complex, a gp4 hexamer forms a spiral arrangement of subunits around ssDNA that in turn adopts a spiral conformation similar to B‐form DNA,43 and a hand‐over‐hand mechanism was proposed with the helicase advances two‐nucleotides per step.
As noted above, helicases are sometimes loaded onto ssDNA by helicase loader proteins. Helicase loaders are members of the AAA+ (ATPases associated with various cellular activities) superfamily of nucleotide hydrolases. In E. coli, helicase loader DnaC is believed to act as a “ring‐breaker,” parting a subunit interface in the DnaB helicase ring. Observed by cryo‐EM, the E. coli DnaB6.DnaC6 complex forms a three‐tiered assembly in which DnaC N‐terminal domain, binding to the DnaB‐CTD adopts a spiral configuration that produces a break in DnaB, allowing mounting onto ssDNA [Fig. 3(d)].44 ATPase activity of DnaC is not required for ring opening or loading of DnaB onto ssDNA.45 Instead, DnaC bound to ATP appears to stabilize the open spiral conformation of the complex. Binding of DnaB6.DnaC6 to ssDNA stimulates ATP hydrolysis by DnaC and leads to closure of the break [Fig. 3(e)] and dissociation of DnaC through an unknown mechanism. Remarkably, the open ring conformation of DnaB observed in the DnaB6.DnaC6 complex is identical to that observed in the complex of DnaB with bacteriophage λ helicase loader (λP).46 Here, five copies of λP bind at the interfaces of the DnaB‐CTD to stabilize the open spiral.
Single‐Stranded DNA‐Binding Protein
Single‐stranded DNA produced by the action of helicase is vulnerable to damage. The SSB protein binds to and protects ssDNA, and prevents the formation of secondary structures that might impede replication. SSB is found as a tetramer with D 2 symmetry; the N‐terminal domain (residues 1–112) adopts the OB fold and is responsible for DNA binding.47 The CTD contains an intrinsically disordered linker region (residues 113–178 in E. coli), terminated by nine highly conserved residues (MDFDDDIPF; “SSB‐Ct”) that mediate interactions between SSB and interaction partners that include DnaG (vide supra) [Fig. 4(a)]. Deinococcus radiodurans SSB is unusual: it is a homodimer in which monomers contain two OB domains.48 Escherichia coli SSB displays at least three modes of binding to ssDNA that are favored under different concentrations of mono‐ and divalent ions. Referred to as (SSB)65, (SSB)56, and (SSB)35, the binding modes differ in the number of nucleotides, (SSB)n bound to the tetramer.49 Favored under high salt conditions (>200 mM NaCl, >10 mM MgCl2), the (SSB)65 mode shows limited positive cooperativity. Electron microscopy shows DNA wrapped in beads of ~140–160 nucleotides around SSB octamers.50 Under low‐salt conditions (<20 mM NaCl, <1 mM MgCl2), the (SSB)35 mode is a highly cooperative binding mode in which SSB tetramers are clustered on ssDNA. However, highly cooperative binding of E. coli SSB to DNA also occurs at physiological salt and glutamate concentrations.51
Figure 4.
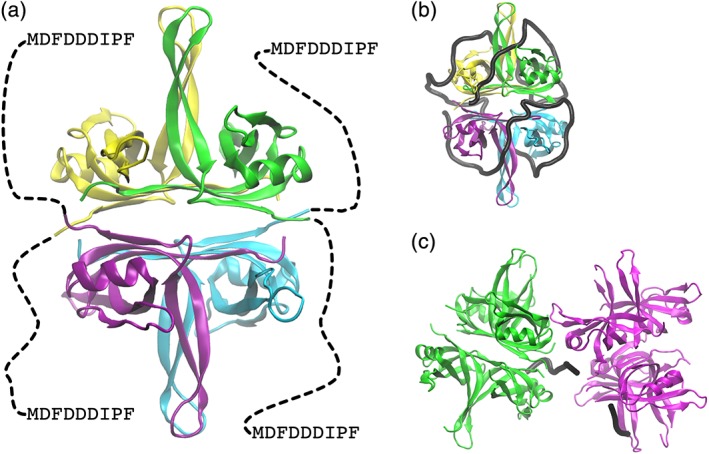
Cartoon representations of SSB. (a) Escherichia coli tetramer (PDB ID 4MZ9). Each monomer is shown in a different color. Disordered C‐terminal region indicated by dashed line. (b) Mode of (SSB)65 complex based on E. coli SSB‐DNA complex (PDB ID 1EYG). DNA is shown as a black trace. (c) Bacillus subtilis SSBA octamer with bridging DNA (black trace) (PDB ID 6BHX).
The structure of E. coli SSB in complex with dC35 polymers revealed a “baseball seam” topology of the DNA [Fig. 4(b)].52 Interaction of SSB with DNA occurs through salt‐bridges with the phosphate backbone, stacking of bases on aromatic side‐chains, and occasional H‐bonds with bases. Crystal structures of SSB homologs from different species with ssDNA, for example, Helicobacter pylori,53 Mycobacterium smegmatis, B. subtilis,54 and Plasmodium falciparum 55 suggest that in other species ssDNA is bound with a “baseball seam” topology similar to E. coli but opposite 5′ → 3′ polarity.
Insight into the arrangement of SSB octamers is provided by a recent crystal structure of a B. subtilis SSB homolog (SSBA) bound to dT35. SSBA tetramers associate via an ssDNA bridge and a conserved tetramer–tetramer interaction surface termed the “bridge interface” [Fig. 4(c)].56
SSB‐Ct is known to bind to at least 14 other proteins involved in genome maintenance including the aforementioned DnaGC. Being essential, interactions of SSB‐Ct with binding partners, such as ExoI,57 PriA,58 and DnaGC,59 have been targeted for the discovery of novel inhibitors for antibacterial development.
Polymerase polIIIα
Bacterial replicative polymerases (PolIIIα in E. coli) comprise the “C family” of DNA polymerases. There are two major forms: PolC (present in low‐GC Gram‐positive bacteria) and DnaE (Fig. 5).60 PolC and DnaE share only ~20% sequence identity and display domain rearrangements. PolC contains a 3′ → 5′ directed proofreading exonuclease domain. In organisms using DnaE as the replicative polymerase, there is a separate exonuclease protein (vide infra). The structure of DnaE from E. coli 61 and Thermus aquaticus 62 was reported in 2006. The structures have been likened to a cupped right hand, with Fingers, Palm, and Thumb domains that form the catalytic core of the enzyme (shared with all other DNA polymerases). In addition, PolC and DnaE homologs contain a polymerase and histidinol phosphatase (PHP) domain adopting a TIM‐barrel topology located near the Thumb domain. In some species (for example, in Thermus thermophilus PolIIIα) the PHP domain is a metal‐dependent nuclease that may play a role in proofreading, and in others is inactive.63 Surprisingly, the structures of DnaE and PolC are unrelated to the eukaryotic replicative polymerases: their palm domain has the topology of the X‐family DNA polymerases, which includes Pol β (a non‐processive eukaryotic polymerase involved in base excision repair).
