Abstract
Micro RNA (miRNAs) is a kind of non coding small RNAs with negative regulation function, which plays an important role in regulating the occurrence and development of tumors. In this study, we analyzed the expression level and role of miRNA-186-5p and Eg5 in neuroblastoma and neuroblastoma cell lines SHSY-5Y, Kelly, NBL-S and SK-N-AS. Results of Real-time PCR and immunohistochemistry showed that the expression level of Eg5 in tumor tissues was higher than that in tumor adjacent tissues, while miRNA-186-5p expression level in tumor tissues was lower than that in tumor adjacent tissues. miRNA-186-5p mimics or Eg5 siRNA was transfected into SHSY-5Y and Kelly cells, CCK-8 and soft agar clone formation tests’ results showed that the cell proliferation was inhibited. Flow cytometry analysis of cell apoptosis and cell cycle showed that overexpression of Mi-186-5p or down-regulation of Eg5 could promote cell apoptosis and lead to arrest cell cycle at G1 phase. Bioinformatics predicts that miRNA-186-5p can bind to the 3’UTR of Eg5. Luciferase reporter gene analysis and Western blot assay also confirmed that microRNA335-5p could target ICAM-1 to inhibit its expression. The tumor growth in nude mice inoculated SHSY-5Y cells with overexpression of miRNA-186-5p was inhibited. In a word, our study found that miR-186-5p could inhibit tumor proliferation by targeting Eg5 in neuroblastoma. This finding will help to better understand the pathogenesis of neuroblastoma and provide new insights into the treatment of tumors.
Keywords: miRNAs, Eg5, miRNA-186-5p, neuroblastoma
Introduction
Neuroblastoma (NB) is a type of sympathetic nervous system tumor, it is the most common extracranial tumor in infancy and the second common extracranial tumor in childhood [1,2]. In recent years, the survival rate of children with neuroblastoma has been greatly improved with the continuous improvement of treatment strategies [3,4]. The survival rate of high-risk neuroblastoma is still 40~50% [5,6]. More than half of high-risk children relapse after receiving standard chemotherapy, these children have poor prognosis and the 10-year survival rate is less than 20% [7]. Therefore, the improvement of therapeutic efficacy in children with high-risk neuroblastoma depends on in-depth study of the pathogenesis and discovery of new therapeutic methods.
Eg5 (kif11, kinesin spindle protein) is a member of the kinesin-5 family, it is a type of dynein with ATP hydrolase activity. In the early stage of mitosis, Eg5 can promote the formation of bipolar spindles and the movement of chromosomes toward the poles, it plays an important role in cell proliferation and division [8,9]. Blocking the expression of Eg5 will destroy the normal movement mechanism of the spindle, interfere the normal separation of the spindle, which induce the cell cycle arrest and cell apoptosis [10]. Eg5 is mainly expressed in proliferative tissues such as bone marrow and tumor tissues, but not in non-proliferative tissues such as the central nervous system. Eg5 has been found to be highly expressed in leukemia [11], breast cancer [12], lung cancer [13], bladder cancer [14], prostate cancer [15], glioma [16] and nephroblastoma [17]. Several inhibitors of Eg5 have been identified, which have antitumor effects in vivo and in vitro [18-20]. Although high expression of Eg5 has been found to be associated with a variety of tumors, the role of Eg5 in neuroblastoma is unclear.
MicroRNA (miRNA) is a class of nonprotein-encoded single-stranded RNA molecules with 19-25 nucleotides encoded by endogenous genes, it is highly conserved among species. miRNA usually acts on one or more messenger RNA (mRNA), and negatively regulates gene expression by degrading mRNA or inhibiting translation level. Many studies have shown that miRNA can be used as a valuable biomarker for the diagnosis and prognosis of cancer. The imbalance of miRNA expression is closely related to the occurrence and progression of neuroblastoma. MiR-186-5p is tumor-specific and plays a carcinogenic or inhibitory role in different tumors. The expression of miR-186-5p is down regulated in non-small cell lung cancer (NSCLC) and ovarian cancer [22]. Overexpression of microRNA-186-5p can inhibit the proliferation and invasion of NSCLC cell lines, and has an inhibitory effect on cancer [21]. In contrast, overexpression of microRNA186-5p in pancreatic cancer and bladder cancer enhanced the migration, invasion and cloning of tumor cells [23,24]. The role of miR-186-5p in neuroblastoma has not been reported.
In this study, we detected the expression of microRNA-186-5p and Eg5 in neuroblastoma tissues. It was found that the expression level of microRNAs-186-5p was down-regulated, while Eg5 expression was up-regulated. Mi186-5p can target Eg5 to inhibit the proliferation of neuroblastoma in vitro.
Materials and methods
Subjects
The resected neuroblastoma tissues and their corresponding paracancerous tissues were collected from the Department of Pediatrics, First Affiliated Hospital of China University of Science and Technology from February 2015 to December 2017. All subjects signed the informed consent forms. This study was approved by the ethics committee of the First Affiliated Hospital of University of Science and Technology of China.
Cell culture
All cells were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were cultured with RPMI1640 medium containing 10% FBS, 100 U/ml penicillin G, 100 U/ml treptomycin sulfate and 2 mM L-Glutamine (Invitrogen-Gibco, Carlsbad, CA, USA) at 37°C with 5% CO2.
Experimental animal
BALB/c-nu nude mice (4~6 week old, weighT 16-20 g) were purchased from Anhui experimental animal center. They were bred in the SPF class barrier system of Anhui experimental animal center, eat and drink freely. All animal experiments were conducted according to Principles of Laboratory Animal Care (National Society for Medical Research).
miRNA and siRNA transfection
All siRNA transfections were performed using Lipofectamine 2000 Reagent (Life Technologies, Carlsbad, CA, USA), according to the manual protocol. The cells were divided into control, miR-186-5p, Eg5 siRNA and siRNA NC groups randomly. miR-186-5p mimics and Eg5 siRNAs were purchased from GenePharma Co., Ltd (Shanghai, China).
