Abstract
Invasive pulmonary aspergillosis (IPA) is a severe infectious disease with high mortality. However, the clinical diagnosis of IPA remains difficult since the microbiological evidence is hard to acquire. The main issue of the current microbiological methods is that they are time-consuming and offer a low yield. Currently, next-generation sequencing (NGS) has become an attractive alternative method for broad-based pathogen discovery due to the rapid turnaround time and high accuracy. This article describes 3 cases of IPA. Two patients had a history of chronic obstructive pulmonary disease (COPD) and asthma, and the other patient had no underlying diseases. The Aspergillus fumigatus gene was found in the bronchoalveolar lavage fluid in all three cases by NGS. This report explores the role of NGS in the diagnosis of IPA and emphasizes that IPA may occur in non-neutropenic patients.
Keywords: Next generation sequencing, invasive pulmonary aspergillosis, lung infection, diagnostic method
Introduction
Invasive pulmonary aspergillosis (IPA) is a life-threatening opportunistic infection. A variety of articles about IPA involve patients with classic risk factors such as prolonged neutropenia and hematopoietic stem cell transplantation [1]. In recent years, however, many cases of IPA have been reported in non-neutropenic patients, especially those with chronic obstructive pulmonary disease (COPD) [2,3]. In addition to direct microscopy and culture, several biomarkers have emerged such as galactomannan (GM), 1,3-β-D-glucan (BDG), and Aspergillus DNA [4]. Unfortunately, the diagnosis of IPA remains difficult. The main issues for traditional microbiological methods are that they are time-consuming and produce a low yield. New circulating biomarkers also have low sensitivity in non-neutropenic patients [4]. Polymerase chain reaction (PCR)-based methods for detecting pathogens in infectious diseases have been widely used since they are more rapid and sensitive than conventional culturing procedures. However, pathogens cannot be detected when specific primer sets are not included.
Aspergillus fumigatus accounts for the majority of IPA cases, but some infections are caused by other Aspergillus species [5]. In 2005, the revolutionary technology called next-generation sequencing (NGS) was able to detect almost every species present in a sample [4], and the complete bacterial genome sequences could be assayed and dissected in days or even hours [6]. NGS has many advantages in finding pathogens [7-10]. An article published in the New England Journal of Medicine reported that a 14-year-old boy with severe combined immunodeficiency had a fever and headache for 4 months, which finally progressed to hydrocephalus and status epilepticus. Leptospira was found in the cerebro-spinal fluid (CSF) by NGS while the other clinical assays were all negative for leptospirosis including the brain biopsy [11]. In this study, we report three cases of severe pneumonia in which NGS led to the identification of Aspergillus fumigatus. Two of them were confirmed by traditional methods while one was only detected by NGS as the other traditional tests were all negative. None of the three patients responded to antibacterial therapy. Then, they received anti-fungal therapy when their NGS results were positive and recovered after solo anti-fungal therapy.
Case 1
A 60-year-old man was admitted to Nanjing Jinling Hospital with a 10-day history of fever, cough, purulent sputum and dyspnea on exertion. The patient had no underlying diseases. He was a smoker of 40 pack-years with a normal lung function test.
At 6 days after onset, the patient was admitted to the local hospital. A chest CT scan showed emphysema, scattered infiltrates in both lungs and multiple small lymph nodes in the mediastinum. His initial laboratory test showed a WBC of 22.6*109/L (normal 4-10*109/L), neutrophil count of 18.6*109/L (normal 2.0-7.5*109/L), C-reactive protein (CRP) of 170.82 mg/L (normal 0-8 mg/L) and procalcitonin (PCT) of 0.223 μg/L (normal <0.046 µg/L). The empirical antibiotic therapy (mezlocillin sodium and sulbactam sodium combined with levofloxacin followed by imipenem and moxifloxacin) did not alleviate his symptoms. Then, the serum G test was positive (252 pg/mL, and the cutoff value is 100 pg/mL).
