Abstract
The biological function of microRNA-4513 (miR-4513) in human cancers is emerging. However, it remains unknown whether miR-4513 has a role in breast cancer (BC). In this study, we analyzed the expression of miR-4513 and tripartite motif containing 3 (TRIM3) in BC cell lines. The biological roles of miR-4513 and TRIM3 in BC were analyzed by cell counting kit-8 assay, colony formation assay, wound healing assay, and transwell invasion assay. The effects of miR-4513 or TRIM3 expression on the overall survival of BC patients were analyzed at Kaplan-Meier plotter website. We found miR-4513 expression was upregulated, whereas TRIM3 expression was downregulated in BC cell lines. Importantly, high miR-4513 or low TRIM3 expression was revealed as predictors for poorer overall survival of BC patients. Luciferase activity assay and western blot assay revealed TRIM3 was a direct target of miR-4513. Moreover, we showed miR-4513 was able to regulate BC cell proliferation, colony formation, cell migration, and cell invasion through regulating TRIM3. Taken together, our results suggested miR-4513 functions as an oncogene in the progression of BC and, therefore, miR-4513 may be validated as a potential therapeutic target in the future.
Keywords: miR-4513, TRIM3, breast cancer, oncogene, cell events
Introduction
Breast cancer (BC) remains a huge threat to women’s health to date [1]. Worldwide, it was estimated there will be 2.1 million female BC patients been diagnosed at 2018 [1]. Even though significantly progresses on the treatment measures of BC have been achieved, however, the treatment of BC remains a challenge because of the lack of effective therapeutic targets [2,3]. Therefore, it is essential to investigate the potential mechanisms underlying the progression of BC to identify novel diagnostic and therapeutic targets for BC.
microRNAs (miRNAs) are non-coding RNAs with 18-24 nucleotides in length [4]. miRNAs can bind to the 3’-untranslated region (3’-UTR) of targeted genes and hence resulted in message RNA degradation or protein translation inhibition [4]. Accumulating evidence has suggested that miRNAs function as crucial roles in human cancers [5]. Moreover, the importance of miRNAs in regulating human cancer progression has been appreciated [6]. It was found miRNAs could regulate cancer cell proliferation, cell fate. And tumorigenesis [6]. Multiple miRNAs, including miR-27a and miR-181 have been reported to be upregulated in BC and thus play an oncogenic role, while the miRNAs including miR-196b-5p and miR-22 were found downregulated and function as tumor suppressive miRNAs [7-10].
miR-4513, a newly identified miRNA, whose polymorphisms in the seed sequence are often associated with human diseases [11,12]. It was found rs2168518: G>A, a genetic variant in the seed region of miR-4513, was found to have a dominant role on lipid and glucose homeostasis, blood pressure, and coronary artery disease [11]. Furthermore, the rs2168518: G>A genetic variant was also found to be associated with age-related macular degeneration [12]. Very recently, miR-4513 was found to have a role in cancer as its expression was closely associated with the prognosis of lung adenocarcinoma after treatment with tyrosine kinase inhibitors targeting epidermal growth factor receptors [13]. However, little is known regarding the expression and molecular mechanism of miR-4513 in BC.
In this study, we investigated the expression patterns of miR-4513 in human BC cell lines, followed by functional analyses in BC cell lines. Our data revealed that miR-4513 was upregulated in BC cell lines. High expression of miR-4513 predicts poor overall survival of BC patients. Moreover, our results clearly suggested that miR-4513 promoted cell growth, colony formation, migration, and invasion in BC cell lines via down-regulating tripartite motif containing 3 (TRIM3) expression, further supporting miR-4513 as a potential therapeutic target in BC.
Materials and methods
Cell lines
Human BC cell lines (MCF-7, T-47D), and the breast epithelial cell line (MCF-10A) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China).
Cell culture and cell transfection
The above-mentioned cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) at a 37°C humidified incubator containing 5% of CO2.