RNA extraction and qRT-PCR
Total RNA was extracted from cells and tissues using a Trizol reagent kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA concentration and purity were detected using a NanoDropTM 1000 spectrophotometer (Thermo Fisher Scientific, Inc). A total of 1 μg RNA was subjected to reverse transcription using a miScript II RT Kit (Qiagen) according to the manual. The expression level of microRNAs-186-5p was detected using commercial primers Taqman Universal Mix II No UNG plus specific PCR primers (Assay #002285) and miScript SYBR Green PCR Kit (Qiagen) on the ABI StepOne Plus system (Applied Biosystems, Waltham, MA). The thermocycling conditions were as follows: Pre-degeneration at 95°C for 10 min, followed by 40 cycles of 95°C for 12 sec and 62°C for 40 sec. GAPDH and U6 genes were used as an internal control. The 2-ΔΔCT method was used for quantification. The primers used in this study are as follows: ICAM-1 (Accession number: NM_004523) forward: 5’-TGTTTGATGATCCCCGTAACAAG-3’ and reverse: 5’-CTGAGTGGGAACGACTAGAGT-3’; GAPDH forward: 5’-ACCTGACCTGCCGTCTAGAA-3’ and reverse: 5’-TCCACCACCCTGTTGCTGTA-3’.
Cell viability detection
The cell viability was detected with Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the kit’s manual protocol. The cells in the logarithmic growth phase were digested with trypsin and inoculated into 96-well plates (2000 cell/well), they were cultured at 37°C overnight. Each well was added 10 μl cck-8 solution, A450 values were detected by Biotek Synergy HT plate reader (Bio Tek, Winooski, VT,USA) every day after culture for 1~5 days to evaluate the viability of cells. 6 wells were repeatedly detected for each group.
Soft agar clone formation experiment
The cells in the logarithmic growth phase were digested with trypsin, 1 × 103 cells were re-suspended with 0.35% soft agar solution configured with complete medium, the cells were evenly spread on the 0.7% solidified agar layer. They were placed in 10 cm cell culture dish, and were continuously cultured at 37°C with 5% CO2 for 12 d. They were stained with 0.1% crystal violet dye solution (Richard-Allan Scientific, San Diego, CA, USA) and observed under the microscope, the clone with a diameter of > 80 m was taken as positive. 6 wells were repeatedly detected for each group.
Cell apoptosis analysis
Cell apoptosis was analyzed by flow cytometry (FACScan; BD Biosciences, San Jose, CA, USA) using AnnexinV-FITC Analysis Kit (Beyotime, Shanghai, China). The operation was carried out according to the instructions. Cells in each group were cultured for 48 h under the same conditions, they were harvested and digested with trypsin. The digested cells were washed with the pre cooled PBS for 3 times, and then they were lightly re-suspended by adding 195 μl Annexin V-FITC binding solution, 5 μl Annexin V-FITC was added into them and mixed gently. They were incubated at room temperature (20-25°C) avoiding light for 10-20 minutes after 10 μl propidium iodide staining solution was added and mixed gently. They were detected using FlowJo 7.6.5 software (BD Biosciences). 3 wells were repeatedly detected for each group.
BrdU proliferation assay
Cell proliferation was analyzed by Cell Proliferation ELISA 5-bromo-2’-deoxyuridine (BrdU) colorimetric kit (Sigma Aldrich, St. Louis, MO) according to the instructions. 5 × 103 cells were inoculated into each well of the 96-well plates and cultured at 37°C with 5% CO2 for 24 h, BrdU was added into them. A370 and A492 values were detected by Biotek Synergy HT plate reader (Bio Tek, Winooski, VT, USA). 6 wells were repeatedly detected for each group.
Cell cycle detection
The cells were inoculated into 25 cm2 cell culture dishes, after the cells of different group were transfected for 24 h, the cell culture supernatant was collected. The adherent cells were digested with pancreatin and the cells were re-suspended with supernatant. They were centrifuged at 200 × g and the supernatant was discarded, the cells were washed with PBS for two times. The cells were fixed with pre cooled 70% ethanol at 4°C overnight. They were washed with PBS for two times and re-suspended with 50 μl PBS containing 100 μg/ml RNase, 200 μl PBS containing 50 μg/ml PI was added into them and incubated on ice avoid light for 30 min. They were detected by flow cytometry (FACScan; BD Biosciences, San Jose, CA, USA) and analyzed with FlowJo 7.6.5 software.
Double luciferase reporter gene analysis
The luciferase reporter gene plasmid pGL3-Eg5 and point mutation of luciferase reporter gene plasmid mut-pGL3-Eg5 were constructed. The 293T cells were inoculated into 24 well plates and cultured overnight, luciferase reporter plasmid, Renilla luciferase and miR-186-5p mimic or control were transfected into 293T cells simultaneously. The cells were split using Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the instructions after culture for 48 h. The results were detected using Panomics Luminometer (Affymetrix, Santa Clara, CA, USA) after the luminescence was added. The sea renin fluorescence was used as internal reference.
Xenograft mouse model
Lentiviral LV-miR-186-5p expressing miR-186-5p and control lentiviral LV-NC (Shanghai Jima Gene Co., Ltd.) were inoculated into cells, the stable cells were screened by puromycin. Each nude mouse was inoculated 5 × 106 cells. Tumor volume was measured every three days (0.5 × length × width2). The mice were sacrificed after transfection for 4 weeks, and the tumor was removed for weight measurement. The tumor tissues were fixed with 4% paraformaldehyde and paraffin section was prepared. The positive rate of Ki67 was observed by immunohistochemical staining to further analyze the growth of tumor.