His body temperature remained as high as 39.2°C; thus, he was transferred to our hospital. At admission, the chest CT scan showed a high-density shadow and a cavity was found in some lesions (Figure 1A). Given the persistence of the symptoms despite therapy, tigecycline was administered and a bronchoalveolar lavage (BAL) was performed after admission. BAL fluid (BALF) was sent for testing using NGS (BGI-Shenzhen). Within 48 h after receipt of the sample, NGS detected sequence reads of Aspergillus fumigatus in the patient’s BALF (Table 1). The BALF GM was positive (4.33 and a cutoff value of 0.7 at our setting [12]).
Figure 1.
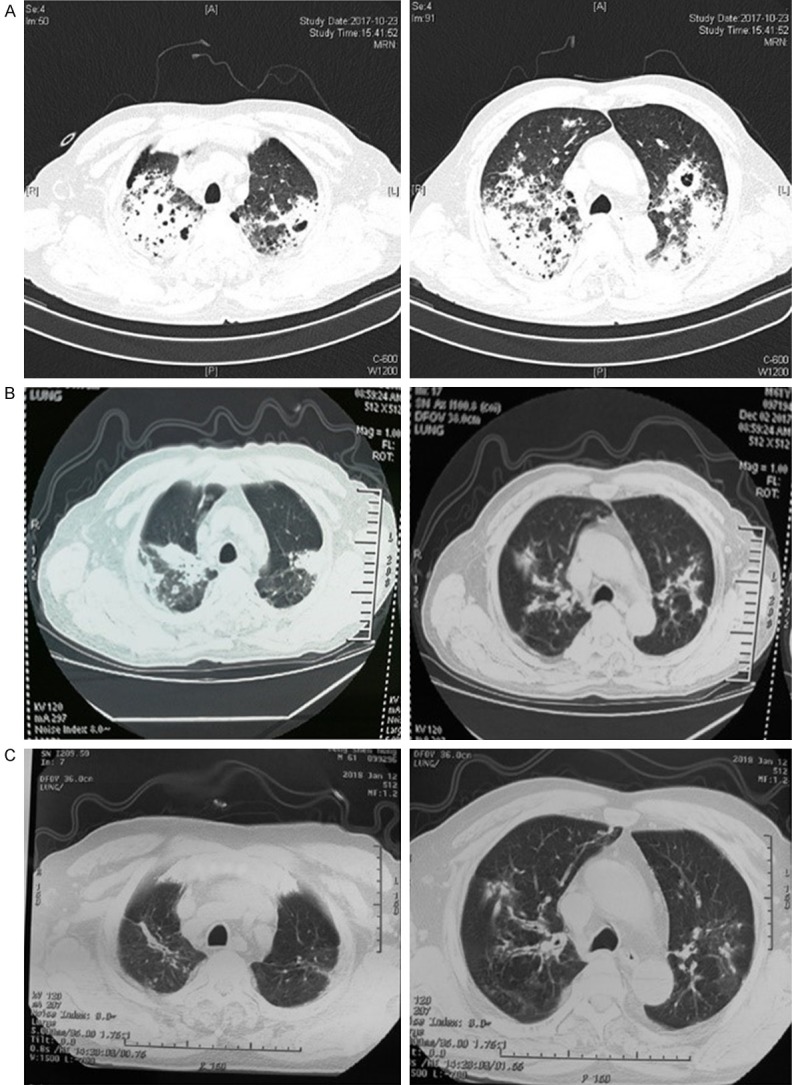
Chest CT of case 1: (A) day 3 after admission, (B) day 43, and (C) day 85.
Table 1.
NGS report of the microorganism in BALF (case 1)
The sequence number of the strict comparison of the microorganism detected at the level of genus/species.
Considering the results of BALF GM and NGS, the patient was immediately treated as IPA. Tigecycline was discontinued, and voriconazole combined with caspofungin was administered. After 10 days of combination therapy, the patient improved significantly and caspofungin was discontinued.
Before discharge, the laboratory test showed a WBC of 10.2*109/L with 81.7% neutrophils and a procalcitonin (PCT) of 0.144 μg/L. At discharge, oral voriconazole (0.2 g, twice a day) was used for after-treatment. The patient had largely recovered 2 months after voriconazole treatment (Figure 1B, 1C).
