Abstract
The DXO family of proteins participates in eukaryotic mRNA 5′-end quality control, removal of non-canonical NAD+ cap and maturation of fungal rRNA precursors. In this work, we characterize the Arabidopsis thaliana DXO homolog, DXO1. We demonstrate that the plant-specific modification within the active site negatively affects 5′-end capping surveillance properties of DXO1, but has only a minor impact on its strong deNADding activity. Unexpectedly, catalytic activity does not contribute to striking morphological and molecular aberrations observed upon DXO1 knockout in plants, which include growth and pigmentation deficiency, global transcriptomic changes and accumulation of RNA quality control siRNAs. Conversely, these phenotypes depend on the plant-specific N-terminal extension of DXO1. Pale-green coloration of DXO1-deficient plants and our RNA-seq data reveal that DXO1 affects chloroplast-localized processes. We propose that DXO1 mediates the connection between RNA turnover and retrograde chloroplast-to-nucleus signaling independently of its deNADding properties.
INTRODUCTION
The DXO family of proteins functions in eukaryotic mRNA 5′-end quality control (5′QC) (1–3), removal of the noncanonical NAD+cap (deNADding) (4,5), and in the processing and degradation of fungal rRNA precursors (6–8). Transcripts synthesized by RNA polymerase II (RNAP II) immediately acquire a methylated guanosine cap structure (m7G) that confers their stability and facilitates further processing, export, turnover and mRNA translation (9). Cap synthesis is subjected to complex regulatory mechanisms (10), which occasionally lead to accumulation of capping intermediates; capped, but unmethylated Gppp-RNAs and uncapped triphosphorylated ppp-RNAs. This may occur during co-transcriptional capping in the nucleus or post-transcriptional re-capping of previously decapped mRNAs in the cytoplasm (9,11–13). Potentially dysfunctional capping intermediates are removed by the 5′QC mechanism mediated by DXO enzymes. While canonical NUDIX decapping proteins (e.g. Dcp2, Nudt16 and Nudt3) specifically release m7Gpp from RNAs with the mature cap, DXO enzymes remove the entire cap structure together with the first transcribed nucleotide from both m7Gppp- and Gppp-RNAs (14). In turn, uncapped ppp-RNAs are cleaved within the triphosphate linkage by the DXO pyrophosphohydrolase (PPH) activity that releases PPi, with an exception of Ashbya gossypii triphosphonucleotide hydrolase (TPH) Rai1 that liberates the entire first nucleotide (15).
DXO proteins also show strong deNADding activity on RNAs with non-canonical NAD+ cap that consists of nicotinamide adenine dinucleotide. NAD+ is either occasionally introduced at the transcription start site with an A at position +1 or possibly also as a posttranscriptional modification (16,17). NAD+-capped fraction constitutes 1–6% of these mRNAs and may connect transcription to a cellular redox state that is reflected by the NAD+/NADH ratio (18). Bacterial NAD+ caps stabilize mRNAs (19,20), whereas in mammals they serve as markers for degradation (4). Importantly, mammalian DXO exhibits ∼6-fold more robust deNADding than 5′QC activity (4). Besides DXO enzymes, NAD+ cap is also removed by NUDIX hydrolases, like prokaryotic NudC and mammalian Nudt12 (19–21), and most likely by other NUDIX enzymes that are broadly represented in eukaryotes (18,22). All hydrolytic activities of DXO proteins produce monophosphorylated p-RNAs that can be further degraded either distributively by DXO or by processive exoribonucleases from the Xrn family.
Biochemical properties of DXO proteins are governed by the phosphodiesterase PD-(D/E)XK active site, but catalytic profiles and substrate specificities may vary among homologs from different organisms (15). Mammalian DXO shows all four activities, although its decapping activity is limited by ribose methylation at the first and second nucleotides of the transcript, a hallmark of mature mRNA in eukaryotic cells (23). Moreover, the efficiency of DXO 5′-3′ exoribonuclease may depend on the 5′-end sequence of the substrate (24) and the activity is negatively affected by adenosine 3′,5′-bisphosphate (PAP), which is also an inhibitor of 5′-3′ exoribonucleases from the Xrn family (25,26). In yeast, biochemical properties are distributed between the two paralogs, Dxo1 and Rai1. Both proteins have deNADding and decapping activities, but Rai1 has a strong preference toward unmethylated caps, while Dxo1 resembles mammalian DXO and is less selective. PPH or TPH activity toward ppp-RNAs is present only in Rai1 homologs, while 5′-3′ exoribonuclease properties are exclusive for Dxo1. Rai1 can instead enhance the function of fungal Rat1 5′-3′ exoribonuclease in rRNA maturation (6–8). Certain DXO homologs show very low activity, e.g. Candida albicans Rai1 with a single amino acid substitution in the active pocket (15), or are catalytically inactive, like Drosophila melanogaster Cutoff that become a player in piRNA synthesis and maturation (27–30).
In this work, we present a structural, biochemical and functional analysis of the DXO1 protein, which is a sole DXO homolog in Arabidopsis thaliana. DXO1 and its homologs from other plant species share high active site conservation with their mammalian and fungal counterparts, as also revealed by our DXO1 crystal structure. Plant DXO proteins, however, contain a point amino acid substitution in the vicinity of the active site that negatively affects the biochemical properties of DXO1. As a result, 5′QC activities are generally reduced in DXO1, except for a weak 5′-3′ exoribonuclease activity. In turn, DXO1 shows very strong deNADding activity in vitro, which is almost unaffected by a plant-specific mutation that severely limits other activities. However, our data clearly show that deNADding by DXO1 is not critical for plant metabolism, suggesting a possible redundancy with one or more NUDIX hydrolases that are represented by multiple homologs in plants (18,22).
Another distinctive feature of DXO1 in plants is an N-terminal extension (NTE) that restrains 5′QC activities, confers RNA-binding properties in vitro, and is crucial for DXO1 function in vivo. Plants with DXO1 knockout display severe morphological and transcriptomic changes that can be reverted only by the full-length DXO1 regardless of the presence of its enzymatic activity. Sequencing of small RNAs in dxo1 revealed the accumulation of RNA quality control siRNA (rqc-siRNA) that is characteristic of Arabidopsis RNA degradation mutants (31–36). However, the 5′-end status of rqc-siRNAs-producing mRNAs suggested that this phenotype does not directly result from the lack of DXO1 enzymatic activity. Based on our RNA-seq studies, we conclude that the observed phenotypes are most likely associated with the plant-specific role of DXO1 in chloroplast functioning that in turn affects nuclear gene expression. Collectively, our data show that, unlike other eukaryotic counterparts, DXO1 probably has a minor or redundant contribution to 5′QC and deNADding. Instead, the most prominent function of DXO1 is independent of its enzymatic activities, relies on the plant-specific N-terminal extension and impacts chloroplast-related processes.