Western blotting test
The neuroblastoma tissues and their corresponding paracancerous tissues (0.5 g) were ground in liquid nitrogen and lysed with RIPA lysis buffer (Sigma-Aldrich, St. Louis, MO, USA). The cells in the logarithmic growth period were harvested and lysed with Cell Lysis Solution (Sigma-Aldrich, St. Louis, MO, USA), then they were centrifuged with 10000 rpm at 4°C for 5 min. The extract supernatants were added protein electrophoresis buffer and loaded on 12% SDS-polyacrylamide gels and transferred onto polyvinylidene fluoride membranes (PVDF, Amersham Biosciences, Piscataway, NJ, USA). The membranes were blocked for 2 h at 37°C with 5% non-fat milk in Tris Buffered Saline with Tween 20 (TBST), and were incubated with primary antibodies (1:500 Eg5, 1:1000 Bcl-2, 1:500 survivin, 1:500 p21, 1:500 p27 and 1:2000 β-actin, Abcam, Cambridge, UK) respectively at 4°C overnight. Following incubation with HRP-conjugated secondary antibody (1:50,000, Abcam, Cambridge, UK) for 1 h at 37°C. The membrane was coated with ECL luminescence (Perkin-Elmer Inc.) after washing the film for 3 times, they were observed using Imagequant LAS4000 (GE Healthcare, Japan). β-actin was used as normalization.
Statistical analysis
The data were expressed as mean ± SD. Statistical analysis was performed using one-way ANOVA or Student’s t-test with SPSS 17.0 software (SPSS, Chicago, IL, USA). P < 0.05 was considered to be statistically significant.
Results
The expression level of miR-186-5p was downregulated, while the expression level of Eg5 was upregulated in neuroblastoma tissues
The expression levels of miR-186-5p and Eg5 were detected in 31 cases of neuroblastoma and corresponding adjacent normal tissues, it was found that the miR-186-5p expression in neuroblastoma tissues was significantly lower than that in adjacent tissues (Figure 1A), while the Eg5 expression in neuroblastoma tissues was significantly higher than that in adjacent tissues (Figure 1B, 1C). In neuroblastoma cell lines SHSY-5Y, Kelly, NBL-S and SK-N-AS, it was also found that the expression of miR-186-5p was down-regulated while that of Eg5 was up-regulated (Figure 1A, 1B). Western blotting results also showed that the Eg5 and Bcl-2 expression levels in neuroblastoma tissues were significantly higher than that in adjacent tissues, while p21 and p27 expression levels in neuroblastoma tissues were significantly lower than that in adjacent tissues (Figure 1D, 1E).
Figure 1.
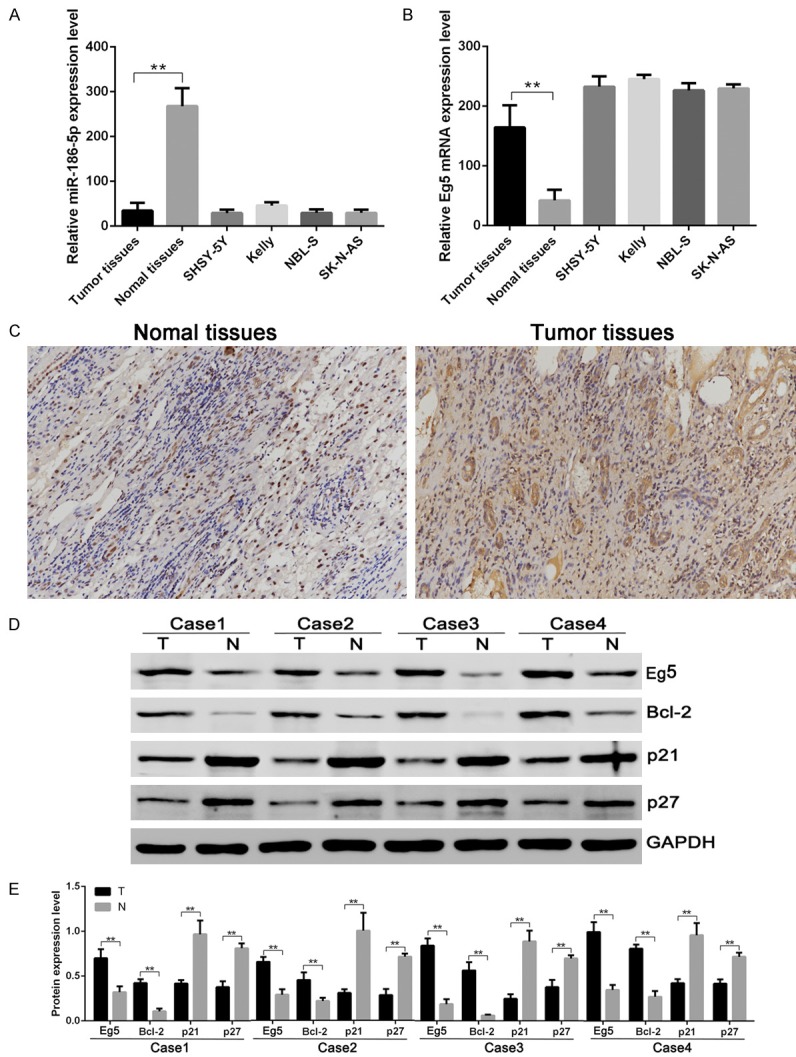
The expression levels of microRNA-186-5p, Eg5, Bcl-2, p21 and p27 in neuroblastoma. A: miR-186-5p mRNA expression in neuroblastoma detected by RT-PCR. B: Eg5 mRNA expression in neuroblastoma detected by RT-PCR. C: Eg5 expression in neuroblastoma and adjacent normal tissues detected by immunohistochemical method. D: Eg5, Bcl-2, p21 and p27 expression in neuroblastoma and adjacent normal tissues detected by Western blotting assay. E: Comparison of relative expression levels of Eg5, Bcl-2, p21 and p27. The Eg5 and Bcl-2 expression levels in neuroblastoma tissues were significantly higher than that in adjacent tissues, while miR-186-5p, p21 and p27 expression levels in neuroblastoma tissues were significantly lower than that in adjacent tissues. **P < 0.01 vs Normal tissue.