Case 2
A 69-year-old man was admitted to Nanjing Jinling Hospital due to fever, cough and expectoration. He had a previous diagnosis of COPD for 1.5 years. His daily medication included inhaled corticosteroids/long acting β2-agonist (ICS/LABA).
Twenty-two days before admission, the patient developed a cough and breathlessness. The symptoms were not relieved after the administration of montelukast, doxofylline, sulperazone and moxifloxacin.
The patient developed fever and expectoration 3 days before admission with body temperatures up to 38°C. The chest CT scan showed deterioration in the bilateral pulmonary infiltrates with cavities in some lesions (Figure 2A).
Figure 2.
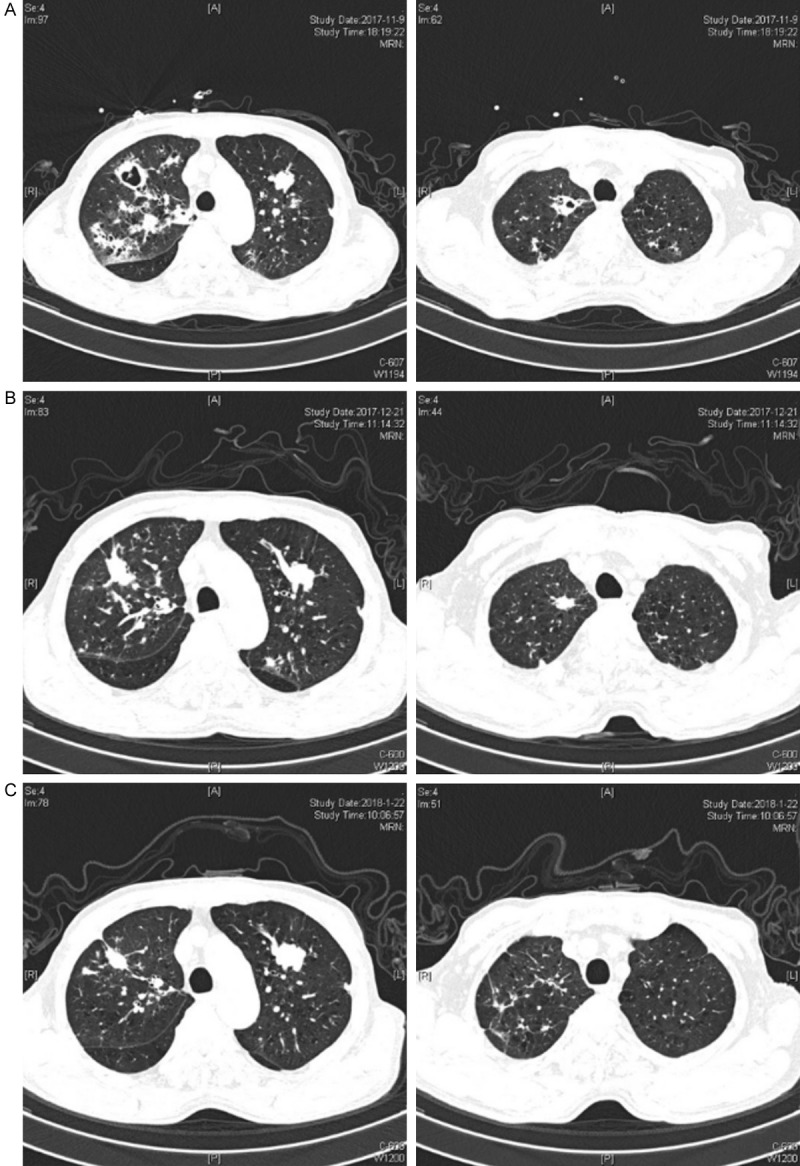
Chest CT of case 2: (A) 3 days before admission, (B) day 41, and (C) day 71.