MATERIALS AND METHODS
Protein crystallization, data collection and structure determination
Arabidopsis thaliana (At) DXO1(ΔN194) was cloned into the pET26b (C-terminal His tag) vector (Novagen). The protein was expressed in Escherichia coli Rosetta DE3 cells at 20°C and purified by Ni-NTA Superflow (Qiagen) and gel filtration (Sephacryl S-300, GE Healthcare) chromatography. The protein was concentrated to 23 mg/ml in a buffer containing 250 mM NaCl, 20 mM Tris (pH 7.5) and 5% (v/v) glycerol, frozen in liquid nitrogen and stored at -80°C. DXO1(ΔN194) crystals were obtained by the sitting-drop vapor diffusion method at 20°C with a reservoir solution containing 0.2 M NH4F and 18% (w/v) PEG3350. X-ray diffraction data were collected at the National Synchrotron Light Source (NSLS) beamline X29A. The diffraction images were processed and scaled using HKL2000 (37). The structure was solved with the molecular replacement method with the program Phaser (38) and the structure of S. pombe Rai1 as the search model. The structure refinement was carried out with the Crystallography and NMR System (CNS) (39) and Refmac (40). The atomic models were built with the Coot program (41). The crystallographic information is summarized in Supplementary Table S1.
RNA substrates
RNA oligonucleotides 3′-labeled with FAM (6-carboxyfluorescein) were purchased from Bio-Synthesis Inc or Metabion (see sequences of RNA substrates in Supplementary Table S2). Radioactively 32P-labeled substrates were obtained by in vitro transcription with T7 RNA polymerase using 300 ng of PCR-generated DNA templates. Templates for guanosine-rich and guanosine-deficient RNAs were prepared from pGEM-T Easy Vector and fragment of the Saccharomyces cerevisiae oriIV-1 DNA, respectively. NAD+-capped substrates were prepared as described (4) with DNA template that contained T7 φ2.5 promoter and was deprived of adenosines except at the transcription start site (see the list of oligonucleotides in Supplementary Table S3). Uniformly labeled RNAs were transcribed in the presence of [α-32P]GTP (Hartmann Analytics) and GMP, AMP or NAD (Sigma) to generate 5′ pG-RNA, pA-RNA or NppA-RNA, respectively. 5′-end labeled pppG-RNA was transcribed in the presence of [γ-32P]GTP and NppA-RNA labeled at adenylate phosphate was transcribed in the presence of [32P]NAD (Perkin Elmer). 5′-end-cap 32P-labeled RNA was generated using 20 pmol of non-radioactive uncapped RNA and ScriptCap™ m7G Capping System (Epicentre) and [α-32P]GTP with or without SAM according to manufacturer's protocol.
RNA in vitro exonuclease and 5′-end structure hydrolysis assays
In vitro assays were carried out at 37°C for indicated times in 20 μl final volume of reaction buffer E (10 mM Tris pH 7.8, 50 mM KOAc, 2 mM MgOAc, 2 mM MnCl2, 1 mM DTT, 0.1 mM spermine, 1 U/μl RiboLock RNase Inhibitor) with 100 nM FAM-labeled RNA or 100 cps of radiolabeled RNA substrates and indicated amount of recombinant protein. Reactions were stopped with 0.05 M EDTA and separated on 8–15% denaturing PAGE (exonuclease assays) or resolved on polyethyleneimine-cellulose TLC plates (hydrolysis assays) in 0.45 M (NH4)2SO4 (m7G- and G-capped substrates), 0.7 M KH2PO4 (triphosphate substrates) or 0.75 M LiCl (NAD+-capped substrates). Gels and plates were analyzed with PhosphorImager Typhoon FLA 9000 (GE Healthcare) and quantitated with ImageJ software.
Dcp2, DXO1 and exoribonuclease treatment
Total plant RNA from three biological replicates was treated with RNase-free TURBO DNase (Ambion) for 60 min at 37°C. 500 ng of DNA-free RNA was incubated for 40 min at 37°C with Dcp2, DXO1 or Terminator exonuclease (Epicentre), or Terminator exonuclease combined with either Dcp2 or DXO1. Samples were purified with phenol-chloroform, ethanol precipitated and reverse transcribed using SuperScript III (Invitrogen). cDNA was diluted 6.66 times and analyzed with qPCR (see Supplementary Materials and Methods). mRNA levels were normalized to Pol III-transcribed capless pre-tRNA-snoRNA (tsnoRNA) that contains G at position +1 (42). Oligonucleotides used for RT-qPCR detection are listed in Supplementary Table S3.
Subcellular localization analysis
Arabidopsis mesophyll protoplasts were prepared and transfected as described (43) with the pGWB654 vectors that carried DXO1(wt) and DXO1(ΔN194) C-terminally fused with RFP under the control of constitutive 35S promoter from the cauliflower mosaic virus (CaMV). Protoplast samples were imaged using a FV1000 confocal system (Olympus) with 60×/1.2 water immersion lens. RFP fluorescence and chlorophyll autofluorescence were excited with 559 and 405 nm laser, respectively, and collected with spectral detectors with appropriate detection windows. Images were analyzed with Fiji software. Chloroplasts were isolated as described (44). Briefly, 3 g of two-week-old seedlings were homogenized on ice in the Chloroplast Isolation Buffer, CIB (20 mM HEPES/KOH, pH 8.0, 0.3 M sorbitol, 5 mM MgCl2, 5 mM EGTA, 5 mM EDTA, 10 mM NaHCO3). Homogenate was filtered through two layers of Miracloth and centrifuged at 1000g for 5 min at 4°C. Pellet was suspended by rotating in 500 μl of residual supernatant and transferred onto the top of the Percoll gradient (13 ml Percoll, 13 ml 2× CIB, 5 mg glutathione; precentrifuged at 43 000g for 30 min at 4°C). Intact chloroplasts were separated by centrifugation at 7800g for 10 min at 4°C, carefully recovered from the 40/85% Percoll interface and washed twice with 10 ml CIB (1000g for 5 min at 4°C). Chloroplast proteins were released in the lysis buffer (10 mM HEPES–KOH, pH 8.0, 5 mM MgCl2, protease inhibitor cocktail) and analyzed by western blotting with mouse monoclonal GFP antibody (Santa Cruz Biotechnology) and rabbit polyclonal PsbO, RbcL and UGPase antibodies (Agrisera). Anti-mouse (Thermo Scientific) or anti-rabbit (Calbiochem) horseradish peroxidase-conjugated antisera were used as secondary antibodies.