For cell transfection, cells were seeded at the density of 6 × 104 cells/well and incubated for 24 h. Then, cells were transfected with miR-4513 inhibitor (5’-AUGGGCCUCCAGCCGUCAGUCU-3’), inhibitor negative control (NC-inhibitor, 5’-GCUCCGGCUUGCGGUAUCACAC-3’), small-interfering RNA targeting TRIM3 (si-TRIM3, 5’-GCUCACUGUCACUACCAAATT-3’), or NC-siRNA (5’-UUCUCCGAACGUGUCACGUTT-3’, all purchased from GenePharma, Shanghai, China) at the final concentration of 100 nmol/l using Lipofectamine 2000 (Invitrogen) according to the supplier’s instruction. After transfection for 48 h, cells were collected for following analyses.
RNA isolation and real-time quantitative PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturers’ protocols. The PrimeScriptTM RT-PCR kit (Takara, Dalian, Liaoning, China) was used to synthesized the first-strand complementary DNA (cDNA). SYBR Green Mater Mix Kit (Applied Biosystems, Foster City, CA, USA) was used for qRT-PCR at the Applied Biosystems 7300 system. U6 small nuclear RNA (U6 snRNA) was used as internal control and the relative miR-4513 expression levels were analyzed using the 2-ΔΔCt method. The primers were as follows: miR-4513: forward, 5’-ACACTCCAGCTGGGAGACTGACGGCTGGAG-3’, reverse, 5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGATGGGC-3’; U6 snRNA: forward, 5’-CTCGCTTCGGCAGCACA-3, reverse, 5’-ACGCTTCACGAATTTGCGT-3’. Experiments were conducted in triplicates.
Protein isolation and western blot
Total protein was extracted from cell lines using RIPA lysis buffer (Beyotime, Haimen, Jiangsu, China) according to the manufacturer’s instructions. 25 µg protein samples were isolated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membranes (Beyotime). After blocked with 5% fat-free milk, membranes were incubated with the primary antibodies (rabbit anti-TRIM3: ab111840, rabbit anti-GAPDH: ab181602; Abcam, Cambridge, MA, USA) for over-night at 4°C, followed by HRP-conjugated secondary antibody (ab6721, Abcam) for 3 h at room temperature. Western blot bands were visualized using BeyoECL kit (Beyotime) and quantified with Image J 1.42 software (NIH, Bethesda, MD, USA). Experiments were conducted in triplicates.
Dual luciferase reporter assay
The potential miR-4513 binding site in the 3’-UTR of TRIM3 was predicted by TargetScan. TRIM3 3’-UTR sequence was amplified from genome and cloned into psiCHECK-2 luciferase vector (Promega, Madison, WI, USA) to generate TRIM3-wt. Site-direct mutagenesis kit (Takara) was used to produce TRIM3-mt. For dual luciferase reporter assay, cells were co-transfected with TRIM3-wt or TRIM3-mt and miR-4513 inhibitor or NC-inhibitor using Lipofectamine 2000 according to the manufacturer’s protocols. After 48 h of transfection, Firefly and Renilla luciferase activities were detected using Dual Luciferase Reporter Assay System (Promega).
Cell counting kit-8 (CCK-8) assay
Cells were seeded at the density of 5 × 103 cells/well and allow to growth for 24 h. Then, CCK-8 solution (Beyotime) was added to the medium at indicated time (0, 24, 48 and 72 h) and further incubation for 2 h. Optical density at 450 nm was measured by a microplate reader (Bio-Rad, Hercules, CA, USA). Experiments were conducted in triplicates.
Colony formation assay
Cells were seeded at the density of 3 × 103 cells/well) and cultured for 14 days at the above-mentioned conditions. Subsequently, cells were fixed with methanol and stained with 0.1% crystal violet (Beyotime). Colony numbers were counted under a light microscope (Eclipse TS100; Nikon, Tokyo, Japan). Experiments were conducted in triplicates.
Wound healing assay
Cells were seeded at density of 6 × 104 cells/well and incubated to 75% confluence. Wounds were created with 10 µl pipette tips at the cell surface. Then, cells were washed with PBS to remove debris and incubated at the above-mentioned conditions. Cell movements in the scratch were observed and photographed at 0 and 48 h. The width of the scratch was detected with Image J 1.42 software (NIH). Experiments were conducted in triplicates.