Figure 5.
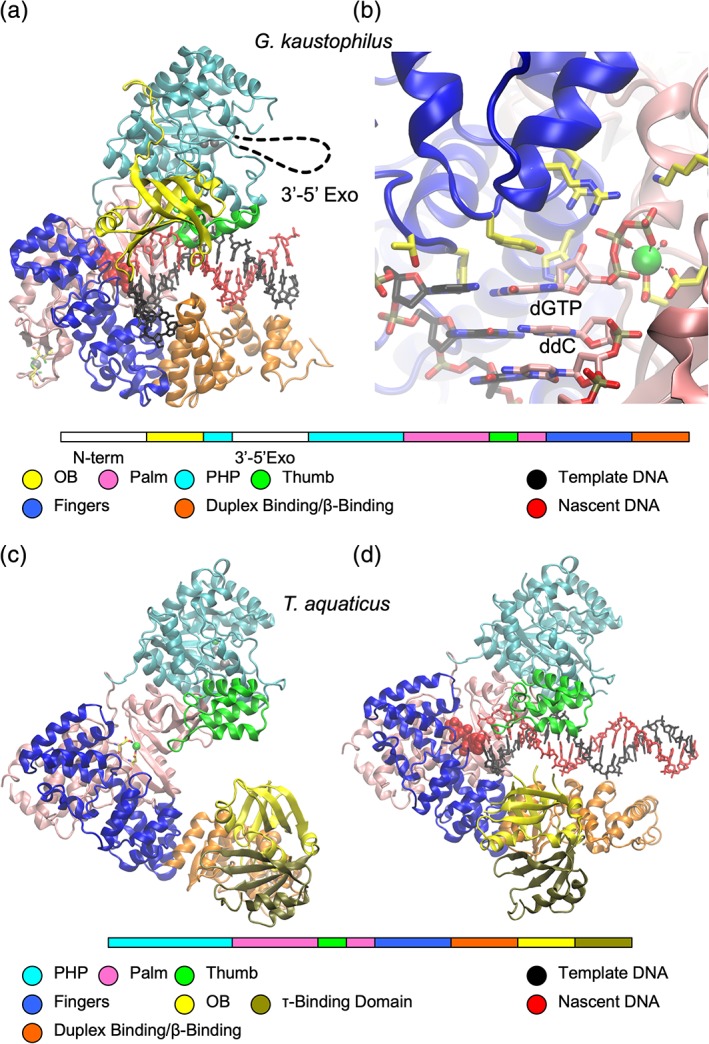
Cartoon representations of bacterial DNA polymerases. (a) PolC from G. kaustophilus with arrangement and color coding of domains shown below (PDB ID 3F2B). (b) PolC catalytic site. Template and nascent DNA strands shown in stick form with carbon atoms black and pink, respectively. (c) Thermus aquaticus DnaE with arrangement and color coding of domains shown below (PDB ID 2HPI). (d) Thermus aquaticus PolIIIα in complex with DNA (PDB ID 3E0D).
The first high‐resolution structure of a replicase‐DNA complex was the Geobacillus kaustophilus PolC in a ternary complex with DNA and dGTP, reported by Evans et al. in 2008.64 In the PolC construct used, the poorly conserved N‐terminal domain was deleted, and the exonuclease domain was removed to protect the DNA from degradation during crystallization. The 3′‐end of the primer strand DNA was terminated with a dideoxynucleoside to prevent ligation of the incoming dGTP by the (still active) protein. The incoming template ssDNA strand enters the active site through a crevice formed between the Fingers and Duplex binding domain. In spite of the OB domains of PolC and DnaE being located in different orders in sequence, they appear to play similar roles in binding ssDNA: guiding the template strand into the catalytic site. The active site lies between the palm and fingers domains [Fig. 5(a)]. In general DNA polymerases use two metal ions, “metal A” and “metal B” to catalyze DNA synthesis. Metal A lowers the pK a of the primer terminal 3′‐OH group and metal B coordinates the incoming nucleoside 5′‐triphosphate. In the PolC structure, an Mg2+ ion was observed at the metal B site coordinated to the dGTP triphosphate and to carboxylic acid groups of two conserved aspartate residues in the palm domain. Evans et al. observed no density for metal A, attributing its apparent absence to the lack of 3′‐OH group in the primer. It should be noted that structural studies of eukaryotic polymerases (including X‐family polymerases) have identified a third metal “metal C” that stabilizes the product PPi. Whether this third metal contributes to the transition state remains controversial.65
In DnaE and PolC structures, the duplex DNA product is held between the thumb and fingers domains. Contacts of these domains are primarily with the phosphate backbone, utilizing electrostatic and hydrogen bonding interactions.
The availability of full‐length α structures from T. aquaticus in the absence62 and presence66 of DNA shows how the presence of DNA induces conformational changes in the polymerase. The β‐clamp binding domain twists by about 20° toward the palm, allowing it to interact with dsDNA [Fig. 5(c,d)]. This rotation appears to bring its OB domain into a position to bind the incoming ssDNA template.
The Proofreading Exonuclease
The 3′ → 5′ directed proofreading function of PolIIIε (encoded by dnaQ) contributes to the extremely high fidelity of bacterial DNA replication. The ε subunit comprises a 185‐residue N‐terminal domain with exonuclease activity, a flexible “Q‐linker” sequence, and a smaller CTD that interacts with PolIIIα. The high mobility of the 22 residue Q‐linker in the αεθβ2 complex was demonstrated by NMR.67
The structure of the catalytic N‐terminal domain of the ε subunit (ε‐NTD) of PolIII from E. coli was determined in 2002 by Hamdan et al.68 The ε‐NTD adopts a five‐stranded mixed β‐sheet topology decorated by α helices. The N‐terminal domain belongs to the “ribonuclease H‐like” superfamily that includes many exonucleases.69 The active site is formed by residues contributed by α‐helices (α4 and α7) and strand β1 (Fig. 6). Two divalent metal cations were observed to be coordinated by three aspartate residues (D12, D103, and D167), and water molecules. The structure contained bound competitive inhibitor, thymidine‐5′‐monophosphate. The hydrolysis reaction is thought to involve a coordinated water molecule (acting as a nucleophile) that is deprotonated by a histidine residue (H162) that acts as a general base.
Figure 6.