RESULTS
Unique structural features of Arabidopsis thaliana DXO homolog
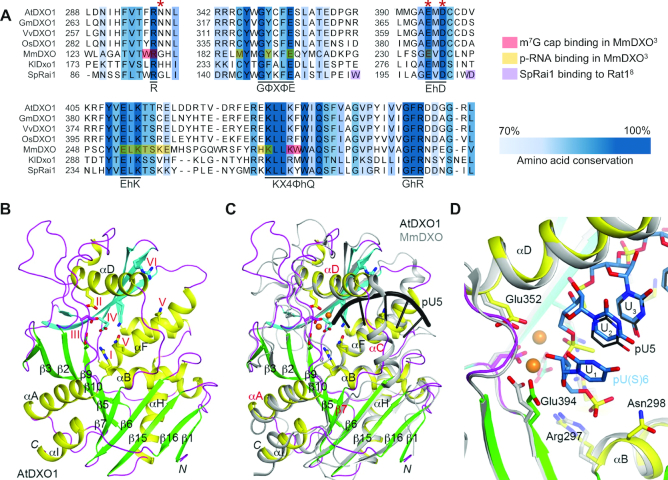
DXO1 (AT4G17620) was identified in BLAST analysis as the only DXO homolog in Arabidopsis. It shows 28% overall sequence identity with the previously studied mammalian DXO (MmDXO), but preserves high conservation within the active site, which is composed of six motifs: R residue (motif I), GΦXΦE (motif II, where Φ is an aromatic or hydrophobic residue and X any residue), EhD (motif III, where h is a hydrophobic residue), EhK (motif IV), KX4ΦhQ (motif V) and GhR (motif VI) (15) (Figure 1A and Supplementary Figure S1A). Amino acids that contact capped mRNAs and capping intermediates are generally conserved in plant homologs, but the region responsible for fungal Rai1 interaction with Rat1 is absent. Accordingly, DXO1 did not complement 5.8S and 25S rRNA maturation defects that result from the deletion of RAI1 in S. cerevisiae (Supplementary Figure S1B). A distinctive feature of plant DXO homologs is the N-terminal extension (NTE), estimated as 194 amino acids long in DXO1 from Arabidopsis. This segment is highly diversified among plants, but shows conservation in the proximity of the Ser119 residue, which may undergo phosphorylation (45) (Supplementary Figure S1A). We decided to remove the N-terminal extension for structural analysis, as it could disturb crystallization due to its high disorder probability (Supplementary Figure S1C).
Figure 1.
Sequence and structure of DXO1 (AT4G17620). (A) Fragment of T-Coffee sequence alignment of DXO1, GmDXO1 (soybean Glycine max), VvDXO1 (wine Vitis vinifera), OsDXO1 (rice Oryza sativa), MmDXO (mouse Mus musculus), SpRai1 (yeast Schizosaccharomyces pombe) and KlDxo1 (yeast Kluyveromyces lactis). Six motifs that form DXO active site are indicated; bold highlight, conserved residues (threshold 0.7); red asterisks, residues modified in this study; the legend below the alignment explains marking used for residues that contact m7G cap and 5′ monophosphate RNA in MmDXO and residues responsible for interaction of SpRai1 with Rat1. (B) Schematic representation of DXO1(ΔN194) crystal structure. Strands in the two β-sheets are colored green and cyan, respectively. Residues in the six conserved motifs are labeled and shown as stick models. (C) Overlay of DXO1(ΔN194) (in color) and mouse DXO (gray) structures in complex with the pU5 oligonucleotide (black). Relevant secondary structure elements are labeled in red. (D) Overlay of DXO1(ΔN194) (in color) and mouse DXO (gray) active sites in complex with the pU5 (black, thin lines) or the pU(S)6 (light blue, sticks) oligonucleotide. See Supplementary Figure S1 for additional information.
Crystal structure obtained for DXO1(ΔN194) free protein at 1.8 Å resolution showed good agreement with the X-ray diffraction data, expected bond lengths and angles, as well as other geometric parameters (Supplementary Table S1). The overall DXO1(ΔN194) structure is similar to mouse MmDXO, as both proteins contain two mixed-β-sheets that are surrounded by α-helices (Figure 1B). Substantial differences were, however, observed within one of the walls positioned close to the pU5 oligoribonucleotide in the active site pocket of the MmDXO structure (3) (Figure 1C). In MmDXO this region is formed by αC, αD helices and their connecting loop, but in DXO1 the αC helix is absent. Instead, there is a long loop that links the β7 strand and αD helix. Residues in this segment are also poorly conserved between the plant DXO1 and mouse MmDXO (Supplementary Figure S1A). In addition, the αA helix in DXO1 (with four turns) is longer than that in MmDXO (three turns); and the loop connecting αA and β5 is shorter in DXO1 than that in MmDXO. In the active site region, the six conserved motifs have essentially the same conformation in DXO1 and MmDXO. We did not observe any metal ions in the structure, although we expect DXO1 to be able to bind metal ions in the active site. We noticed that DXO1 and homologs from other plant species contain an asparagine residue that directly follows motif I in the αB helix (Asn298 in Arabidopsis), which is an expected binding site for the 5′ segment of the RNA substrate (Figure 1D). The majority of mammalian and fungal homologs contain glycine in the corresponding position (2) (Figure 1A and Supplementary Figure S1A), but modification to glutamic acid in this position is present in Candida albicans Rai1 (CaRai1), resulting in reduced catalytic activity (15). It is possible that the presence of a large amino acid side chain in the immediate vicinity of the active site could interfere with substrate binding through steric repulsion, which could also be the case for plant DXO1.
Features present in the sequence and structure of DXO1, as well as in published data on the DXO family of proteins, prompted us to generate a set of DXO1 variants that were further used for in vitro and in vivo studies. This set included DXO1(ΔN194) with the deletion of the N-terminal extension, DXO1(N298G) with asparagine-to-glycine modification in the active site and DXO1(E394A/D396A) that we expected to be catalytically inactive based on earlier mutagenesis in yeast and mouse cells (1–4).
Plant-specific features negatively affect DXO1 biochemical activity in vitro
We set out to determine whether DXO1 shows enzymatic properties characteristic of its fungal and mammalian homologs (1–5). We studied the activity of different DXO1 variants toward in vitro generated short (30 nucleotides) and long (70–100 nucleotides) substrates that represented RNAs with canonical cap (5′ m7Gppp-RNA), capping intermediates (5′ ppp-RNA, 5′ Gppp-RNA), decapped RNAs (5′ p-RNA) and molecules containing non-canonical NAD+cap (5′ NppA-RNA) (Supplementary Table S2).
As predicted from the sequence and structure analysis, DXO1 had reduced 5′QC biochemical activities, which was due to the presence of Asn298 residue. DXO1(wt) showed no enzymatic activity toward capped RNAs and capping intermediates, but it retained a weak 5′-3′ exoribonuclease activity observed at increased 4 μM concentration (Figure 2A). This activity relied on the conserved DXO active site and was abolished in DXO1(E394A/D396A). Deletion of the N-terminal extension in DXO1(ΔN194) enhanced the activity toward 5′ p-RNA, slightly improved the removal of 5′ ppp-RNA and 5′ Gppp-RNA, but did not restore the activity toward 5′ m7Gppp-RNA. The most pronounced improvement was observed when the plant-specific asparagine residue was modified to glycine in DXO1(N298G), regardless of the presence of NTE. DXO1(ΔN194) and DXO1(N298G) variants showed properties of distributive exoribonucleases, as they were limited by the high guanosine content of the substrate and arrested degradation at the guanosine located 7-nt from the RNA 3′ end (Supplementary Figure S2A–C).