Transwell invasion assay
Cell invasion ability was analyzed by transwell invasion assay. Cells were seeded into the upper chamber and supplemented with DMEM. The lower chamber was supplemented with DMEM supplemented with FBS. The membrane was pre-coated with Matrigel (Franklin Lakes, NJ, USA). The cells were incubated for 48 h and the invasion cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet solution (Beyotime). Invasion numbers were counted under microscope. Experiments were conducted in triplicates.
Kaplan-Meier survival analysis
Kaplan-Meier plotter (www.kmplot.com) was used to assess the effect of miR-4513 or TRIM3 expression on the overall survival of BC. Cut-off value was auto-selected in the algorithm. Log-rank test was used to calculate the difference between low or high expression groups.
Statistical analysis
Data were presented as mean ± standard deviation (SD) and analyzed using SPSS 21.0 (IBM, Armonk, NY, USA). The differences were assessed using a two-tailed Student’s t-test or one-way ANOVA and Tukey post-hoc test for two or above groups, respectively. P<0.05 was considered to indicate a statistically significant.
Results
Upregulate expression of miR-4513 in BC
The expression of miR-4513 in BC cell lines was analyzed using qRT-PCR. As shown in Figure 1A, miR-4513 expression was significantly increased in BC cell lines (MCF-7 and T-47D) investigated compared with breast epithelial cell line (MCF-10A). Moreover, the analysis of overall survival data obtained from 1061 BC patients in the KM plotter website showed high miR-4513 predicts worse overall survival of BC patients (Figure 1B).
Figure 1.
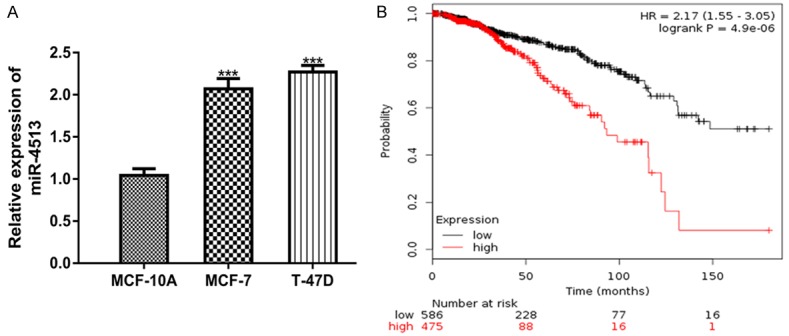
Overexpression of miR-4513 in BC. A. qRT-PCR to analyze miR-4513 in BC cell lines (MCF-7 and T-47D) and breast epithelial cell line (MCF-10A). B. Effects of miR-4513 expression on the overall survival of BC patients. (***P<0.001) qRT-PCR: real-time quantitative PCR; BC: breast cancer; miR-4513: microRNA-4513.
Downregulate expression of TRIM3 in BC
The expression of TRIM3 in BC cell lines was analyzed using western blot. As displayed in Figure 2A, TRIM3 expression was dramatically decreased in MCF-7 and T-47D cell lines compared to MCF-10A cell line. The overall survival analysis on 3951 BC patients showed that low TRIM3 was correlated with poor overall survival od BC patients (Figure 2B).
Figure 2.
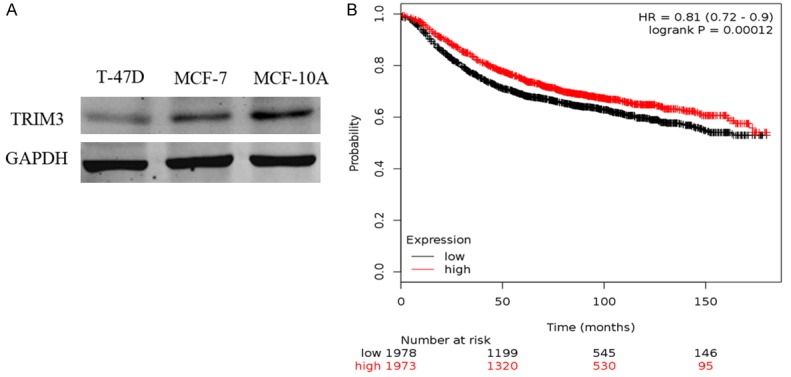
Downregulate expression of TRIM3 in BC. A. Western blot to analyze TRIM3 in BC cell lines (MCF-7 and T-47D) and breast epithelial cell line (MCF-10A). B. Effects of TRIM3 expression on the overall survival of BC patients. BC: breast cancer; TRIM3: tripartite motif containing 3.