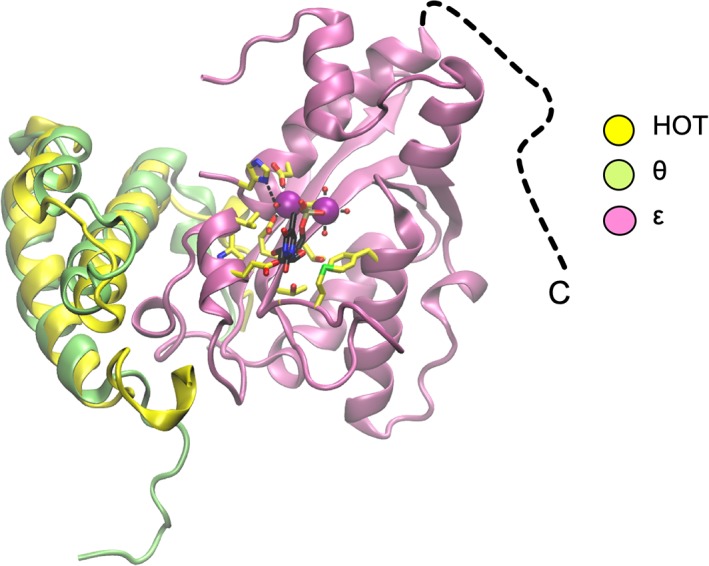
Cartoon representations of polIIIε NTD in complex with HOT (PDB ID 2IDO). Superposed on HOT is the NMR structure of polIIIθ from the polIIIεθ complex (PDB ID 2AXD). The disordered C‐terminal region of ε is indicated by a dashed line. Catalytic residues and bound dTMP are shown (yellow and black carbon atoms, respectively). Catalytic Mn2+ ions are shown as magenta spheres.
In E. coli, ε is stabilized by, and its efficiency enhanced by, polIIIθ (encoded by holE). However, it is not essential to the function of ε and is not present in most bacteria.9 The structure of ε‐NTD was solved in complex with HOT, a θ homolog from bacteriophage P1.70 Furthermore, the structure of 13C/15N‐labeled θ in complex with unlabeled ε‐NTD has been determined by multidimensional NMR spectroscopy.71 The structure was refined using pseudocontact shifts that resulted from the use of lanthanide ions bound to the active site of ε‐NTD. Both HOT and θ form three‐helix bundles (Fig. 6) and interact with an edge of the β‐sheet and helix α1, across which the nucleotide substrate lies. Thus, HOT and θ appear to stabilize the active site of ε.
The Sliding Clamp
PolIIIβ is the sliding clamp that serves as a processivity‐promoting factor in DNA replication. It acts as a mobile tether on DNA and prevents dissociation of the other components of the polymerase core (αεθ). It is not only utilized in DNA replication, but also by repair polymerases, DNA ligases, exonucleases, and the mismatch repair protein MutS. First reported in 1992,72 the structure is deceptively simple: the protein is a dimer, each monomer consisting of three “DNA clamp” domains (I–III), and the overall structure is of a torus that surrounds DNA [Fig. 7(a)]. The monomers interact in a head‐to‐tail fashion that imparts C2 symmetry on the functional protein. (Clamps from archaea, eukaryota, and some viruses are C3 trimers with each monomer containing two repeats for the DNA‐clamp domain.) Sliding clamps are very stable on closed‐circular but not linear dsDNA: the half‐life of the E. coli sliding clamp bound to circular DNA is 72 min at 37°C,73 but dissociates rapidly from linear DNA.74 These observations provided the first indication that clamps could slide freely on dsDNA.
Figure 7.
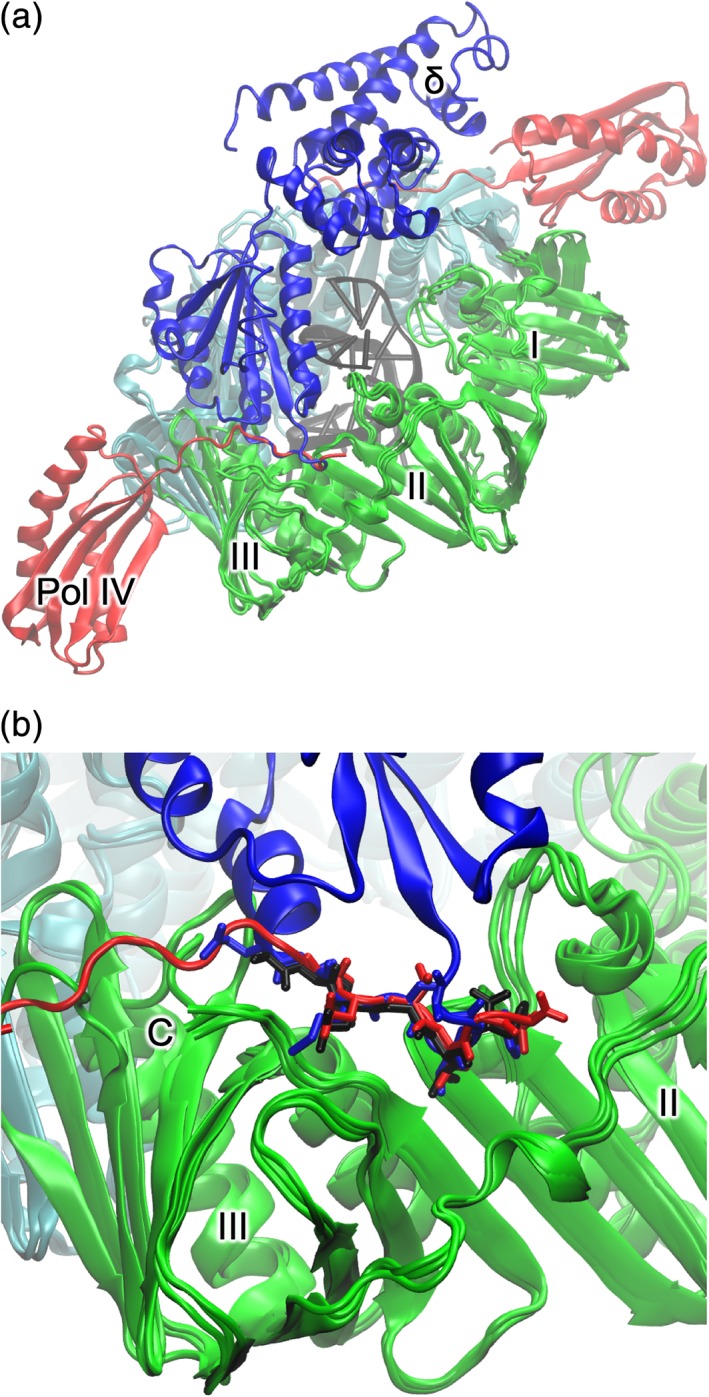
Cartoon representations of complexes of the β‐sliding clamp. β‐subunits are shown in green and cyan. Domains I–III are labeled. (a) Superposed are β‐sliding clamp in complex with polIIIδ (blue) (PDB ID 1JQJ), Pol IV little‐finger domain (red) (PDB ID 1UNN), and DNA (black) (PDB ID 3BEP). (b) Close‐up of the common binding site of β‐clamp binding partners.
The sliding clamp contains a pocket for recognizing and binding specific clamp binding motifs (CBMs; with consensus sequence QL[S/D]LF or QLxLx[L/F]) in binding partner proteins.75 Several structures of β‐clamps in complex with peptides and proteins from binding partners have been reported. The Pol IV “little finger” domain,76 peptides derived from Pol II and PolIIIα77 and PolIIIδ78 bind to the β‐clamp with a common interaction with a conserved binding pocket in domain III [Fig. 7(b)].