Figure 2.
Plant-specific features affect DXO1 biochemical activity in vitro. (A) Activity toward 30-nt ppp-RNA, Gppp-RNA, m7Gppp-RNA and p-RNA with 3′-end FAM fluorophore. Reaction products were resolved by urea-PAGE. (B) Activity toward in vitro transcribed 5′ triphosphate RNA radioactively labeled at gamma phosphate (marked with asterisk). Reaction products were resolved by TLC in 0.75 M KH2PO4 buffer and identified based on the migration for MmDXO (lanes 13–14) and markers (lanes 15–16). (C–E) Activities toward in vitro transcribed RNA substrates with unmethylated or methylated cap radioactively labeled at gamma phosphate (marked with asterisk). Reaction products were resolved by TLC in 0.45 M (NH4)2SO4 and identified based on the migration for MmDXO and Dcp2 (E). (F, G) Activity toward NAD+-capped RNA substrate labeled at 5′ end (marked with asterisk). Reaction products were resolved by TLC in 0.75 M LiCl (F) and 15% urea-PAGE (G). (B–F) Average activity [%] was calculated from three replicates. See Supplementary Figure S2 for additional information.
We also applied thin-layer chromatography (TLC) to increase the detection sensitivity and identify molecules that were released from the 5′ ends of the RNA substrates. As in exonuclease assays, DXO1(wt) was inactive toward 5′ triphosphate RNA (Figure 2B) and capped substrates (Figure 2C). DXO1(ΔN194) released both PPi and GTP from the γ-labeled 5′ triphosphate RNA, with a slight tendency toward pyrophosphate, while MmDXO produced only PPi, according to its reported PPH activity (3) (Figure 2B). N298G modification in the full-length and N-terminally truncated DXO1 strongly favored GTP production, which suggests that the architecture of the active site may affect the balance between PPH and TPH activities. DXO1 variants with enhanced activity preferentially acted on substrates with unmethylated caps rather than mature methylated caps (Figure 2D), while MmDXO was less selective (Figure 2E), as previously reported (3). Based on the identical migration of cleavage products released from capped RNAs by DXO1 and MmDXO, we infer that DXO1 variants with enhanced activity hydrolyze unmethylated or methylated caps together with the first transcribed nucleotide (GpppN or m7GpppN, respectively), as shown for MmDXO (3). This mode of decapping is characteristic of DXO proteins, and contrasts with the canonical decapping enzyme Dcp2 that cleaves between the α and β phosphates of the cap structure (14) (Figure 2E). However, taking into account that wild-type DXO1 has hardly any of these activities, we envisage that decapping by DXO1 in vivo is not biologically relevant.
Although DXO1(wt) 5′QC activities were inhibited by the plant-specific features, the wild-type enzyme retained strong activity toward RNA with non-canonical NAD+ cap (Figure 2F, G and Supplementary Figure S2D). DXO1(wt) was already active on this substrate at 10 nM concentration, while its weak 5′-3′ exoribonuclease properties were only revealed at increased 4 μM concentration (Figure 2A). Accordingly, a shift in migration corresponding to NppA hydrolysis was detected on polyacrylamide gel, but no further degradation was observed in the case of DXO1(wt) (Figure 2G). Since NTE and Asn298 had only moderate inhibitory effect on deNADding by DXO1, these plant-specific elements may function cooperatively to modulate specificity toward NAD+-capped RNAs instead of the canonical capping intermediates.
Altogether, our biochemical studies show that 5′-3′ exoribonuclease, PPH/TPH and decapping activities of DXO1 are greatly reduced by the Asn298 residue, and to a lower extent by NTE. However, provided that the inhibitory influence of the N-terminal region was alleviated in vivo, it is still possible that these activities may in some way contribute to 5′QC mechanisms in Arabidopsis. In turn, deNADding is a predominant activity of DXO1 in vitro and is only slightly affected by the plant-specific features of the protein.
Cooperation between Asn298 and NTE in the selective inhibition of DXO1 activity
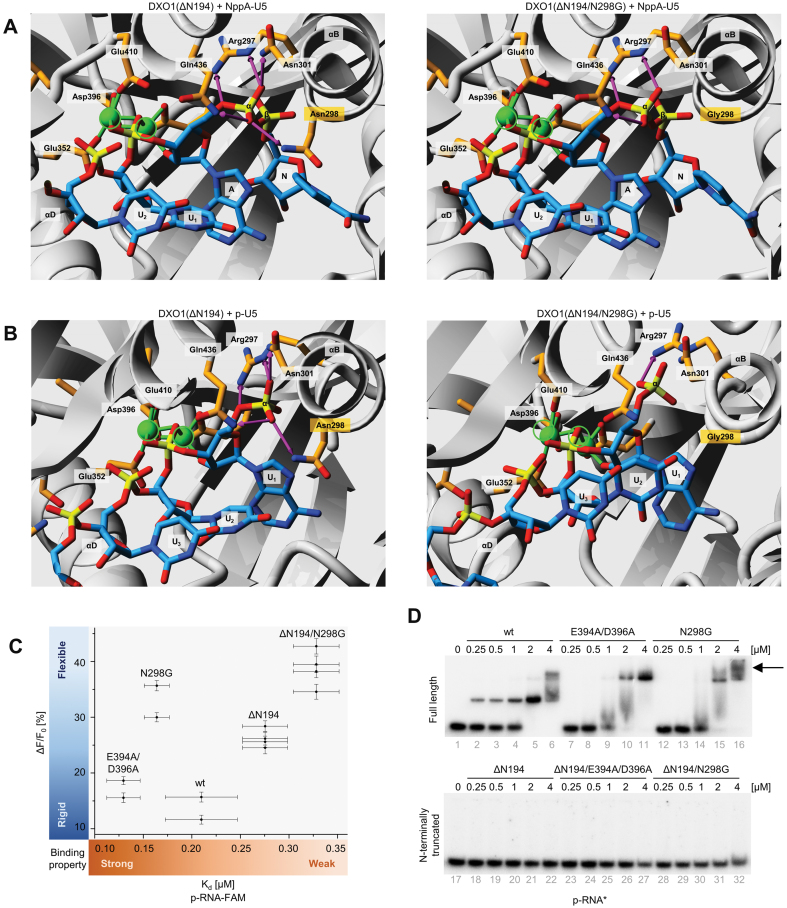
To understand how Asn298 selectively inhibits DXO1 5′QC activities with little impact on deNADding, we modeled structure of DXO1 with various RNA substrates. We observed that positioning of the NppA-RNA is almost unaltered upon modification of Asn298 to Gly298 (Figure 3A). This correlates well with efficient hydrolysis of the non-canonical NAD+ cap by both DXO1(wt) and DXO1(N298G). In contrast, phosphate moieties present in ppp- and m7Gppp-RNAs are strongly attracted by Arg297, Asn298, Asn301 and Gln436 that together bind the triphosphate and pull the substrate away from the two catalytic magnesium ions coordinated by the side-chain carboxyl groups of Glu352, Glu394, Asp396 and Glu410 (Supplementary Figure S3A–C). Here, Asn298 substitution to Gly298 substantially reduces the capacity for hydrogen bonding between phosphate moieties and functional amino acids. This increases the flexibility of interaction and alters the orientation of the scissile phosphate bond relative to magnesium ions, which results in RNA positioning that is optimal for catalysis.