TRIM3 was a direct target of miR-4513
Considering the importance of miR-4513 and TRIM3, we are interested to investigate the relationship of them. The TargetScan prediction algorithm showed that TRIM3 contains a binding site for miR-4513 in its 3’-UTR (Figure 3A). Luciferase activity reporter assay showed that miR-4513 inhibitor transfection increased the luciferase activity in cells transfected with TRIM3-wt, while it did not have significance influence on the luciferase activity of cells transfected with TRIM3-mt (Figure 3B).
Figure 3.
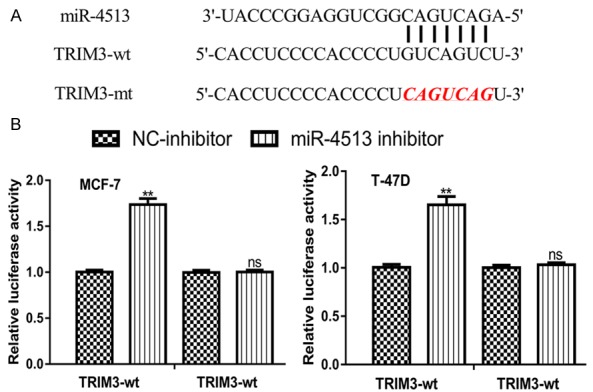
TRIM3 was a direct target of miR-4513. A. Binding site between miR-4513 and the 3’-UTR of TRIM3. B. Luciferase activity reporter assay to measure luciferase activity in cells transfected with TRIM3-wt or TRIM3-mt and miR-4513 inhibitor or NC-inhibitor. (ns not significant, **P<0.01) BC: breast cancer; TRIM3: tripartite motif containing 3; miR-4513: microRNA-4513; UTR: untranslated region; wt: wild-type: mt: mutant; NC-inhibitor: negative control for miR-4513 inhibitor.
miR-4513 regulates BC cell proliferation, colony formation, cell migration, and cell invasion through targeting TRIM3
The above results have illustrated the importance of miR-4513 and TRIM3 in BC, we then interested to investigate the biological roles of miR-4513 and TRIM3. The transfection of miR-4513 inhibitor significantly decreased the expression of miR-4513 (Figure 4A). In the meantime, the transfection of si-TRIM3 significantly decreased the expression of TRIM3 (Figure 4B). Meanwhile, the stimulation effects of miR-4513 inhibitor on TRIM3 expression can be reversed by si-TRIM3 (Figure 4B). CCK-8 assay showed that cell proliferation was inhibited by miR-4513 inhibitor but promoted by si-TRIM3 (Figure 4C). Colony formation assay confirmed the results of CCK-8 assay, which showed colony numbers were increased in si-TRIM3 group but decreased in miR-4513 group (Figure 4D). Wound-healing assay revealed that cell migration can be repressed by miR-4513 inhibitor but enhanced by si-TRIM3 (Figure 4E). Transwell invasion assay showed that cell invasion was inhibited by miR-4513 inhibitor but promoted by si-TRIM3 (Figure 4F). Furthermore, si-TRIM3 transfection could partially reversed the inhibitory effects of miR-4513 inhibitor on BC cell proliferation, colony formation, cell migration, and cell invasion in vitro (Figure 4C-F).
Figure 4.