At admission, his initial laboratory tests showed a WBC of 15.6*109/L with 92.5% neutrophils, a C-reactive protein (CRP) of 84.4 mg/L, and a procalcitonin (PCT) of 0.226 μg/L. The patient was treated with intravenous biapenem. The β-1,3-glucan test and GM test of his serum were both negative, while the sputum culture of Aspergillus fumigatus was positive. The BALF GM test was positive (3.82). NGS of the BALF detected Aspergillus fumigatus and several other pathogens (Table 2).
Table 2.
NGS report of the microorganisms in BALF (case 2)
| Genus | Species | ||
|---|---|---|---|
|
| |||
| Name | Sequence numbera | Name | Sequence numbera |
| Rothia | 198 | Rothia mucilaginosa# | 197 |
| Streptococcus | 58 | Streptococcus mitis# | 5 |
| Streptococcus pseudopneumoniae | 4 | ||
| Haemophilus | 12 | Haemophilus parainfluenzae | 11 |
| Enterococcus | 6 | Enterococcus faecium | 6 |
| Aspergillus | 62 | Aspergillus fumigatus | 57 |
| Human herpesvirus 1 | 15 | ||
The sequence number of the strict comparison of the microorganisms detected at the level of genus/species.
Normal flora present in the oral sample.
Considering the history of COPD, clinical symptoms, chest imaging, the result of BALF GM, the sputum culture and the NGS results, the patient was clinically diagnosed as IPA. Biapenem was discontinued and voriconazole was administered. At 12 days after admission, the patient had no fever and the symptoms were relieved. The laboratory tests showed a WBC of 7.2*109/L with 82.4% neutrophils, and the procalcitonin (PCT) levels decreased from 0.226 to 0.101 μg/L. The patient was discharged and took the oral voriconazole (0.2 g, twice a day). One month after discharge, the CT (Figure 2B) scan showed that the lesions in right lung were gradually absorbed, but the nodule in the left lung was enlarged. Two months after discharge, infected lesions in the right lung were further absorbed but the nodule in the left lung was further enlarged. Therefore, lung cancer was suspected for the left upper lung nodule. Unfortunately, the patient declined further examination since he felt much better.
Case 3
A 65-year-old man was admitted to Nanjing Jinling Hospital due to the fever, cough and expectoration. He had a history of asthma for 50 years.
At 18 days before admission, the patient developed a cough and expectoration. He developed fever with body temperatures up to 38.7°C at 9 days before admission. The symptoms were not relieved after the antibiotic therapy for a week in a local hospital.
At admission, his initial laboratory test showed a WBC of 10.5*109/L with 84.9% neutrophils, a C-reactive protein (CRP) of 154.5 mg/L and a procalcitonin (PCT) of 0.124 μg/L. The chest CT scan showed nodules scattered in the lung (Figure 3A). Moxifloxacin, ceftazidime and tazobactam sodium were administered. Further examination results showed the anti-Mycoplasma pneumoniae IgG antibody (ELISA) was positive (2+), and the serum GM test and BALF GM test were both negative. The sputum culture was also negative for fungi. Even with the therapy, the patient still had a fever, especially at night. Finally, the NGS of BALF detected Aspergillus fumigatus and several other pathogens (Table 3).
Figure 3.