Figure 3.
Catalytic site and NTE cooperate in RNA binding by DXO1. (A, B) Structural modeling of DXO1(ΔN194) and DXO1(ΔN194/N298G) with NAD+-capped (A) and 5′ monophosphate (B) pU5 substrates. RNA oligonucleotide and functional residues are labeled and shown as stick models. Magnesium ions are depicted as green circles and their positions as in DXO1(ΔN194) are marked in both structures. Hydrogen bonds are represented as pink arrows. N, nicotinamide; A, adenine. (C) Parameters of RNA binding determined with fluorescence polarization. Kd, binding affinity was fitted globally to several repetitions; dF/Fo, reflects the change in fluorescence upon RNA binding to protein relative to the fluorescence of free RNA. Values were fitted individually for each experiment. (D) Electrophoretic mobility shift assay (EMSA) of DXO1 variants and radioactively labeled 5′ monophosphate RNA. The arrow indicates additional complexes for DXO1(N298G). Variants and concentrations of recombinant proteins are indicated. Assays were repeated at least three times. See Supplementary Figure S3 for additional information.
Single phosphate in p-RNA also forms numerous hydrogen bonds with Arg297, Asn298, Asn301 and Gln436 that are diminished when Asn298 is modified to Gly298. Although location of p-RNA is driven the by a similar mechanism as ppp-RNA and m7Gppp-RNA, this interaction is presumably weaker than for ppp- and m7Gppp-RNAs, because single phosphate forms fewer hydrogen bonds with the surrounding acidic amino acid side chains (Figure 3B). Among all studied substrates, 5′ monophosphate RNA was also the most readily degraded upon deletion of the N-terminal extension (Figure 2A and Supplementary Figure S2C). As an intrinsically disordered fragment, NTE interferes with the structural approach, so we used fluorescence polarization (FP) to study its impact on binding to a short fluorescent 5′ p-RNA (Figure 3C). Signal from the fluorescent RNA (ΔF/F0 factor) correlated well with the strength of the exoribonucleolytic properties of each protein. N298G and ΔN194 DXO1 variants had increased ΔF/F0 signal, which may reflect higher flexibility or faster p-RNA dissociation. This is in line with the proposed model of the complex, in which N298G substitution causes weaker binding of the 5′-end phosphate/s due to the reduced number of formed hydrogen bonds. Reorientation of the substrate toward a more flexible position may increase active site accessibility and/or protein mobility along the RNA molecule. RNA binding was also weaker for N-terminally truncated variants, which was reflected by higher dissociation constants, suggesting that NTE stabilizes protein-RNA interaction.
We therefore performed electrophoretic mobility shift assays (EMSAs) to analyze the binding of the full-length and truncated DXO1 variants to radioactively labeled 5′ monophosphate RNA (Figure 3D), as well as capping intermediates and RNAs capped with the canonical m7G structure or non-canonical NAD+ cap (Supplementary Figure S3D). In these experiments, magnesium was substituted with calcium that does not support catalysis (4). As expected from the FP analysis, EMSA revealed an additional mode of RNA association through the N-terminal extension of DXO1. This interaction varied somehow for different 5′-end structures and DXO1 variants (Figure 3D and Supplementary Figure S3D), which suggests a cooperation between NTE and the catalytic site. In the case of DXO1(wt), NTE-dependent binding was weaker for p- and NppA-RNAs, molecules that can be degraded by this protein. We also noticed that DXO1(wt) and DXO1(N298G) differed in the pattern of higher order complexes, which may reflect substrate rearrangement within the active site that abolishes the inhibitory influence of NTE and leads to an increased activity of DXO1(N298G). It is possible that NTE-mediated stabilization of the RNA body counteracts its movement trough the active site, but is not sufficiently strong to inhibit 5′QC activities when the substrate adopts an optimal position upon N298G substitution. NTE deletion has a stimulating effect on the 5′-3′ exoribonuclease activity, but only weakly enhances 5′QC activities, possibly because binding between the p-RNA exoribonuclease substrate and the active site is already more flexible due to fewer hydrogen bonds than in the case of ppp-RNA and m7Gppp-RNAs. Interaction between the enzyme and substrates with three phosphate moieties is more fixed and therefore lack of NTE is not sufficient to relieve these constraints. These observations show that the catalytic site and NTE cooperate in selective inhibition of DXO1 activities. While the inhibitory effect of Asn298 is most likely immutable in vivo, the activity on p-RNA (and possibly also ppp-RNA, see Supplementary Figure S3B and C) may be regulated by the RNA stabilizing capacity of NTE, for example through post-translational modifications. Together, these data explain well a strong deNADding and weak 5′-3′ exoribonuclease activities of DXO1, with a possibility of NTE-mediated regulation of the latter.
Chloroplast-associated phenotype of DXO1 deficiency
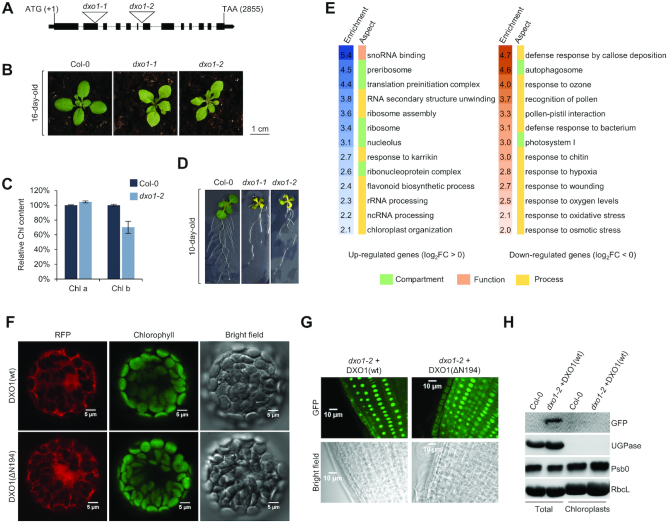
To assess in vivo effects of DXO1 deficiency, we obtained Arabidopsis dxo1-1 (SALK_103157) and dxo1-2 (SALK_032903) lines that contain T-DNA insertions in the second exon and the fifth intron of the AT4G17620 gene, respectively (Figure 4A). Both lines showed growth retardation, pale-green pigmentation (Figure 4B and Supplementary Figure S4A) that may result from reduced chlorophyll b content (Figure 4C), defects in lateral root formation (Figure 4D) and changes in the expression of several mRNAs (Supplementary Figure S4B). We chose dxo1-2 for further studies, as dxo1-1 exhibited increased transcription downstream of the T-DNA insertion that could potentially cause undefined phenotypic effects (Supplementary Figure S4C).