Sliding clamps from numerous bacterial species have been reported including pathogens M. tuberculosis,79 H. pylori,80 and Pseudomonas aeruginosa.81 The structures are highly conserved with respect to the E. coli structure. The conserved nature of the binding pocket and interacting peptides have suggested the β‐clamp as an antibiotic target.77, 82, 83, 84 Of note is the structure of the M. tuberculosis clamp bound to the antibiotic griselimycin—a compound effective at killing resistant M. tuberculosis by blocking the peptide‐binding site on the β‐clamp.79
Putting the Core Together
Several approaches have been used to obtain clues concerning the structure of the replicase core, β‐clamp, and DNA. The structure of the E. coli β‐clamp in complex with dsDNA with an ssDNA extension has been reported.85 The dsDNA component passes through the center of the torus at a pronounced angle (22°) [Fig. 7(b)]. The ssDNA forms a crystal contact with an adjacent β subunit where it binds the protein‐binding pocket of the sliding clamp. The structure of the C‐terminal region of ε (ε‐CTS) interacting with the PHP‐domain of α was reported.67 Furthermore, the linker region of ε contains a weak (K D = 210 ± 50 μM) but important CBM.86 The crystal structure of the CTD of an α‐τ chimera has also been determined (Prof. Nicholas Dixon & Dr. Zhi‐Qiang Xu, unpublished results).
Recent cryo‐EM studies have provided 7–8 Å structures of the polymerase α of E. coli in complexes with the polymerase‐binding domain (V) of the clamp loader τ (residues 500–643; vide infra) β‐clamp, proofreading exonuclease ε, and DNA [Fig. 8(a,b)].87 Structures of three complexes were generated: αεβ2τ, αεβ2τ with DNA bound, and αεβ2 with DNA. The structures show how DNA interacts with α and pass through the β‐clamp, and how the proofreading exonuclease ε is positioned in the complex. A cryo‐EM structure of the catalytic core in the editing mode was determined.88 The presence of a mismatch in the DNA caused fraying and enabled the nascent strand to reach the exonuclease active site, and the polymerase thumb domain acted as a wedge that separated the two DNA strands [Fig. 8(c)].
Figure 8.
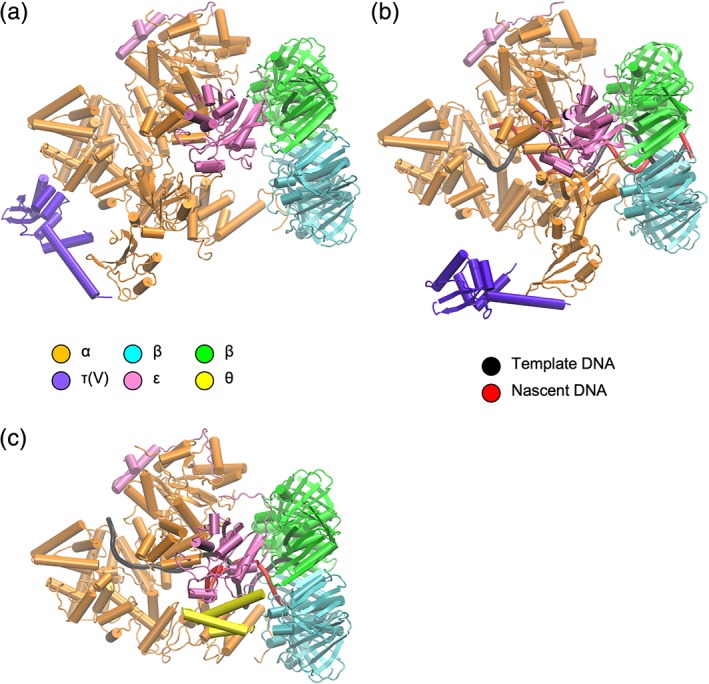
Cartoon representations of the polIII replicase cores in different complexes. Color coding of different subunits in the complexes is indicated. (a) Without DNA (PDB ID 5FKU). (b) With DNA (PDB ID 5FKV). (c) With DNA in proofreading mode (PDB ID 5M1S).
The Clamp Loader Complex
Sliding clamps are actively loaded onto primed template DNA by ATP‐dependent clamp loader complexes.89 The E. coli CLC comprises seven subunits: δτnγ(3 − n)δ'–ψχ (n = 0–3; CLCs in the replisome have n ≥ 2). The δ and δ' subunits (encoded by holA and holB) together with three copies of γ and/or τ (encoded by dnaX) form a heteropentamer. The χ and ψ subunits (encoded by holC and holD) are not required for clamp‐loading activity, but serve to bridge the CLC with SSB. The γ subunit is a truncated (residues 1–431) form of τ (residues 1–643) resulting from a programmed frameshift during translation of dnaX mRNA.90, 91, 92 The apo‐structure of the E. coli δγ3δ' complex was first determined by Jeruzalmi et al. in 2001,93 and later in complex with ADP or ATPγS94 and DNA.95 Common to the δ, γ/τ, and δ' subunits are three conserved domains (I–III). The first two (I and II) are related to the nucleotide‐binding domains of AAA+ ATPases, although only the γ/τ subunits support ATPase activity. The third domain (III) forms a “helical collar” that supports the pentameric arrangement of subunits.93 The τ subunit consists of an additional two domains: IV, which binds DnaB helicase;96 and V, which binds PolIIIα [Fig. 9(a)].97 NMR‐based studies of domains IV and V have shown that only a 14 kDa fragment of domain V is structured in the absence of binding partners.98 This fragment (residues 500–643) was observed in the above complex with α. The structure of the complex of DnaB helicase with domain IV of τ remains unknown.
Figure 9.