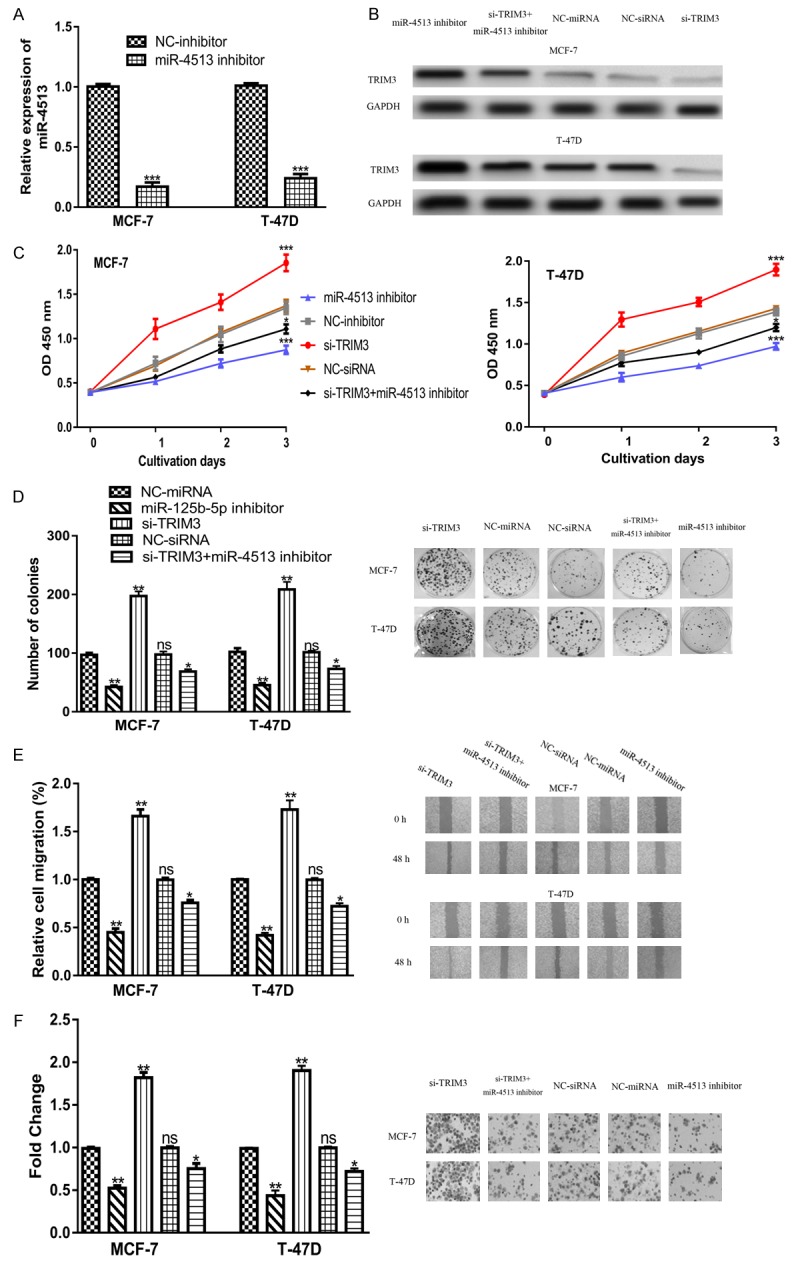
miR-4513 regulates BC cell proliferation, colony formation, and cell migration through targeting TRIM3. (A) qRT-PCR to analyze miR-4513 expression in cells after miR-4513 inhibitor or NC-inhibitor transfection. (B) TRIM3 expression, (C) Cell proliferation, (D) Colony formation, (E) Cell migration, and (F) Cell invasion in cells transfected with si-TRIM3, NC-siRNA, miR-4513 inhibitor, NC-inhibitor, or miR-4513 inhibitor and si-TRIM3 co-transfection. (ns not significant, *P<0.05, **P<0.01, ***P<0.001) BC: breast cancer; TRIM3: tripartite motif containing 3; miR-4513: microRNA-4513; qRT-PCR: real-time quantitative PCR; NC-inhibitor: negative control for miR-4513 inhibitor; NC-siRNA: negative control small interfering RNA.
Discussion
TRIM3 located at chromosome 11p15.5, a region that has been reported to contain multiple cancer-related genes [14,15]. Current studies have demonstrated that TRIM3 expression was reduced in several human cancers including liver cancer, gastric cancer, cervical cancer, and colorectal cancer [16-19]. TRIM3 was also revealed to regulate cell behaviors including cell proliferation, metastasis, cell migration, and cell invasion [16-19]. Upregulation of TRIM3 expression inhibited these cell malignancy behaviors, while the downregulation of TRIM3 promoted cell malignancy [16-19]. Importantly, TRIM3 was reported to function as tumor suppressor through regulating p21 (WAF1/CIP1) to prevent the accumulation of cyclin D1-cdk4 [20].