Figure 4.
Chloroplast-associated phenotype of DXO1 deficiency. (A) Structure of the DXO1 (AT4G17620) gene. Exons are represented with black bars and localization of T-DNA insertions are indicated. (B) 16-day-old wild-type Col-0, dxo1-1 and dxo1-2 plants. (C) Relative content of chlorophyll a (chl a) and chlorophyll b (chl b) in leaves of Col-0 and dxo1-2 plants. Results are the mean of three independent biological replicates with error bars representing standard deviation. (D) Roots of 10-day-old Col-0, dxo1-1 and dxo1-2 plants. (E) GO term analysis of protein-coding nuclear genes with affected expression (FDR < 0.05, |log2FC| > 0) in the dxo1-2 mutant based on RNA-seq data (only selected categories are shown, see complete GO analysis in Supplementary Dataset 2). (F–G) Nuclear-cytoplasmic localization of DXO1(wt) and DXO1(ΔN194) fused to GFP observed by confocal microscopy in transiently transformed Arabidopsis protoplasts (F) and root-tips of stably transformed plants (G). (H) Western blot analysis of total and chloroplast fraction of Col-0 and dxo1-2 stably transformed with DXO1(wt) fused to GFP. UGPase and PsbO were used as cytoplasm and chloroplast markers, respectively. RbcL, loading control.
High-throughput RNA sequencing revealed that 4847 genes were down-regulated and 4878 were up-regulated in dxo1-2 plants (DESeq2, |log2FC| > 0, FDR < 0.05), among which 993 and 1009 were strongly down-regulated and up-regulated, respectively (|log2FC| > 1) (Supplementary Figure S4D and Supplementary Dataset 1). Down-regulated genes (log2FC < 0) were often associated with the defense response, photorespiration and oxidative stress, while up-regulated genes (log2FC > 0) were repeatedly involved in ribosome biogenesis and RNA processing (Figure 4E and Supplementary Dataset 2). Among the up-regulated genes we noticed CBP20, eIF4E, SERRATE, LSM complex components and translation initiation factors. Most notably, we observed down-regulation of 61 chloroplast-encoded mRNAs and only one case of up-regulation. These results suggest that DXO1 could connect the expression of nuclear-encoded proteins to chloroplast-related processes.
Since dxo1-2 plants exhibit chloroplast-associated phenotypes, we examined the presence of DXO1 in this organelle. We used Arabidopsis protoplasts transiently transformed with RFP-tagged DXO1 and root tips of stable plant lines expressing GFP-tagged DXO1 (Figure 4F and G). Both approaches showed nuclear-cytoplasmic localization of DXO1 and no detectable fluorescent signal from the chloroplasts, regardless of the presence of the NTE. We also did not observe DXO1 in chloroplasts by western blotting of protein extract isolated from intact chloroplasts of plants expressing the full-length DXO-GFP (Figure 4H). Although we cannot exclude that DXO1 becomes translocated to chloroplasts in certain conditions, its default localization is predominantly nuclear and, to a lower extent, cytoplasmic. The absence of DXO1 in chloroplasts suggests that it may affect the function of this organelle through a metabolic coupling to the nucleus and/or other cellular compartments, as a component of anterograde (to chloroplasts) or retrograde (from chloroplasts) signaling.
RNA quality control siRNAs (rqc-siRNAs) accumulate in dxo1 plants
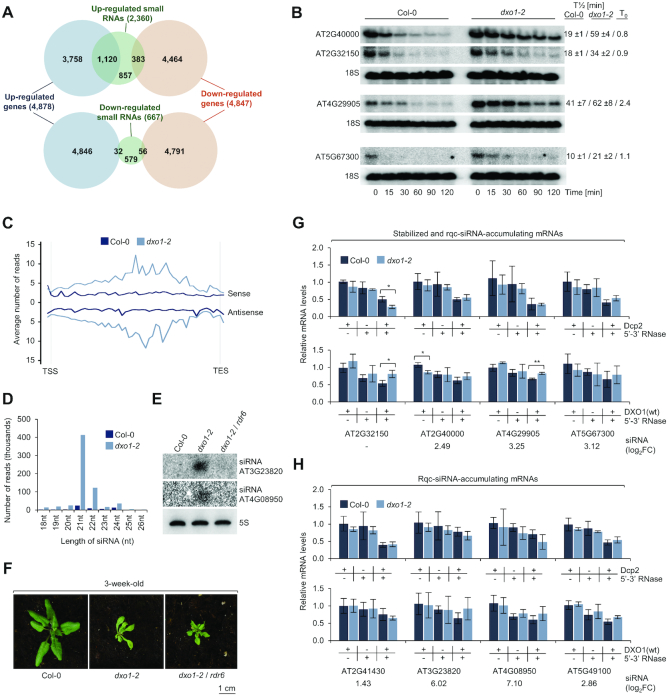
Based on biochemical activities of DXO1 and its homologs from other organisms, we expected that it may be involved in RNA degradation. In Arabidopsis, RNA degradation mutants exhibit increased biogenesis of RNA quality control small interfering RNAs (rqc-siRNA) derived from sense and antisense strands of mRNAs that would otherwise undergo rapid elimination (33–35,46,47). Sequencing of dxo1-2 small RNAs (sRNA-seq) revealed that generation of siRNAs was strongly increased for 2360 genes, while only 667 genes showed a strong siRNA decrease (DESeq2, |log2FC| > 1; FDR < 0.05) (Supplementary Dataset 3). Notably, 47.5% of the genes that were highly enriched in siRNAs also exhibited increased expression in the whole transcriptome sequencing data (1120 genes, 23% of all up-regulated genes), while only 16.2% siRNA-enriched genes were down-regulated (383 genes, 7.9% of all down-regulated genes) (Figure 5A). These siRNAs are likely to be derived from mRNAs that are not properly degraded in the absence of DXO1, as several of these mRNAs were also stabilized in the dxo1-2 mutant following transcription inhibition by cordycepin (Figure 5B and Supplementary Figure S5A).
Figure 5.