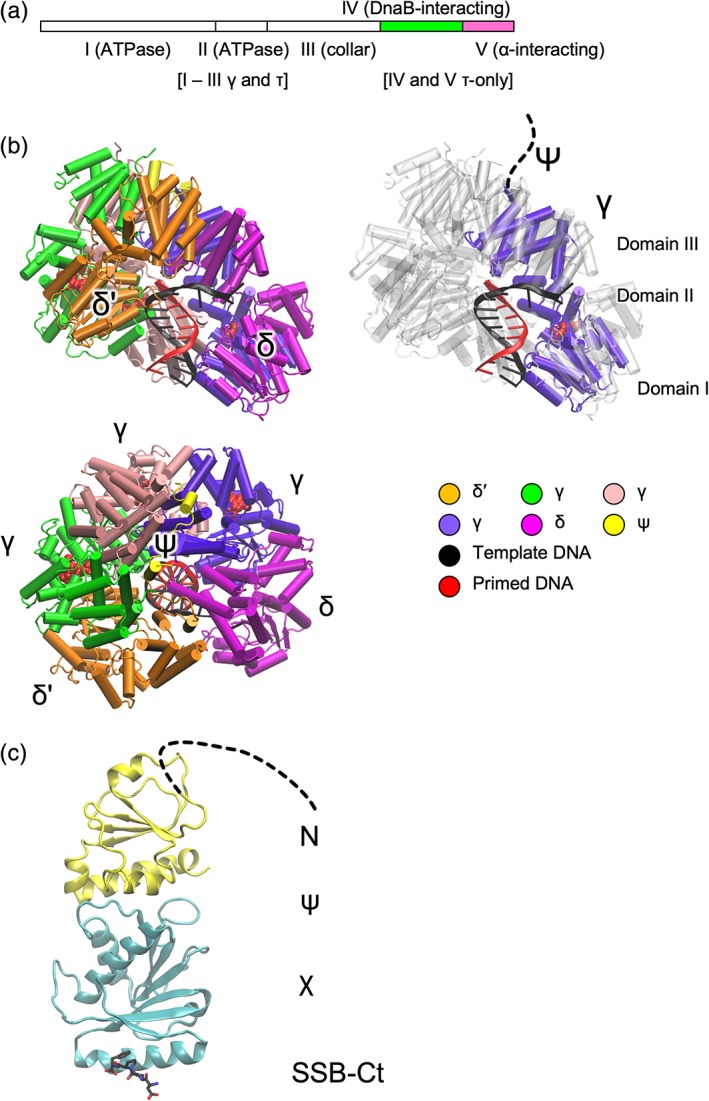
Representations of clamp loader complex proteins. (a) Organization of τ domains; the truncated version γ comprises domains I–III. (b) Cartoon representations of E. coli CLC in complex with primed template DNA (PDB ID 3GLI). Views perpendicular and parallel to the axis of DNA are shown. The view on the right shows one of the γ‐subunits (domains I–III) with others in gray. (c) Cartoon representation of the ψ:χ complex with SSB‐Ct peptide (black carbon atoms) (PDB ID 3SXU).
The δγ3δ' complex in the apo, ADP‐, or ATPγS‐bound states are nearly identical.94 While oligomeric AAA+ ATPases typically bind ATP at the interface of adjacent monomers,99 the δγ3δ' complexes with bound nucleotide showed no nucleotide‐mediated interactions in the interfaces. However, in the δγ3δ' complex bound to DNA, an N‐terminal segment of ψ and ATP analogue ADP·BeF3, domains I and II shift significantly to form a spiral that tracks the primed‐DNA template strand and brings the “arginine fingers” (R169 in γ; R158 in δ') into contact with BeF3 (analogous to the γ‐phosphate) in adjacent domains [Fig. 9(b)].95 Without bound DNA, the nucleotide‐binding domains do not adopt this spiral arrangement.93 This accords with the observation that the CLCs use ATP hydrolysis to trigger release of the complex from DNA. Hinging motions between the collar and ATPase domains accommodate the different arrangements of each set of domains. On release from DNA, the CLC can exchange bound ADP for ATP.89 The collar domain of the δ subunit recognizes primers: the side chain of Y316 stacks on the nucleotide at the 3′ end of the primer strand. The emerging template strand follows a groove on the δ subunit surface. The N‐terminal 28 residues of ψ bind to across the collar domains of all three γ subunits [Fig. 9(b)] and is sufficient to promote clamp‐loading activity. The only change induced to the collar by ψ peptide binding is the rotation of the collar domain of the C‐terminal tail of one of the γ subunits and the formation of a β‐sheet with ψ. DNA‐ and ψ‐binding appear independently to induce the same conformational change in the collar domain subunit.
In E. coli, the χ and ψ subunits serve to link the clamp loader complex and SSB, with χ binding to SSB. Through its interaction with the CLC and SSB, the χψ complex plays an important role in the processivity of Okazaki fragment synthesis. The structure of E. coli χψ100 revealed that the folds of χ and ψ are similar to mononucleotide and dinucleotide binding proteins, respectively. The N‐terminal 26 residues of ψ (that bind to the CLC‐collar) are disordered in this structure. The E. coli χψ complex with SSB‐Ct [Fig. 9(c)] shows binding of the peptide in a pocket on χ opposite the disordered N‐terminal end of ψ.101
The δ subunit is able to function as a clamp‐opener in isolation, binding to the sliding clamp ring and opening it. The crystal structure of the β:δ complex shows that δ binds to β such that one of its dimer interfaces is destabilized.78
To date there is no reported structure of a complex of a bacterial CLC with DNA and β‐clamp. However, the structure of the bacteriophage T4 clamp loader in complex with ATP, open clamp, and primer‐template DNA shows that both the CLC and open clamp adopt spiral conformations that match the helical symmetry of DNA.102
Concluding Remarks
With structures of all DNA‐replication components determined, attention is moving toward understanding higher‐order assemblies and dynamic structural changes. The past ~7 years has seen the emergence of techniques for the generation of near‐atomic resolution structures by cryoelectron microscopy in what has been called the “resolution revolution.”103 With many subcomplexes of the replisome refractory to crystallization, it appears likely that this technique will be used to observe novel subcomplexes and previously determined complexes in novel arrangements and functional states not previously observed. This is exemplified by the recent determination of the structure of the complex of polIIIαεβ2τ(V) with dsDNA in polymerization87 and proofreading modes.88 The detailed mechanisms by which the CLC and DnaB helicase transduce the energy of NTP hydrolysis into structural changes remain to be elucidated, and new structures of these complexes in different states could illuminate these essential processes.
In cases where structures of homologs of nanomachines such as DnaB helicase were determined from different species in different structural states, there is difficulty in their interpretation. Much of the available biochemical data pertain to the E. coli DnaB and it is not clear how much is relevant to G. stearothermophilus, A. aeolicus, and phage SPP1 DnaB homologs. New structures of these proteins in different structural states will allow comparative structural biology and assist in the elucidation of species‐specific differences in function.
Other areas where new structural biology will inform understanding of function may include complexes involving the replisome encountering “road‐blocks” such as active gene transcription, DNA lesions, and replisomes traveling in opposite directions as would occur at termination of replication.
Acknowledgments
I would like to thank Dr. Gökhan Tolun, Dr. Zhi‐Qiang Xu, Dr. Slobodan Jergic, and Prof. Nicholas Dixon for helpful suggestions.