Overexpression of miR-186-5p or downregulation of Eg5 expression in neuroblastoma cell lines can inhibit cell proliferation
To further analyze the effects of miR-186-5p and Eg5 on neuroblastoma, we selected SHSY-5Y and Kelly cell lines for study. CCK-8 detected results showed that transfection of miR-186-5p mimic or Eg5 siRNA could inhibit cell proliferation. Soft agar clone formation assay showed that both transfection of microRNA-186-5p mimic and Eg5 siRNA could inhibit cell clone formation. These results indicated that miR-186-5p and Eg5 could affect the proliferation of neuroblastoma cells (Figure 2).
Figure 2.
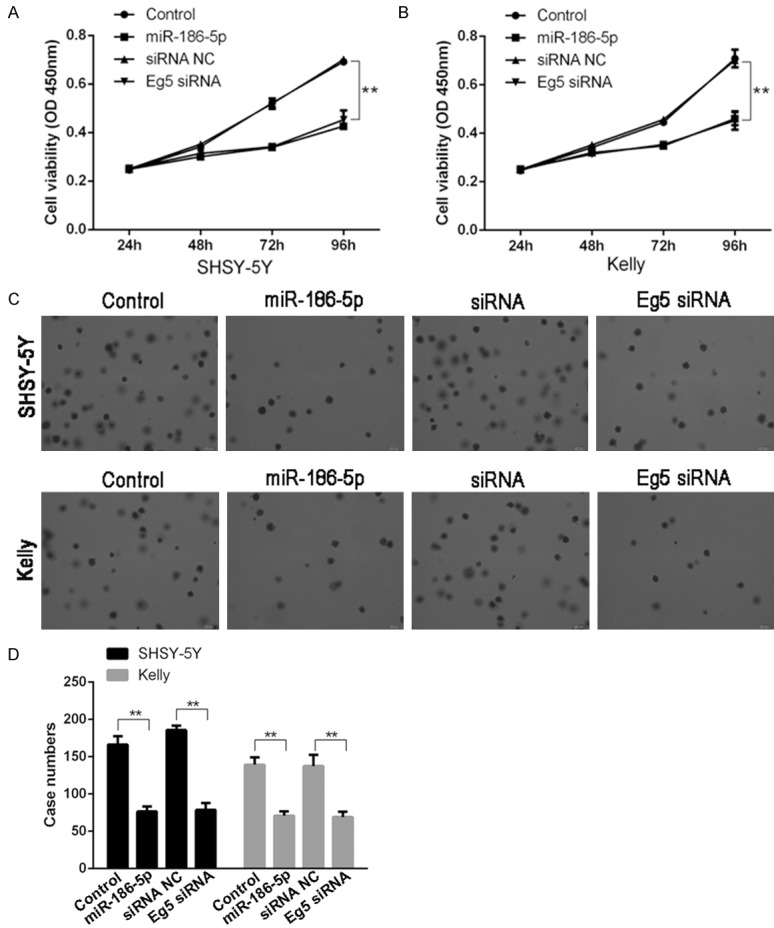
Overexpression of miR-186-5p or downregulation of Eg5 expression in neuroblastoma cell lines could inhibit cell proliferation. A: The SHSY-5Y cell viability in miR-186-5p overexpression group and Eg5 siRNA group was lower than that of control group; B: The Kelly cell viability in miR-186-5p overexpression group and Eg5 siRNA group was lower than that of control group; C, D: The SHSY-5Y and Kelly cell clones in miR-186-5p overexpression group and Eg5 siRNA group was lower than that of control group. **P < 0.01 vs Control. Overexpression of miR-186-5p or downregulation of Eg5 expression could inhibit the proliferation and clone formation of SHSY-5Y and Kelly cells.
Overexpression of miR-186-5p or down-regulation of Eg5 in neuroblastoma cell lines could promote cell apoptosis and induce G1 cell cycle arrest
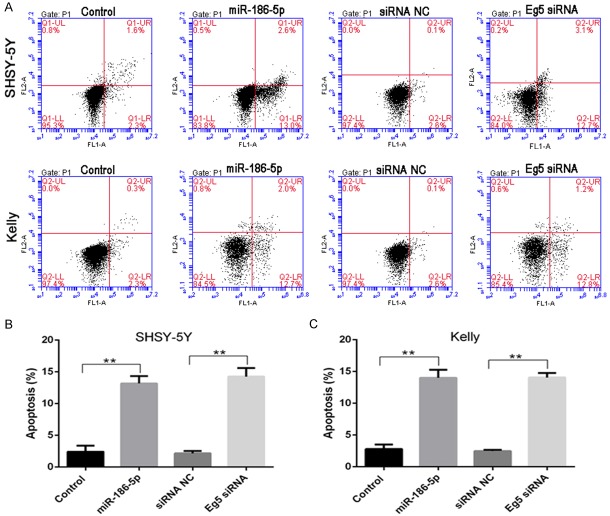
Flow cytometry analysis showed that up-regulation of miR-186-5p or down-regulation of Eg5 in neuroblastoma cell lines SHSY-5Y and Kelly could promote apoptosis (Figure 3). To analyze whether miR-186-5p and Eg5 affected cell proliferation by altering cell cycle, we detected the cell cycle changes of neuroblastoma cell lines after transfection of miR-186-5p or Eg5 siRNA. It was found that overexpression of miR-186-5p or down-regulation of Eg5 in SHSY-5Y and Kelly cell lines could increase the number of G1 phase cells and decrease the number of S phase and G2/M phase cells (Figure 4). BrdU proliferation assay results also showed that overexpression of miR-186-5p or down regulation of Eg5 could inhibit DNA synthesis of G1 phase. Therefore, in addition to inducing apoptosis, miR-186-5p could also inhibit cell proliferation through G1 phase arrest.