RNA quality control siRNAs (rqc-siRNAs) accumulate in dxo1 plants. (A) Overlap between genes with affected expression (FDR < 0.05, |log2FC| > 0) and genes with affected sRNA levels in dxo1-2 (FDR < 0.05, |log2FC| > 1). Numbers represent genes identified in RNA-seq and sRNA-seq experiments and overlapping genes. (B) Half-lives of selected mRNAs by northern blot after transcriptional inhibition by cordycepin. T1/2, mean values of transcript levels relative to untreated plants; T0, relative level of transcript in dxo1-2 versus Col-0 at zero time point; based on three biological replicates. 18S rRNA; loading control. (C) Small RNAs in dxo1-2 originate from both mRNA strands. Average read counts over protein-coding nuclear genes with siRNA up-regulation (FDR < 0.05, |log2FC| > 1, n = 2,288 loci). (D) Up-regulated siRNAs divided according to length. (E) Small RNA detection by northern blotting; 5S rRNA, loading control. (F) Three-week-old Col-0, dxo1-2 and dxo1-2/rdr6 plants. (G, H) Stabilized (G) and rqc-siRNA-enriched (H) genes do not show defects in mRNA deNADding and capping. Total RNA treated with indicated combinations of DXO1, Dcp2 and Terminator was analyzed by RT-qPCR. mRNA levels were normalized to Pol III-transcribed pre-tRNA-snoRNA (tsnoRNA) negative control and calculated relative to untreated Col-0 and dxo1-2. Results are the mean of three biological replicates, error bars represent standard deviation; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (t-test); siRNA accumulation from sRNA-seq (log2FC) is indicated below the graph. See Supplementary Figure S5 for additional information.
Up-regulated siRNAs were derived from both sense and antisense strands of mRNA precursors (Figure 5C) and were predominantly 21-nt and 22-nt long (Figure 5D and Supplementary Figure S5B). This suggests the involvement of RNA-dependent RNA polymerase 6 (RDR6) in their biogenesis, as reported for rqc-siRNAs in dcp2-1 and vcs-6 decapping mutants (33). Indeed, in the dxo1-2/rdr6 double mutant rqc-siRNA biogenesis was inhibited (Figure 5E), but in contrast to a reverted seedling lethality of dcp2-1/rdr6 and vcs-6/rdr6 plants, the dxo1-2/rdr6 mutant had similar morphology to dxo1-2 (Figure 5F). This observation suggests that rqc-siRNA accumulation is a secondary outcome of DXO1 knockdown and possibly represents a mechanism that serves as a protection from excessive mRNA accumulation. Rqc-siRNAs detected in dxo1-2 partially overlapped with those in dcp2-1 and vcs-6 mutants (Supplementary Figure S5C) despite different growth conditions and developmental stages (33). Similar repertoire of rqc-siRNA-accumulating mRNAs in dxo1-2 and decapping mutants implies that DXO1 indirectly affects canonical 5′-3′ RNA degradation.
Considering that DXO1 has a strong deNADding activity and its knockdown may lead to accumulation of NAD+-capped RNAs, we analyzed the 5′-end status of stabilized and rqc-siRNA-accumulating mRNAs using different combinations of 5′-3′ exoribonuclease (Terminator), Dcp2 and DXO1 on total RNA from Col-0 and dxo1-2 plants. RNase treatment leads to degradation of p-RNAs, Dcp2/RNase combination eliminates m7Gppp-RNAs, while DXO1/RNase, based on our in vitro studies, specifically removes NAD+-capped RNAs at a low DXO1 concentration. Using this approach we did not detect enrichment of either NAD+-capped, improperly capped or decapped mRNAs for any of the nine stabilized (Figure 5G and Supplementary Figure S5D) and other rqc-siRNA-accumulating mRNAs (Figure 5H and Supplementary Figure S5E), including AT3G23820 and AT4G08950 that were among the most highly-enriched in rqc-siRNAs. Based on this low-throughput selective degradation approach, we conclude that little if any changes in the 5′-end status cannot be accountable for mRNA stabilization and possible subsequent incorporation into siRNA pathways in the dxo1-2 mutant. We also note that Arabidopsis contains several NUDIX enzymes (22) that could act redundantly with DXO1 or even overcompensate its absence. This may be the case for AT2G32150 mRNA with a significant increase in the properly capped fraction accompanied with a reduction in NAD+-capped RNAs, which is a somehow unexpected phenotype that is against biochemical properties of DXO1. We also performed NAD capture (19) and analyzed selected mRNAs by RT-qPCR (Supplementary Figure S5F). Previously analyzed mRNAs were not captured efficiently enough to be detected with this approach, but other tested mRNAs were either unaltered or decreased in the NAD+-capped fraction, which again may reflect overcompensation by other deNADding enzymes. Alternatively, mRNA stabilization and rqc-siRNA accumulation in dxo1-2 is independent of biochemical activities of DXO1 and redundant enzymes. In this case, it could basically reflect the secondary outcome of the regulation of gene expression, especially that rqc-siRNAs in dxo1-2, in contrast to dcp2-1 and vcs-6 mutants, do not appear to impair plant development.
The N-terminal extension is crucial for DXO1 function in chloroplast-related processes
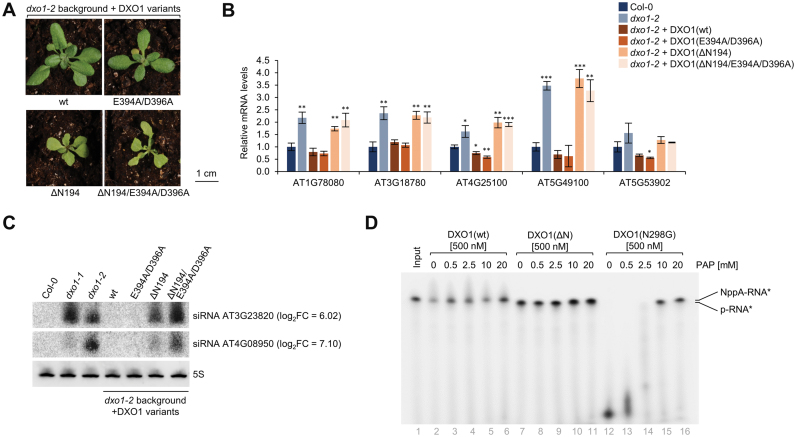
To assess the contribution of the catalytic site and plant-specific N-terminal extension to the function of DXO1 in vivo, we generated transgenic dxo1-2 lines expressing DXO1(wt), DXO1(E394A/D396A), DXO1(ΔN194) and DXO1(ΔN194/E394A/D396A) fused to GFP. Expression of DXO1(wt) in the dxo1-2 background (Supplementary Figure S6A) restored the wild-type morphology (Figure 6A), the levels of several mRNAs affected in dxo1-2 plants (Figure 6B) and abolished the accumulation of rqc-siRNAs (Figure 6C), confirming that these features are due to the loss of DXO1 function. Complementation also occurred upon the expression of the catalytically inactive DXO1(E394A/D396A) variant, but not in dxo1-2 lines expressing N-terminally truncated forms, namely catalytically active DXO1(ΔN194) and inactive DXO1(ΔN194/E394A/D396A), although DXO1(ΔN194) partially reduced the levels of rqc-siRNAs. This observation is in accordance with the interpretation that enzymatic action of DXO1 is either redundant with other enzymes or irrelevant to observed phenotypes. Although the cellular contribution of DXO1 biochemical activity remains elusive at this point, the outcome of the NTE deletion is hardly negligible, especially in the context of chloroplast-associated processes.