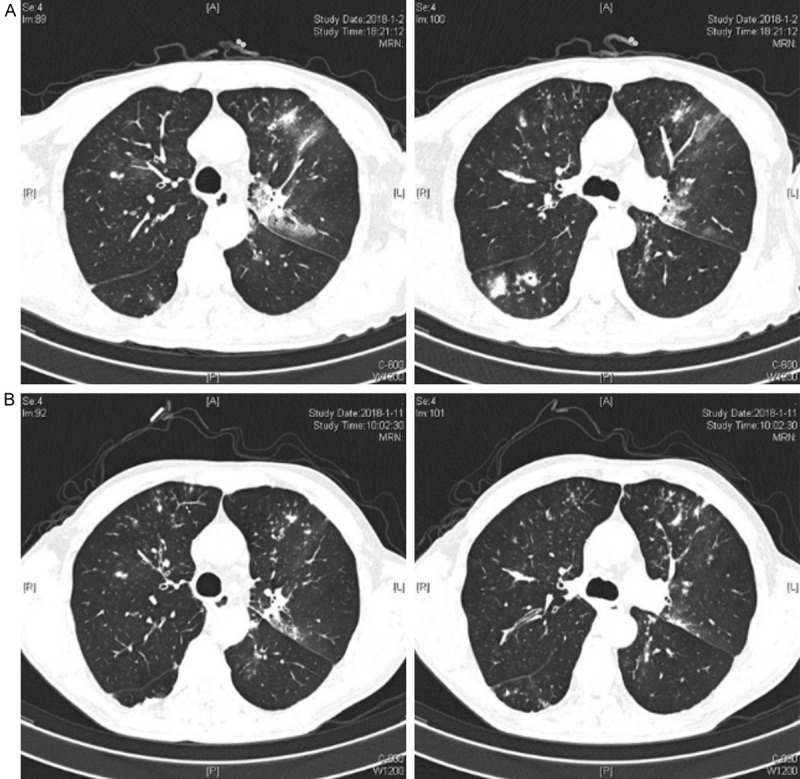
Chest CT of case 3: (A) day 1 and (B) day 10.
Table 3.
NGS report of the microorganisms in BALF (case 3)
| Genus | Species | ||
|---|---|---|---|
|
| |||
| Name | Sequence numbera | Name | Sequence numbera |
| Streptococcus | 400 | Streptococcus parasanguinis# | 36 |
| Streptococcus mitis# | 33 | ||
| Prevotella | 326 | Prevotella melaninogenica# | 231 |
| Prevotella oralb | 21 | ||
| Veillonella | 88 | Veillonella parvula# | 88 |
| Rothia | 58 | Rothia mucilaginosa# | 58 |
| Propionibacterium | 54 | Propionibacterium acnes | 51 |
| Atopobium | 26 | Atopobium parvulum | 26 |
| Bacteroides | 20 | Bacteroides salanitronisb | 13 |
| Campylobacter | 15 | Campylobacter concisus | 15 |
| Aspergillus | 25 | Aspergillus fumigatus | 24 |
| Malassezia | 4 | Malassezia globosa | 4 |
| Human herpesvirus 1 | 267 | ||
| Human herpesvirus 4 | 17 | ||
The sequence number of the strict comparison of the microorganisms detected at the level of genus/species.
Only translated to the genus name.
Normal flora present in the oral sample.
Considering the clinical symptoms, chest imaging, resistance to antibiotics and the NGS results, the patient could be clinically diagnosed as IPA. Moxifloxacin, ceftazidime and tazobactam sodium were discontinued while voriconazole was administered. With the antifungal therapy, the patient rapidly defervesced. At day 15 before discharge, the cough and expectoration were also relieved.
Discussion
For those with infectious diseases, determining the pathogen is of the utmost importance. Culturing is a traditional and common method to identify microorganisms. This process takes time and many microorganisms cannot be isolated in this way. In addition, serological tests and specific polymerase chain reaction (PCR) only target some specific pathogens [13]. The excessive use of empirical treatment leads to increased resistance to antibiotics. Thus, the timely identification of pathogens is vital to the accurate treatment and decrease of the antibiotic-resistance and mortality rate.
Invasive aspergillosis is a fatal opportunistic fungal infection that often occurs in patients with hematologic malignancies. However, many cases of IPA have been reported in nontraditional hosts, especially patients with chronic obstructive pulmonary disease (COPD) [14]. Misdiagnosis occurs much easier in these non-neutropenic patients [15]. The mortality was 90% if the treatment was delivered 10 days after the first clinical or radiological sign of the disease, but it fell to 40% when treatment was administered early [16]. As a result, early diagnosis of invasive aspergillosis is of great significance. If IPA happens in non-neutropenic patients, the fungal load is low in the lesions and there is a low chance that the antigen appears in the blood stream [17]. Hence, serum GM is often negative in non-neutropenic patients [18,19].