Figure 3.
Overexpression of miR-186-5p or down-regulation of Eg5 in neuroblastoma cell lines could promote cell apoptosis. A: Flow cytometry results of SHSY-5Y and Kelly cells; B: The apoptosis proportion of SHSY-5Y cells in different groups; C: The apoptosis proportion of Kelly cells in different groups. **P < 0.01 vs Control. Overexpression of miR-186-5p or down-regulation of Eg5 could promote the apoptosis of SHSY-5Y and Kelly cells.
Figure 4.
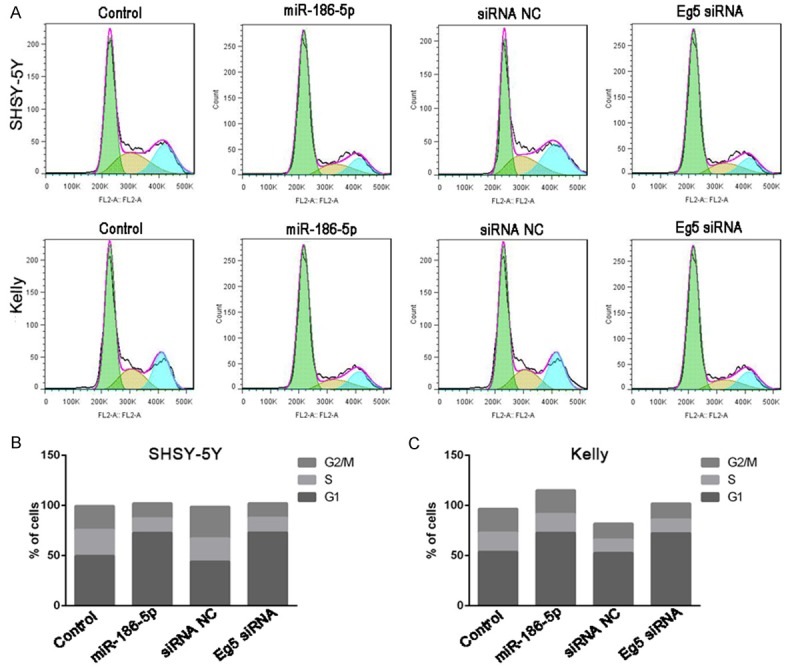
Overexpression of miR-186-5p or down-regulation of Eg5 in neuroblastoma cell lines could induce G1 cell cycle arrest. A: Cell cycle was detected by flow cytometry; B: Cell cycle distribution of SHSY-5Y cells in different groups; C: Cell cycle distribution of Kelly cells in different groups. miR-186-5p overexpression or down-regulation of Eg5 in SHSY-5Y and Kelly cell lines could increase the number of G1 phase cells and decrease the number of S phase and G2/M phase cells.
miR-186-5p could target 3’UTR of Eg5 and inhibit Eg5 expression
RT-PCR and western blotting results showed that miR-186-5p could directly down regulate the expression level of Eg5. Bioinformatics prediction indicated that miR-186-5p could directly act on the 3’UTR of Eg5. Double luciferase reporter gene analysis showed that miR-186-5p could directly bind to 3’UTR of Eg5 to inhibit the expression of luciferase. The inhibitory effect of miR-186-5p disappeared when point mutation was applied to the 3’UTR domain of Eg5 (Figure 5).
Figure 5.
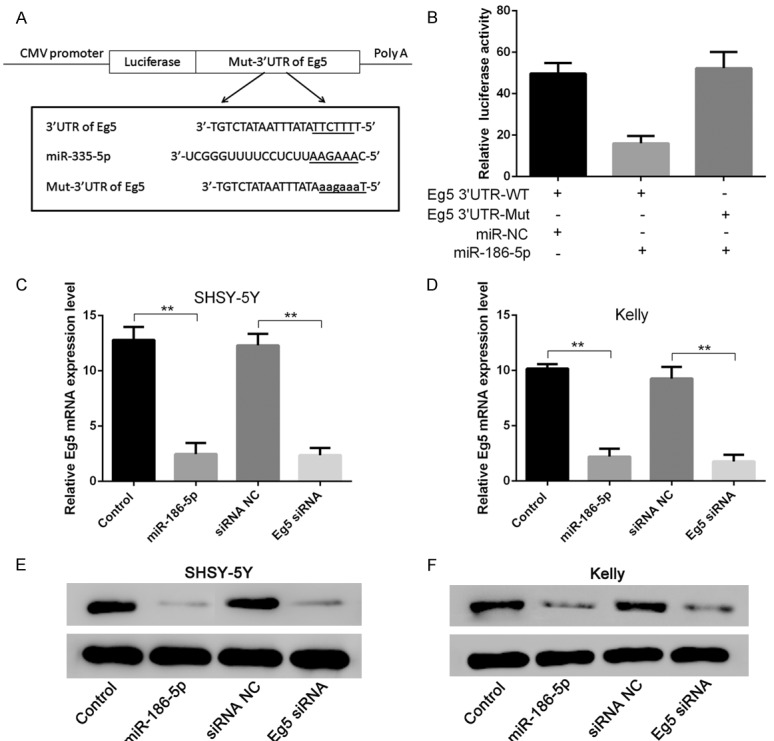
miR-186-5p could target 3’UTR of Eg5 and inhibit Eg5 expression. A: Construction of WT-Eg5 3’UTR and Mut-Eg5 3’UTR vector; B: Detection results of luciferase reporter gene; C, E: overexpression of miR-186-5p and Eg5 siRNA could inhibit Eg5 expression in SHSY-5Y cells; D, F: overexpression of miR-186-5p and Eg5 siRNA could inhibit Eg5 expression in Kelly cells. **P < 0.01 vs Control. miR-186-5p could directly down regulate the expression level of Eg5, but the inhibitory effect disappeared when point mutation was applied to the 3’UTR domain of Eg5.