References
- 1. Lee H, Popodi E, Tang H, Foster PL (2012) Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole‐genome sequencing. Proc Natl Acad Sci U S A 109:E2774–E2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robinson A, Causer RJ, Dixon NE (2012) Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr Drug Targets 13:352–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Eijk E, Wittekoek B, Kuijper EJ, Smits WK (2017) DNA replication proteins as potential targets for antimicrobials in drug‐resistant bacterial pathogens. J Antimicrob Chemother 72:1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yeeles JTP (2014) Discontinuous leading‐strand synthesis: a stop‐start story. Biochem Soc Trans 42:25–34. [DOI] [PubMed] [Google Scholar]
- 5. Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE (2006) A molecular mousetrap determines polarity of termination of DNA replication in E. coli . Cell 125:1309–1319. [DOI] [PubMed] [Google Scholar]
- 6. Elshenawy MM, Jergic S, Xu ZQ, Sobhy MA, Takahashi M, Oakley AJ, Dixon NE, Hamdan SM (2015) Replisome speed determines the efficiency of the Tus‐Ter replication termination barrier. Nature 525:394–398. [DOI] [PubMed] [Google Scholar]
- 7. Dewar JM, Walter JC (2017) Mechanisms of DNA replication termination. Nat Rev Mol Cell Biol 18:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lewis JS, Jergic S, Dixon NE (2016) The E. coli DNA replication fork. Enzyme 39:31–88. [DOI] [PubMed] [Google Scholar]
- 9. Taft‐Benz SA, Schaaper RM (2004) The θ subunit of Escherichia coli DNA polymerase III: a role in stabilizing the ε proofreading subunit. J Bacteriol 186:2774–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao N, O'Donnell M (2016) Bacterial and eukaryotic replisome machines. JSM Biochem Mol Biol 3:1013. [PMC free article] [PubMed] [Google Scholar]
- 11. Windgassen TA, Wessel SR, Bhattacharyya B, Keck JL (2018) Mechanisms of bacterial DNA replication restart. Nucleic Acids Res 46:504–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marians KJ (1992) Prokaryotic DNA replication. Annu Rev Biochem 61:673–719. [DOI] [PubMed] [Google Scholar]
- 13. Frick DN, Richardson CC (2001) DNA primases. Annu Rev Biochem 70:39–80. [DOI] [PubMed] [Google Scholar]
- 14. Pan H, Wigley DB (2000) Structure of the zinc‐binding domain of Bacillus stearothermophilus DNA primase. Structure 8:231–239. [DOI] [PubMed] [Google Scholar]
- 15. Krishna SS, Majumdar I, Grishin NV (2003) Structural classification of zinc fingers: survey and summary. Nucleic Acids Res 31:532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keck JL, Roche DD, Lynch AS, Berger JM (2000) Structure of the RNA polymerase domain of E. coli primase. Science 287:2482–2486. [DOI] [PubMed] [Google Scholar]
- 17. Aravind L, Leipe DD, Koonin EV (1998) Toprim‐‐a conserved catalytic domain in type IA and II topoisomerases, DnaG‐type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res 26:4205–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rymer RU, Solorio FA, Tehranchi AK, Chu C, Corn JE, Keck JL, Wang JD, Berger JM (2012) Binding mechanism of metal·NTP substrates and stringent‐response alarmones to bacterial DnaG‐type primases. Structure 20:1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Corn JE, Pease PJ, Hura GL, Berger JM (2005) Crosstalk between primase subunits can act to regulate primer synthesis in trans. Mol Cell 20:391–401. [DOI] [PubMed] [Google Scholar]
- 20. Khopde S, Biswas EE, Biswas SB (2002) Affinity and sequence specificity of DNA binding and site selection for primer synthesis by Escherichia coli primase. Biochemistry 41:14820–14830. [DOI] [PubMed] [Google Scholar]
- 21. Corn JE, Pelton JG, Berger JM (2008) Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat Struct Mol Biol 15:163–169. [DOI] [PubMed] [Google Scholar]
- 22. Hou C, Biswas T, Tsodikov OV (2018) Structures of the catalytic domain of bacterial primase DnaG in complexes with DNA provide insight into key priming events. Biochemistry 57:2084–2093. [DOI] [PubMed] [Google Scholar]
- 23. Oakley AJ, Loscha KV, Schaeffer PM, Liepinsh E, Pintacuda G, Wilce MC, Otting G, Dixon NE (2005) Crystal and solution structures of the helicase‐binding domain of Escherichia coli primase. J Biol Chem 280:11495–11504. [DOI] [PubMed] [Google Scholar]
- 24. Su XC, Schaeffer PM, Loscha KV, Gan PH, Dixon NE, Otting G (2006) Monomeric solution structure of the helicase‐binding domain of Escherichia coli DnaG primase. FEBS J 273:4997–5009. [DOI] [PubMed] [Google Scholar]
- 25. Syson K, Thirlway J, Hounslow AM, Soultanas P, Waltho JP (2005) Solution structure of the helicase‐interaction domain of the primase DnaG: a model for helicase activation. Structure 13:609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdul Rehman SA, Verma V, Mazumder M, Dhar SK, Gourinath S (2013) Crystal structure and mode of helicase binding of the C‐terminal domain of primase from Helicobacter pylori . J Bacteriol 195:2826–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naue N, Beerbaum M, Bogutzki A, Schmieder P, Curth U (2013) The helicase‐binding domain of Escherichia coli DnaG primase interacts with the highly conserved C‐terminal region of single‐stranded DNA‐binding protein. Nucleic Acids Res 41:4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakayama N, Arai N, Kaziro Y, Arai K (1984) Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA‐dependent ATP hydrolysis and for protein association in the primosome. J Biol Chem 259:88–96. [PubMed] [Google Scholar]
- 29. Story RM, Weber IT, Steitz TA (1992) The structure of the E. coli recA protein monomer and polymer. Nature 355:318–325. [DOI] [PubMed] [Google Scholar]
- 30. Ye J, Osborne AR, Groll M, Rapoport TA (2004) RecA‐like motor ATPases‐‐lessons from structures. Biochim Biophys Acta 1659:1–18. [DOI] [PubMed] [Google Scholar]
- 31. Jezewska MJ, Kim US, Bujalowski W (1996) Binding of Escherichia coli primary replicative helicase DnaB protein to single‐stranded DNA. Long‐range allosteric conformational changes within the protein hexamer. Biochemistry 35:2129–2145. [DOI] [PubMed] [Google Scholar]
- 32. San Martin MC, Stamford NP, Dammerova N, Dixon NE, Carazo JM (1995) A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol 114:167–176. [DOI] [PubMed] [Google Scholar]
- 33. Yu X, Jezewska MJ, Bujalowski W, Egelman EH (1996) The hexameric E. coli DnaB helicase can exist in different quaternary states. J Mol Biol 259:7–14. [DOI] [PubMed] [Google Scholar]
- 34. Seitz H, Weigel C, Messer W (2000) The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli . Mol Microbiol 37:1270–1279. [DOI] [PubMed] [Google Scholar]
- 35. San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles CS, Dixon NE, Carazo JM (1998) Three‐dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure 6:501–509. [DOI] [PubMed] [Google Scholar]
- 36. Weigelt J, Brown SE, Miles CS, Dixon NE, Otting G (1999) NMR structure of the N‐terminal domain of E. coli DnaB helicase: implications for structure rearrangements in the helicase hexamer. Structure 7:681–690. [DOI] [PubMed] [Google Scholar]
- 37. Fass D, Bogden CE, Berger JM (1999) Crystal structure of the N‐terminal domain of the DnaB hexameric helicase. Structure 7:691–698. [DOI] [PubMed] [Google Scholar]