NGS is an assay that can sequence the entire DNA/RNA of a sample, and this process does not need any primers or probes. It holds the promise of identifying most of the pathogens. Furthermore, NGS can generate billions of DNA/RNA sequences per run, and this enables metagenomic analysis [10]. In the analyses of intestinal microbiota, NGS is now comparable in culturomics [20]. Over the years, the cost of NGS has been reduced, and it has become an attractive and alternative method for broad-based pathogen discovery with rapid turnaround time and high accuracy.
We utilize NGS (BGISEQ-500/100 sequencing platform) to explore the microbiome in lung-based infectious diseases. With the advantages of being broad spectrum, low time consumption and high accuracy, the NGS method has made it easier to detect pathogens in patients with respiratory tract infection within 3 days. Notably, in case 3, Aspergillus fumigatus was only detected by NGS while the other clinical assays were all negative. These attempts strongly suggest that NGS can be an alternative diagnostic method or a complement method to help clinicians make an appropriate decision.
Finding specific microorganisms by NGS is only the first step. However, not all microorganisms determined by NGS are associated with diseases, such as oral colonizing bacteria (e.g., Streptococcus parasanguinis, Streptococcus mitis, and Prevotella melaninogenica). Collectively, clinicians should identify the real pathogen according to both NGS and other examinations.
IPA is an opportunistic infection with a high health care cost and high mortality. More vigorous actions should be taken to determine whether it exists, especially in non-neutropenic patients. The threshold of the abundance of the pathogen should be further explored to determine whether it is colonized or not. Random control trials are needed to verify its value and to identify IPA in future studies.
Conclusion
This case report highlights the feasibility of deploying NGS of BALF as a rapid and sensitive diagnostic assay for IPA. It may help clinicians to make a precise diagnosis of infectious diseases, such as IPA, in the near future.
Acknowledgements
This work was supported by the Project of National Natural Science Foundation of China [Grant number 81873400].
Disclosure of conflict of interest
None.
References
- 1.Segal BH, Walsh TJ. Current approaches to diagnosis and treatment of invasive aspergillosis. Am J Respir Crit Care Med. 2006;173:707–717. doi: 10.1164/rccm.200505-727SO. [DOI] [PubMed] [Google Scholar]
- 2.Cunha C, Carvalho A. Toward the identification of a genetic risk signature for pulmonary aspergillosis in COPD. Clin Infect Dis. 2018;66:1153–1154. doi: 10.1093/cid/cix944. [DOI] [PubMed] [Google Scholar]
- 3.Pittet D, Huguenin T, Dharan S, Sztajzel-Boissard J, Ducel G, Thorens JB, Auckenthaler R, Chevrolet JC. Unusual cause of lethal pulmonary aspergillosis in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1996;154:541–544. doi: 10.1164/ajrccm.154.2.8756836. [DOI] [PubMed] [Google Scholar]
- 4.Alanio A, Bretagne S. Challenges in microbiological diagnosis of invasive aspergillus infections. F1000Res. 2017:6. doi: 10.12688/f1000research.10216.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes PD, Marr KA. Aspergillosis: spectrum of disease, diagnosis, and treatment. Infect Dis Clin North Am. 2006;20:545–561. vi. doi: 10.1016/j.idc.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 7.Yao M, Zhou JL, Zhu YC, Zhang YX, Lv X, Sun RX, Shen A, Ren HT, Cui LY, Guan HZ, Wu HL. Detection of listeria monocytogenes in CSF from three patients with meningoencephalitis by next-generation sequencing. J Clin Neurol. 2016;12:446–451. doi: 10.3988/jcn.2016.12.