miR-186-5p suppressed neuroblastoma tumorigenesis in vivo
We further analyzed whether miR-186-5p affected tumor growth in vivo through a nude mouse model of tumorigenesis. SHSY-5Y cells infected with LV-miR-186-5p and LV-NC were inoculated subcutaneously to nude mice. The tumor volume and weight decreased significantly in LV-miR-186-5p group compared with LV-NC group 30 days later. The expression level of Ki67 in LV-miR-186-5p group was also significantly lower than that of LV-NC group. These results indicated that miR-186-5p could inhibit the proliferation of neuroblastoma in vivo (Figure 6).
Figure 6.
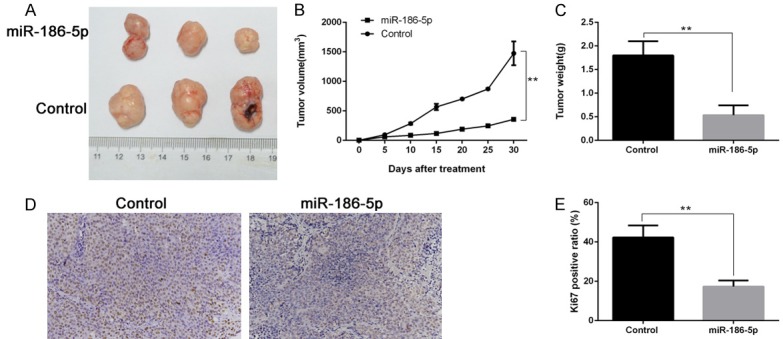
miR-186-5p inhibited neuroblastoma growth in vivo. A: miR-186-5p could inhibit subcutaneous tumor growth; B: Changes of subcutaneous tumor volume; C: Subcutaneous tumor weight; D, E: The proportion of Ki67 positive cells in different groups detected by immunohistochemical method. **P < 0.01 vs Control. miR-186-5p could inhibit the proliferation of neuroblastoma in vivo.
Discussion
MiRNA is one of the important factors that affect gene regulation and participate in tumor pathogenesis. In this study, we detected 31 cases of neuroblastoma and corresponding adjacent tissues and found that the miR-186-5p expression in neuroblastoma tissues was significantly lower than that in adjacent tissues, in some neuroblastoma cell lines SHSY-5Y, Kelly, NBL-S and SK-N-AS, it was also found that the expression of miR-186-5p was down-regulated. Up regulation of miR-186-5p expression in SHSY-5Y and Kelly cells could inhibit cell proliferation. These results suggested that miR-186-5p may be a tumor suppressor gene in neuroblastoma. Previous studies have shown that miR-186-5p is a tumor suppressor gene in non-small cell lung cancer [21], ovarian cancer [22], oral squamous cell carcinoma [25], multiple myeloma [26], cervical cancer [27], esophageal cancer [28], g astric cancer [29], liver cancer [30], renal cancer [31] and glioblastoma [32]. In contrast, it was reported that inhibition of the expression of miR-186-5p in bladder cancer [23], pancreatic cancer [24] and colon cancer cells [33] could reduce their proliferation and invasion, miR-186-5p was characterized by oncogenes. As far as we know, the role of miR-186-5p in neuroblastoma was not reported.
Eg5 is mainly related to chromosomal location, centrosome isolation and bipolar spindle formation and separation. Eg5 is expressed in normal cells, but it is highly expressed in tumor tissues and cells. Previous studies have shown that the high expression of Eg5 has a certain correlation with the occurrence and development of tumors [11-17]. The high expression of EG5 interferes with the normal assembly of spindle and energy balance related to its function, eventually leading to spindle loss, genomic instability and cancer occurrence [34]. Eg5 was upregulated in chronic myeloid leukemia cells, inhibition of Eg5 expression by specific siRNA could induce cell cycle arrest in G2/M phase and promote cell apoptosis [35]. The high expression of Eg5 in astrocytoma [16], nephroblastoma [17] and breast cancer [12] is a high risk factor for poor prognosis. Eg5 inhibitor K858 in glioblastoma can inhibit the proliferation and invasion of tumor cells [18].
In this study, we confirmed that Eg5 was highly expressed in neuroblastoma tissues and cell lines and was associated with the proliferation of neuroblastoma, the expression level of miR-186-5p was opposite to that of Eg5. The expression level of Eg5 in neuroblastoma tissues was higher than that in paracancerous tissues, and the expression trend of Eg5 was consistent with that of Bcl-2, which inhibited apoptosis of tumor cells. However, it was contrary to the expression trend of p21 and p27. Overexpression of miR-186-5p or Eg5 siRNA in SHSY-5Y and Kelly cell lines could inhibit cell proliferation and induce cell cycle G1 phase arrest. Double luciferase reporter gene analysis showed that miR-186-5p could directly act on 3’UTR of Eg5 to inhibit the expression of luciferase. Overexpression of miR-186-5p in SHSY-5Y cells could significantly inhibit the proliferation of subcutaneous solid tumors in nude mice. These results suggested that Eg5 was involved in the progression of neuroblastoma, and that miR-186-5p could target Eg5 as an oncogene.