- 38. Bailey S, Eliason WK, Steitz TA (2007) Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318:459–463. [DOI] [PubMed] [Google Scholar]
- 39. Biswas T, Tsodikov OV (2008) Hexameric ring structure of the N‐terminal domain of Mycobacterium tuberculosis DnaB helicase. FEBS J 275:3064–3071. [DOI] [PubMed] [Google Scholar]
- 40. Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS (2008) The structure of a DnaB‐family replicative helicase and its interactions with primase. Nat Struct Mol Biol 15:94–100. [DOI] [PubMed] [Google Scholar]
- 41. Strycharska MS, Arias‐Palomo E, Lyubimov AY, Erzberger JP, O'Shea VL, Bustamante CJ, Berger JM (2013) Nucleotide and partner‐protein control of bacterial replicative helicase structure and function. Mol Cell 52:844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA (2012) The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell 151:267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao Y, Cui Y, Fox T, Lin S, Wang H, de Val N, Zhou ZH, Yang W (2019) Structures and operating principles of the replisome. Science 363:eaav7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arias‐Palomo E, Puri N, O'Shea Murray VL, Yan Q, Berger JM (2019) Physical basis for the loading of a bacterial replicative helicase onto DNA. Mol Cell 74(1):173–184. 10.1016/j.molcel.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davey MJ, Fang L, McInerney P, Georgescu RE, O'Donnell M (2002) The DnaC helicase loader is a dual ATP/ADP switch protein. EMBO J 21:3148–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chase J, Catalano A, Noble AJ, Eng ET, Olinares PD, Molloy K, Pakotiprapha D, Samuels M, Chait B, des Georges A, Jeruzalmi D (2018) Mechanisms of opening and closing of the bacterial replicative helicase. eLife 7:e41140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Raghunathan S, Ricard CS, Lohman TM, Waksman G (1997) Crystal structure of the homo‐tetrameric DNA binding domain of Escherichia coli single‐stranded DNA‐binding protein determined by multiwavelength X‐ray diffraction on the selenomethionyl protein at 2.9‐Å resolution. Proc Natl Acad Sci U S A 94:6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. George NP, Ngo KV, Chitteni‐Pattu S, Norais CA, Battista JR, Cox MM, Keck JL (2012) Structure and cellular dynamics of Deinococcus radiodurans single‐stranded DNA (ssDNA)‐binding protein (SSB)‐DNA complexes. J Biol Chem 287:22123–22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bujalowski W, Lohman TM (1986) Escherichia coli single‐strand binding protein forms multiple, distinct complexes with single‐stranded DNA. Biochemistry 25:7799–7802. [DOI] [PubMed] [Google Scholar]
- 50. Griffith JD, Harris LD, Register J 3rd (1984) Visualization of SSB‐ssDNA complexes active in the assembly of stable RecA‐DNA filaments. Cold Spring Harb Symp Quant Biol 49:553–559. [DOI] [PubMed] [Google Scholar]
- 51. Kozlov AG, Shinn MK, Weiland EA, Lohman TM (2017) Glutamate promotes SSB protein‐protein interactions via intrinsically disordered regions. J Mol Biol 429:2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Raghunathan S, Kozlov AG, Lohman TM, Waksman G (2000) Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol 7:648–652. [DOI] [PubMed] [Google Scholar]
- 53. Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ (2009) Single‐stranded DNA‐binding protein complex from Helicobacter pylori suggests an ssDNA‐binding surface. J Mol Biol 388:508–519. [DOI] [PubMed] [Google Scholar]
- 54. Yadav T, Carrasco B, Myers AR, George NP, Keck JL, Alonso JC (2012) Genetic recombination in Bacillus subtilis: a division of labor between two single‐strand DNA‐binding proteins. Nucleic Acids Res 40:5546–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Antony E, Weiland EA, Korolev S, Lohman TM (2012) Plasmodium falciparum SSB tetramer wraps single‐stranded DNA with similar topology but opposite polarity to E. coli SSB. J Mol Biol 420:269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dubiel K, Myers AR, Kozlov AG, Yang O, Zhang J, Ha T, Lohman TM, Keck JL (2019) Structural mechanisms of cooperative DNA binding by bacterial single‐stranded DNA‐binding proteins. J Mol Biol 431:178–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lu D, Bernstein DA, Satyshur KA, Keck JL (2010) Small‐molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance. Proc Natl Acad Sci U S A 107:633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Voter AF, Killoran MP, Ananiev GE, Wildman SA, Hoffmann FM, Keck JL (2018) A high‐throughput screening strategy to identify inhibitors of SSB protein‐protein interactions in an academic screening facility. SLAS Discov 23:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chilingaryan Z, Headey SJ, Lo ATY, Xu ZQ, Otting G, Dixon NE, Scanlon MJ, Oakley AJ (2018) Fragment‐based discovery of inhibitors of the bacterial DnaG‐SSB interaction. Antibiotics 7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Timinskas K, Balvociute M, Timinskas A, Venclovas C (2014) Comprehensive analysis of DNA polymerase III α subunits and their homologs in bacterial genomes. Nucleic Acids Res 42:1393–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J (2006) Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell 126:881–892. [DOI] [PubMed] [Google Scholar]
- 62. Bailey S, Wing RA, Steitz TA (2006) The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell 126:893–904. [DOI] [PubMed] [Google Scholar]
- 63. Barros T, Guenther J, Kelch B, Anaya J, Prabhakar A, O'Donnell M, Kuriyan J, Lamers MH (2013) A structural role for the PHP domain in E. coli DNA polymerase III. BMC Struct Biol 13:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Evans RJ, Davies DR, Bullard JM, Christensen J, Green LS, Guiles JW, Pata JD, Ribble WK, Janjic N, Jarvis TC (2008) Structure of PolC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci U S A 105:20695–20700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsai MD (2019) Catalytic mechanism of DNA polymerases‐two metal ions or three? Protein Sci 28:288–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wing RA, Bailey S, Steitz TA (2008) Insights into the replisome from the structure of a ternary complex of the DNA polymerase III α‐subunit. J Mol Biol 382:859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ozawa K, Horan NP, Robinson A, Yagi H, Hill FR, Jergic S, Xu ZQ, Loscha KV, Li N, Tehei M, Oakley AJ, Otting G, Huber T, Dixon NE (2013) Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits α, ε, θ and β reveals a highly flexible arrangement of the proofreading domain. Nucleic Acids Res 41:5354–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE (2002) Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure 10:535–546. [DOI] [PubMed] [Google Scholar]