In this study, we found TRIM3 expression was significantly reduced in BC cell lines compared with the normal cell line. Then, we are interested to discovery the clinical significance of TRIM3 in BC and therefore we analyzed the overall survival data from 3951 BC patients. Notably, we found low TRIM3 expression was closely associated with poor overall survival of BC patients. These results illustrated that TRIM3 also functions as tumor suppressor in BC, which is in consistent with the role of TRIM3 in other human cancers [16-19].
As a type of potential regulator, miRNAs serve a key role in the progression of BC [7-10]. The role of miR-4513 in human cancers is rarely investigated except a recent study showed the importance of miR-4513 in lung adenocarcinoma [13]. By measuring miR-4513 expression in BC cell lines, we found its expression was remarkedly enhanced in BC cell lines. Kaplan-Meier curve showed that high miR-4513 expression was correlated poorer overall survival of BC patients. These results showed miR-4513 might has an oncogenic role in the progression of BC. Then, functional assays were conducted to investigate the role of miR-4513 in BC. It was found downregulation of miR-4513 inhibits BC cell proliferation, colony formation, cell migration, and cell invasion.
Then, software algorithms were performed to search for the possible connection between miR-4513 and TRIM3. Importantly, TargetScan analysis showed that miR-4513 can directly bind to the 3’-UTR of TRIM3. Luciferase activity reporter assay and western blot assay confirmed TRIM3 was a direct target of miR-4513. Recue experiments showed that downregulation of TRIM3 partially reversed the inhibitory effects of miR-4513 inhibitor on BC cell behaviors, which demonstrated that TRIM3 was a functional target of miR-4513 in BC. We also searched the targets of miR-4513 in miRTarBase, a database contains the experimentally validated miRNA targets, and we found TRIM3 was not validated as target of miR-4513 to date. Therefore, the current study provided an experimental validated novel target of miR-4513, which will help us to understand the function of miR-4513 in human cancers. The limitation of this study was that we analyzed the effects of miR-4513 and TRIM3 on the overall survival of BC patients but we did not analyze the associations of miR-4513 and TRIM3 expression with clinicopathological parameters. We will continue this work by recruiting BC patients to validate the value of miR-4513 and TRIM3 expression on the overall survival of BC patients in our local cohort. Moreover, the biological roles of miR-4513 and TRIM3 should be investigated in animal model in our following work.
Conclusion
In conclusion, the present study demonstrated that miR-4513 acted as an oncogenic role, whereas TRIM3 functions as a tumor suppressive role in BC cells. miR-4513 downregulation suppressed cell proliferation, colony formation, cell migration, and cell invasion through targeting TRIM3. Therefore, miR-4513 or TRIM3 may be potential novel therapeutic targets for BC.
Acknowledgements
This study was supported in part by grants from the Natural Science Foundation of Henan Province, China (General Program No. 162300410220), the Educational Commission of Henan Province, China (No. 16A310002, No. 18A310004), the Science and Technique Foundation of Henan Province, China (No. 201403133).