In a word, we demonstrated that miR-186-5p could target 3’UTR of Eg5 and inhibit the proliferation of neuroblastoma by down-regulating the Eg5 expression in this study, miR-186-5p may be a potential small molecule drug for the treatment of neuroblastoma.
Disclosure of conflict of interest
None.
References
- 1.Nakagawara A, Li Y, Izumi H, Muramori K, Inada H, Nishi M. Neuroblastoma. Jpn J Clin Oncol. 2018;48:214–241. doi: 10.1093/jjco/hyx176. [DOI] [PubMed] [Google Scholar]
- 2.Speleman F, Park JR, Henderson TO. Neuroblastoma: a tough nut to crack. Am Soc Clin Oncol Educ Book. 2016;35:e548–557. doi: 10.1200/EDBK_159169. [DOI] [PubMed] [Google Scholar]
- 3.Jeison M, Yaniv I, Ash S. Genetic stratification of neuroblastoma for treatment tailoring. Future Oncol. 2011;7:1087–1099. doi: 10.2217/fon.11.87. [DOI] [PubMed] [Google Scholar]
- 4.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR, Valteau-Couanet D, Pearson AD, Cohn SL. Advances in risk classification and treatment strategies for neuroblastoma. J. Clin. Oncol. 2015;33:3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owens C, Irwin M. Neuroblastoma: the impact of biology and cooperation leading to personalized treatments. Crit Rev Clin Lab Sci. 2012;49:85–115. doi: 10.3109/10408363.2012.683483. [DOI] [PubMed] [Google Scholar]
- 6.Moreno L, Caron H, Geoerger B, Eggert A, Schleiermacher G, Brock P, Valteau-Couanet D, Chesler L, Schulte JH, De Preter K, Molenaar J, Schramm A, Eilers M, Van Maerken T, Johnsen JI, Garrett M, George SL, Tweddle DA, Kogner P, Berthold F, Koster J, Barone G, Tucker ER, Marshall L, Herold R, Sterba J, Norga K, Vassal G, Pearson AD. Accelerating drug development for neuroblastoma - new drug development strategy: an innovative therapies for children with cancer, European network for cancer research in children and adolescents and international society of paediatric oncology Europe neuroblastoma project. Expert Opin Drug Discov. 2017;12:801–811. doi: 10.1080/17460441.2017.1340269. [DOI] [PubMed] [Google Scholar]
- 7.Fischer J, Pohl A, Volland R, Hero B, Dübbers M, Cernaianu G, Berthold F, von Schweinitz D, Simon T. Complete surgical resection improves outcome in INRG high-risk patients with localized neuroblastoma older than 18 months. BMC Cancer. 2017;17:520. doi: 10.1186/s12885-017-3493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- 9.Kwok BH, Yang JG, Kapoor TM. The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol. 2004;14:1783–1788. doi: 10.1016/j.cub.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu M, Ishii H, Ogo N, Unno Y, Matsuno K, Sawada J, Akiyama Y, Asai A. S-trityl-L-cysteine derivative induces caspase-independent cell death in K562 human chronic myeloid leukemia cell line. Cancer Lett. 2010;298:99–106. doi: 10.1016/j.canlet.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Hansen GM, Justice MJ. Activation of hex and meg5 by retroviral insertion may contribute to mouse b-cellleukemia. Oncogene. 1999;18:6531–6539. doi: 10.1038/sj.onc.1203023. [DOI] [PubMed] [Google Scholar]
- 12.Planas-Silva MD, Filatova IS. Estrogen-dependent regulation of Eg5 in breast cancer cells. Anticancer Drugs. 2007;18:773–779. doi: 10.1097/CAD.0b013e3280a02f2b. [DOI] [PubMed] [Google Scholar]
- 13.Saijo T, Ishii G, Ochiai A, Yoh K, Goto K, Nagai K, Kato H, Nishiwaki Y, Saijo N. Eg5 expression is closely correlated with the response of advanced non-small cell lung cancer to antimitotic agents combined with platinum chemotherapy. Lung Cancer. 2006;54:217–225. doi: 10.1016/j.lungcan.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Ding S, Xing N, Lu J, Zhang H, Nishizawa K, Liu S, Yuan X, Qin Y, Liu Y, Ogawa O, Nishiyama H. Overexpression of eg5 predicts unfavorable prognosis in non-muscle invasive bladder urothelial carcinoma. Int J Urol. 2011;18:432–438. doi: 10.1111/j.1442-2042.2011.02751.x. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi N, Koller E, Fazli L, Gleave ME. Effects of eg5 knockdown on human prostate cancer xenograft growth and chemosensitivity. Prostate. 2008;68:1283–1295. doi: 10.1002/pros.20783. [DOI] [PubMed] [Google Scholar]
- 16.Lu M, Zhu H, Wang X, Zhang D, Xiong L, Xu L, You Y. The prognostic role of Eg5 expression in laryngeal squamous cell carcinoma. Pathology. 2016;48:214–218. doi: 10.1016/j.pathol.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Sun D, Lu J, Ding K, Bi D, Niu Z, Cao Q, Zhang J, Ding S. The expression of Eg5 predicts a poor outcome for patients with renal cell carcinoma. Med Oncol. 2013;30:476. doi: 10.1007/s12032-013-0476-0. [DOI] [PubMed] [Google Scholar]
- 18.Nakai R, Iida S, Takahashi T, Tsujita T, Okamoto S, Takada C, Akasaka K, Ichikawa S, Ishida H, Kusaka H, Akinaga S, Murakata C, Honda S, Nitta M, Saya H, Yamashita Y. K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res. 2009;69:3901–3909. doi: 10.1158/0008-5472.CAN-08-4373. [DOI] [PubMed] [Google Scholar]