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye M, Wei W, Yang Z, Li Y, Cheng S, Wang K, Zhou T, Sun J, Liu S, Ni N, Jiang H, Jiang H. Rapid diagnosis of propionibacterium acnes infection in patient with hyperpyrexia after hematopoietic stem cell transplantation by next-generation sequencing: a case report. BMC Infect Dis. 2016;16:5. doi: 10.1186/s12879-015-1306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long Y, Zhang YX, Gong YP, Sun RX, Su LX, Lin X, Shen A, Zhou JL, Caiji ZM, Wang XY, Li DF, Wu HL, Tan HD. Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in ICU patients. Arch Med Res. 2016;47:365–371. doi: 10.1016/j.arcmed.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Guan H, Shen A, Lv X, Yang X, Ren H, Zhao Y, Zhang Y, Gong Y, Ni P, Wu H, Zhu Y, Cui L. Detection of virus in CSF from the cases with meningoencephalitis by next-generation sequencing. J Neurovirol. 2016;22:240–5. doi: 10.1007/s13365-015-0390-7. [DOI] [PubMed] [Google Scholar]
- 11.Wilson MR, Naccache SN, Samayoa E, Biagtan M, Bashir H, Yu G, Salamat SM, Somasekar S, Federman S, Miller S, Sokolic R, Garabedian E, Candotti F, Buckley RH, Reed KD, Meyer TL, Seroogy CM, Galloway R, Henderson SL, Gern JE, DeRisi JL, Chiu CY. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370:2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W, Li H, Zhang Y, Huang M, He Q, Li P, Zhang F, Shi Y, Su X. Diagnostic value of galactomannan antigen test in serum and bronchoalveolar lavage fluid samples from patients with nonneutropenic invasive pulmonary aspergillosis. J Clin Microbiol. 2017;55:2153–2161. doi: 10.1128/JCM.00345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu CY. Viral pathogen discovery. Curr Opin Microbiol. 2013;16:468–478. doi: 10.1016/j.mib.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guinea J, Torres-Narbona M, Gijon P, Munoz P, Pozo F, Pelaez T, de Miguel J, Bouza E. Pulmonary aspergillosis in patients with chronic obstructive pulmonary disease: incidence, risk factors, and outcome. Clin Microbiol Infect. 2010;16:870–877. doi: 10.1111/j.1469-0691.2009.03015.x. [DOI] [PubMed] [Google Scholar]
- 15.Cornillet A, Camus C, Nimubona S, Gandemer V, Tattevin P, Belleguic C, Chevrier S, Meunier C, Lebert C, Aupee M, Caulet-Maugendre S, Faucheux M, Lelong B, Leray E, Guiguen C, Gangneux JP. Comparison of epidemiological, clinical, and biological features of invasive aspergillosis in neutropenic and nonneutropenic patients: a 6-year survey. Clin Infect Dis. 2006;43:577–584. doi: 10.1086/505870. [DOI] [PubMed] [Google Scholar]
- 16.von Eiff M, Zuhlsdorf M, Roos N, Hesse M, Schulten R, van de Loo J. Pulmonary fungal infections in patients with hematological malignancies--diagnostic approaches. Ann Hematol. 1995;70:135–141. doi: 10.1007/BF01682033. [DOI] [PubMed] [Google Scholar]
- 17.Stergiopoulou T, Meletiadis J, Roilides E, Kleiner DE, Schaufele R, Roden M, Harrington S, Dad L, Segal B, Walsh TJ. Host-dependent patterns of tissue injury in invasive pulmonary aspergillosis. Am J Clin Pathol. 2007;127:349–55. doi: 10.1309/UJRV9DLC11RM3G8R. [DOI] [PubMed] [Google Scholar]
- 18.Cordonnier C, Botterel F, Ben Amor R, Pautas C, Maury S, Kuentz M, Hicheri Y, Bastuji-Garin S, Bretagne S. Correlation between galactomannan antigen levels in serum and neutrophil counts in haematological patients with invasive aspergillosis. Clin Microbiol Infect. 2009;15:81–6. doi: 10.1111/j.1469-0691.2008.02122.x. [DOI] [PubMed] [Google Scholar]
- 19.Verweij PE, Weemaes CM, Curfs JH, Bretagne S, Meis JF. Failure to detect circulating aspergillus markers in a patient with chronic granulomatous disease and invasive aspergillosis. J Clin Microbiol. 2000;38:3900–1. doi: 10.1128/jcm.38.10.3900-3901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiergeist A, Glasner J, Reischl U, Gessner A. Analyses of intestinal microbiota: culture versus sequencing. ILAR J. 2015;56:228–240. doi: 10.1093/ilar/ilv017. [DOI] [PubMed] [Google Scholar]