- 69. Lovett ST (2011) The DNA exonucleases of Escherichia coli . EcoSal Plus 4 10.1128/ecosalplus.4.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kirby TW, Harvey S, DeRose EF, Chalov S, Chikova AK, Perrino FW, Schaaper RM, London RE, Pedersen LC (2006) Structure of the Escherichia coli DNA polymerase III epsilon‐HOT proofreading complex. J Biol Chem 281:38466–38471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Keniry MA, Park AY, Owen EA, Hamdan SM, Pintacuda G, Otting G, Dixon NE (2006) Structure of the θ subunit of Escherichia coli DNA polymerase III in complex with the ε subunit. J Bacteriol 188:4464–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kong XP, Onrust R, O'Donnell M, Kuriyan J (1992) Three‐dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell 69:425–437. [DOI] [PubMed] [Google Scholar]
- 73. Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, Pan ZQ, Hurwitz J, O'Donnell M (1996) Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells 1:101–113. [DOI] [PubMed] [Google Scholar]
- 74. Stukenberg PT, Studwell‐Vaughan PS, O'Donnell M (1991) Mechanism of the sliding β‐clamp of DNA polymerase III holoenzyme. J Biol Chem 266:11328–11334. [PubMed] [Google Scholar]
- 75. Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA (2001) A universal protein‐protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A 98:11627–11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bunting KA, Roe SM, Pearl LH (2003) Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β‐clamp. EMBO J 22:5883–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Georgescu RE, Yurieva O, Kim SS, Kuriyan J, Kong XP, O'Donnell M (2008) Structure of a small‐molecule inhibitor of a DNA polymerase sliding clamp. Proc Natl Acad Sci U S A 105:11116–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J (2001) Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106:417–428. [PubMed] [Google Scholar]
- 79. Kling A, Lukat P, Almeida DV, Bauer A, Fontaine E, Sordello S, Zaburannyi N, Herrmann J, Wenzel SC, Konig C, Ammerman NC, Barrio MB, Borchers K, Bordon‐Pallier F, Bronstrup M, Courtemanche G, Gerlitz M, Geslin M, Hammann P, Heinz DW, Hoffmann H, Klieber S, Kohlmann M, Kurz M, Lair C, Matter H, Nuermberger E, Tyagi S, Fraisse L, Grosset JH, Lagrange S, Muller R (2015) Antibiotics. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 348:1106–1112. [DOI] [PubMed] [Google Scholar]
- 80. Pandey P, Tarique KF, Mazumder M, Rehman SA, Kumari N, Gourinath S (2016) Structural insight into β‐clamp and its interaction with DNA ligase in Helicobacter pylori . Sci Rep 6:31181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wolff P, Amal I, Olieric V, Chaloin O, Gygli G, Ennifar E, Lorber B, Guichard G, Wagner J, Dejaegere A, Burnouf DY (2014) Differential modes of peptide binding onto replicative sliding clamps from various bacterial origins. J Med Chem 57:7565–7576. [DOI] [PubMed] [Google Scholar]
- 82. Burnouf DY, Olieric V, Wagner J, Fujii S, Reinbolt J, Fuchs RP, Dumas P (2004) Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J Mol Biol 335:1187–1197. [DOI] [PubMed] [Google Scholar]
- 83. Yin Z, Wang Y, Whittell LR, Jergic S, Liu M, Harry E, Dixon NE, Kelso MJ, Beck JL, Oakley AJ (2014) DNA replication is the target for the antibacterial effects of non‐steroidal anti‐inflammatory drugs. Chem Biol 21:481–487. [DOI] [PubMed] [Google Scholar]
- 84. Yin Z, Whittell LR, Wang Y, Jergic S, Liu M, Harry EJ, Dixon NE, Beck JL, Kelso MJ, Oakley AJ (2014) Discovery of lead compounds targeting the bacterial sliding clamp using a fragment‐based approach. J Med Chem 57:2799–2806. [DOI] [PubMed] [Google Scholar]
- 85. Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O'Donnell M (2008) Structure of a sliding clamp on DNA. Cell 132:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jergic S, Horan NP, Elshenawy MM, Mason CE, Urathamakul T, Ozawa K, Robinson A, Goudsmits JM, Wang Y, Pan X, Beck JL, van Oijen AM, Huber T, Hamdan SM, Dixon NE (2013) A direct proofreader‐clamp interaction stabilizes the Pol III replicase in the polymerization mode. EMBO J 32:1322–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fernandez‐Leiro R, Conrad J, Scheres SHW, Lamers MH (2015) Cryo‐EM structures of the E. coli replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and τ. eLife 4:e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fernandez‐Leiro R, Conrad J, Yang JC, Freund SMV, Scheres SHW, Lamers MH (2017) Self‐correcting mismatches during high‐fidelity DNA replication. Nat Struct Mol Biol 24:140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hedglin M, Kumar R, Benkovic SJ (2013) Replication clamps and clamp loaders. Cold Spring Harb Perspect Biol 5:a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Blinkowa AL, Walker JR (1990) Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III γ subunit from within the τ subunit reading frame. Nucleic Acids Res 18:1725–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Flower AM, McHenry CS (1990) The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proc Natl Acad Sci U S A 87:3713–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Tsuchihashi Z, Kornberg A (1990) Translational frameshifting generates the γ subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci U S A 87:2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jeruzalmi D, O'Donnell M, Kuriyan J (2001) Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106:429–441. [DOI] [PubMed] [Google Scholar]
- 94. Kazmirski SL, Podobnik M, Weitze TF, O'Donnell M, Kuriyan J (2004) Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp‐loader complex. Proc Natl Acad Sci U S A 101:16750–16755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J (2009) The mechanism of ATP‐dependent primer‐template recognition by a clamp loader complex. Cell 137:659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gao D, McHenry CS (2001) τ binds and organizes Escherichia coli replication proteins through distinct domains. Domain III, shared by γ and τ, binds δδ’ and χψ. J Biol Chem 276:4447–4453. [DOI] [PubMed] [Google Scholar]
- 97. Gao D, McHenry CS (2001) τ binds and organizes Escherichia coli replication proteins through distinct domains. Partial proteolysis of terminally tagged τ to determine candidate domains and to assign domain V as the α binding domain. J Biol Chem 276:4433–4440. [DOI] [PubMed] [Google Scholar]
- 98. Su XC, Jergic S, Keniry MA, Dixon NE, Otting G (2007) Solution structure of domains IVa and V of the τ subunit of Escherichia coli DNA polymerase III and interaction with the α subunit. Nucleic Acids Res 35:2825–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wendler P, Ciniawsky S, Kock M, Kube S (2012) Structure and function of the AAA+ nucleotide binding pocket. Biochim Biophys Acta 1823:2–14. [DOI] [PubMed] [Google Scholar]
- 100. Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J (1996) Structure of the C‐terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87:297–306. [DOI] [PubMed] [Google Scholar]
- 101. Marceau AH, Bahng S, Massoni SC, George NP, Sandler SJ, Marians KJ, Keck JL (2011) Structure of the SSB‐DNA polymerase III interface and its role in DNA replication. EMBO J 30:4236–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kelch BA, Makino DL, O'Donnell M, Kuriyan J (2011) How a DNA polymerase clamp loader opens a sliding clamp. Science 334:1675–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kuhlbrandt W (2014) Biochemistry. The resolution revolution. Science 343:1443–1444. [DOI] [PubMed] [Google Scholar]