- 19.Ye XS, Fan L, Van Horn RD, Nakai R, Ohta Y, Akinaga S, Murakata C, Yamashita Y, Yin T, Credille KM, Donoho GP, Merzoug FF, Li H, Aggarwal A, Blanchard K, Westin EH. A novel Eg5 inhibitor (LY2523355) causes mitotic arrest and apoptosis in cancer cells and shows potent antitumor activity in Xenograft tumor models. Mol Cancer Ther. 2015;14:2463–2472. doi: 10.1158/1535-7163.MCT-15-0241. [DOI] [PubMed] [Google Scholar]
- 20.Wakui H, Yamamoto N, Kitazono S, Mizugaki H, Nakamichi S, Fujiwara Y, Nokihara H, Yamada Y, Suzuki K, Kanda H, Akinaga S, Tamura T. A phase 1 and dose-finding study of LY2523355 (litronesib), an Eg5 inhibitor, in japanese patients with advanced solid tumors. Cancer Chemother Pharmacol. 2014;74:15–23. doi: 10.1007/s00280-014-2467-z. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N, Liang S, Lu S, Liu Y, Zhang J, Li F, Li W, Liu F, Sun L, Qi Y. PTTG1 promotes migration and invasion of human non-small cell lung cancer cells and is modulated by miR-186. Carcinogenesis. 2013;34:2145–2155. doi: 10.1093/carcin/bgt158. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Shen H, Yin X, Long L, Xie C, Liu Y, Hui L, Lin X, Fang Y, Cao Y, Xu Y, Li M, Xu W, Li Y. miR-186 regulation of Twist1 and ovarian cancer sensitivity to cisplatin. Oncogene. 2016;35:323–332. doi: 10.1038/onc.2015.84. [DOI] [PubMed] [Google Scholar]
- 23.Zhang ZL, Bai ZH, Wang XB, Bai L, Miao F, Pei HH. miR-186 and 326 predict the prognosis of pancreatic ductal adenocarcinoma and affect the proliferation and migration of cancer cells. PLoS One. 2015;10:e0118814. doi: 10.1371/journal.pone.0118814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Yuan D, Li J, Zheng S, Wang B. miR-186 downregulates protein phosphatase PPM1B in bladder cancer and mediates G1-S phase transition. Tumour Biol. 2016;37:4331–4341. doi: 10.1007/s13277-015-4117-4. [DOI] [PubMed] [Google Scholar]
- 25.Ries J, Vairaktaris E, Agaimy A, Kintopp R, Baran C, Neukam FW, Nkenke E. miR-186, miR-3651 and miR-494: potential biomarkers for oral squamous cell carcinoma extracted from whole blood. Oncol Rep. 2014;31:1429–1436. doi: 10.3892/or.2014.2983. [DOI] [PubMed] [Google Scholar]
- 26.Liu Z, Zhang G, Yu W, Gao N, Peng J. miR-186 inhibits cell proliferation in multiple myeloma by repressing Jagged1. Biochem Biophys Res Commun. 2016;469:692–697. doi: 10.1016/j.bbrc.2015.11.136. [DOI] [PubMed] [Google Scholar]
- 27.Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sultmann H, Scheffner M, Hoppe-Seyler K, Hoppe-Seyler F. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV positive tumor cells. PLoS Pathog. 2015;11:e1004712. doi: 10.1371/journal.ppat.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He W, Feng J, Zhang Y, Wang Y, Zang W, Zhao G. microRNA-186 inhibits cell proliferation and induces apoptosis in human esophageal squamous cell carcinoma by targeting SKP2. Lab Investig. 2016;96:317–324. doi: 10.1038/labinvest.2015.134. [DOI] [PubMed] [Google Scholar]
- 29.Cao C, Sun D, Zhang L, Song L. miR-186 affects the proliferation, invasion and migration of human gastric cancer by inhibition of Twist1. Oncotarget. 2016;7:79956–79963. doi: 10.18632/oncotarget.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan T, He X, Yu J, Hang Z. MicroRNA-186 targets yes-associated protein 1 to inhibit hippo signaling and tumorigenesis in hepatocellular carcinoma. Oncol Lett. 2016;11:2941–2945. doi: 10.3892/ol.2016.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao D, Wu M, Ji L, Liu F, Liu Y. microRNA-186 suppresses cell proliferation and metastasis through targeting Sentrin specific protease 1 in renal cell carcinoma. Oncol Res. 2018;26:249–259. doi: 10.3727/096504017X14953948675430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Jiang H, Wang S, Chen B. Dual functional MicroRNA-186-5p targets both FGF2 and RelA to suppress tumorigenesis of glioblastoma multiforme. Cell Mol Neurobiol. 2017;37:1433–1442. doi: 10.1007/s10571-017-0474-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Islam F, Gopalan V, Vider J, Wahab R, Ebrahimi F, Lu CT, Kasem K, Lam AKY. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017;357:260–270. doi: 10.1016/j.yexcr.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 34.Castillo A, Morse HC 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- 35.Carter BZ, Mak DH, Shi Y, Schober WD, Wang RY, Konopleva M, Koller E, Dean NM, Andreeff M. Regulation and targeting of eg5, a mitotic motor protein in blast crisis cml: overcoming imatinib resistance. Cell Cycle. 2006;5:2223–2239. doi: 10.4161/cc.5.19.3255. [DOI] [PubMed] [Google Scholar]