Disclosure of conflict of interest
None.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ha R, Chow D, Wynn R. Global trend in breast cancer imaging research 1992-2012: bibliometric study. Am J Roentgenol. 2014;202:696–697. doi: 10.2214/AJR.13.11993. [DOI] [PubMed] [Google Scholar]
- 3.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang G, Shi W, Fang H, Zhang X. miR-27a promotes human breast cancer cell migration by inducing EMT in a FBXW7-dependent manner. Mol Med Rep. 2018;18:5417–5426. doi: 10.3892/mmr.2018.9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Fu X, Li Q, Wang Y, Fan D, Zhou Q, Kuang W, Shen L. MicroRNA-181 serves an oncogenic role in breast cancer via the inhibition of SPRY4. Mol Med Rep. 2018;18:5603–5613. doi: 10.3892/mmr.2018.9572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhu X, Rao X, Yao W, Zou X. Downregulation of MiR-196b-5p impedes cell proliferation and metastasis in breast cancer through regulating COL1A1. Am J Transl Res. 2018;10:3122–3132. [PMC free article] [PubMed] [Google Scholar]
- 10.Song YK, Wang Y, Wen YY, Zhao P, Bian ZJ. MicroRNA-22 suppresses breast cancer cell growth and increases paclitaxel sensitivity by targeting NRAS. Technol Cancer Res Treat. 2018;17:1533033818809997. doi: 10.1177/1533033818809997. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Ghanbari M, de Vries PS, de Looper H, Peters MJ, Schurmann C, Yaghootkar H, Dörr M, Frayling TM, Uitterlinden AG, Hofman A, van Meurs JB, Erkeland SJ, Franco OH, Dehghan A. A genetic variant in the seed region of miR-4513 shows pleiotropic effects on lipid and glucose homeostasis, blood pressure, and coronary artery disease. Hum Mutat. 2014;35:1524–1531. doi: 10.1002/humu.22706. [DOI] [PubMed] [Google Scholar]
- 12.Ghanbari M, Erkeland SJ, Xu L, Colijn JM, Franco OH, Dehghan A, Klaver CCW, Meester-Smoor MA. Genetic variants in microRNAs and their binding sites within gene 3’UTRs associate with susceptibility to age-related macular degeneration. Hum Mutat. 2017;38:827–838. doi: 10.1002/humu.23226. [DOI] [PubMed] [Google Scholar]
- 13.Zhang N, Li Y, Zheng Y, Zhang L, Pan Y, Yu J, Yang M. miR-608 and miR-4513 significantly contribute to the prognosis of lung adenocarcinoma treated with EGFR-TKIs. Lab Invest. 2018;99:568–576. doi: 10.1038/s41374-018-0164-y. [DOI] [PubMed] [Google Scholar]
- 14.Koi M, Johnson LA, Kalikin LM, Little PF, Nakamura Y, Feinberg AP. Tumor cell growth arrest caused by subchromosomal transferable DNA fragments from chromosome 11. Science. 1993;260:361–364. doi: 10.1126/science.8469989. [DOI] [PubMed] [Google Scholar]
- 15.El-Husseini AE, Fretier P, Vincent SR. Cloning and characterization of a gene (RFN22) encoding a novel brain expressed ring finger protein (BERP) that maps to human chromosome 11p15.5. Genomics. 2000;71:363–367. doi: 10.1006/geno.2000.6452. [DOI] [PubMed] [Google Scholar]
- 16.Huang XQ, Zhang XF, Xia JH, Chao J, Pan QZ, Zhao JJ, Zhou ZQ, Chen CL, Tang Y, Weng DS, Zhang JH, Xia JC. Tripartite motif-containing 3 (TRIM3) inhibits tumor growth and metastasis of liver cancer. Chin J Cancer. 2017;36:77. doi: 10.1186/s40880-017-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu H, Yang H, Zhang X, Wang B, Mao J, Li X, Wang M, Zhang B, Sun Z, Qian H, Xu W. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J Exp Clin Cancer Res. 2018;37:162. doi: 10.1186/s13046-018-0825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Guo Q, Gao S, Hua K. Tripartite motif-containing protein 3 plays a role of tumor inhibitor in cervical cancer. Biochem Biophys Res Commun. 2018;498:686–692. doi: 10.1016/j.bbrc.2018.03.046. [DOI] [PubMed] [Google Scholar]
- 19.Piao MY, Cao HL, He NN, Xu MQ, Dong WX, Wang WQ, Wang BM, Zhou B. Potential role of TRIM3 as a novel tumour suppressor in colorectal cancer (CRC) development. Scand J Gastroenterol. 2016;51:572–582. doi: 10.3109/00365521.2015.1124285. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Raheja R, Yeh N, Ciznadija D, Pedraza AM, Ozawa T, Hukkelhoven E, Erdjument-Bromage H, Tempst P, Gauthier NP, Brennan C, Holland EC, Koff A. TRIM3, a tumor suppressor linked to regulation of p21 (Waf1/Cip1) Oncogene. 2014;33:308–315. doi: 10.1038/onc.2012.596. [DOI] [PMC free article] [PubMed] [Google Scholar]