Figure 6.
The N-terminal extension is crucial for DXO1 function in chloroplast-related processes. (A) Two-week-old dxo1-2 transgenic plants. (B) RT-qPCR of selected mRNAs in dxo1-2 and dxo1-2 expressing DXO1 variants. Gene identifiers are indicated below the graph. Results are the mean of three independent biological replicates with error bars representing standard deviation; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (t-test). (C) Small RNA detection by northern blotting; siRNA accumulation from sRNA-seq (log2FC) is indicated; 5S rRNA, loading control. (D) Activity toward NAD+-capped RNA substrate in the presence of PAP. Reaction products after 30 min incubation were resolved by 8% urea-PAGE. See Supplementary Figure S6 for additional information.
We also noticed that dxo1-2 RNA-seq data showed a very high overlap with wild-type plants subjected to the high light acclimation treatments and with the stn7/psad1-1 mutant with defect in the redox-dependent retrograde signaling (48) (Supplementary Figure S6B and Supplementary Table S4). Another link between DXO1 and chloroplast function is provided by 3′-phosphoadenosine 5′-phosphate (PAP), an inhibitor of mammalian DXO and 5′-3′ exoribonucleases from the Xrn family (25,26). PAP functions as a chloroplast retrograde signal that modulates nuclear gene expression in response to high light and drought stress, possibly via an inhibition of 5′-3′ RNA processing and degradation (49,50). Using an in vitro assay we show that PAP inhibits both 5′-3′ exoribonuclease and deNADding activities of DXO1 (Figure 6D). It is therefore possible that DXO1 is controlled by a mechanism mediated by PAP regulators, including FRY1, a nuclear-encoded protein that is localized in chloroplasts and mitochondria, and dephosphorylates PAP (49,50). These observations open the possibility for the existence of an autoregulatory feedback loop, whereby DXO1 affects chloroplast retrograde signaling that in turn modulates its function.
Since we did not observe any direct contribution of the active site of DXO1 to RNA turnover and chloroplast-associated phenotypes, we conclude that enzymatic properties of DXO1 are not relevant to these processes. Instead, the N-terminal extension with its RNA-stabilizing capacity is indispensable for DXO1 cellular functions. However, strong deNADding and weak 5′-3′ exoribonuclease activities of DXO1 in vitro imply that it has additional functions in the nucleus and/or cytoplasm that may be redundant with other enzymes.
DISCUSSION
In this work, we show that Arabidopsis DXO1 has a distinct function from its fungal and mammalian homologs. Our biochemical analyses show that properties characteristic of mRNA 5′-end quality control (5′QC) (1–3), but not the removal of the non-canonical NAD+ cap (4,5), are adversely affected by the plant-specific asparagine residue (Asn298) that is located in the immediate vicinity of the DXO1 active site pocket. A glycine residue (Gly) is prevalent in this position in biochemically active DXO homologs, while a large asparagine side chain present in plant counterparts affects substrate binding and positioning within the active site. With the exception of the Gly298Asn mutation, the structure of the DXO1 catalytic domain is generally similar to mammalian MmDXO and the wild-type DXO1 retains a weak 5′-3′ exoribonuclease activity. Remarkably, Asn298 has almost no impact on DXO1 deNADding activity that is governed by the same active site. This can be explained by structural modeling of DXO1 with the NAD+-capped substrate, where modification from Asn298 to Gly298 generates only minor structural rearrangements. Another feature present exclusively in plant DXO homologs is the large and disordered N-terminal extension (NTE) that stabilizes the RNA body and probably restrains its movement within the active site. The N-terminally truncated DXO1 shows enhanced 5′-3′ exoribonuclease activity and moderate improvement in the hydrolysis of RNAs with 5′ triphosphate and unmethylated cap, so some extent of 5′QC activities is possible, if the inhibitory effect of NTE is alleviated in vivo.
Although biochemical properties of DXO1 are adversely affected by the plant-specific features, dxo1 plants show severe morphological and molecular phenotypes, including global gene expression changes that appear to link defense response, chloroplast functioning and RNA processing. These defects are reverted exclusively by the full-length protein regardless of the presence of the functional active site. We conclude that DXO1 may perform an auxiliary role in Arabidopsis cap quality control and deNADding, but its major function is not associated with these activities. Molecular phenotypes directly connected to DXO1 enzymatic properties may also be masked by potential redundancy with known plant XRNs, as well as NUDIX enzymes that were already reported to hydrolyze mRNA cap and free NADH molecule (22).
Plants lacking DXO1 also accumulate mRNAs that enter RNAi pathways, resulting in the production of RNA quality control siRNAs (rqc-siRNAs). The presence of rqc-siRNAs is characteristic of Arabidopsis decapping and RNA degradation mutants (33,35). In these plants, mRNAs that would otherwise be degraded become the source of small RNAs in the RDR6-dependent RNAi pathway (33,46,51). Rqc-siRNAs synthesis in dxo1-2 is indeed mediated by RDR6, as it is inhibited in the dxo1-2/rdr6 double mutant. However, rqc-siRNAs are not the cause of the severe dxo1-2 phenotype, in contrast to the small RNAs that accumulate in dcp2 and vcs decapping mutants and result in seedling lethality. This suggests a distinct origin of rqc-siRNAs in dxo1-2 that may arise as a secondary outcome of another DXO1 function. This notion is supported by the 5′-end status of rqc-siRNA-producing mRNAs that do not show significantly altered decapping, 5′QC and deNADding in dxo1-2 plants. There are however other RNA quality control mechanisms that enhance reporter silencing (31,34,35,51) and may cause stabilization of mRNAs that become the source of rqc-siRNAs upon DXO1 knockdown. In addition, the accumulation of some rqc-siRNAs may be an indirect effect of DXO1 disruption, as intron-less mRNAs are enriched in the rqc-siRNA-accumulating fraction (36.6% compared to 18.9% in the genome) and a lack of introns was shown to promote siRNA production (52).
Our data provide a strong evidence for the chloroplast-related function of DXO1. These include the pale-green pigmentation, reduced chlorophyll b content, functional categorization of dxo1-2 RNA-seq data and global down-regulation of chloroplast-encoded transcripts. A role of DXO1 in these processes depends on its N-terminal extension, with a minor if any involvement of the active site. Although our localization studies did not reveal a substantial amount of DXO1 in chloroplasts under standard conditions, this does not discount the possibility of DXO1 re-localization to chloroplasts when required. In turn, a predominant nuclear and cytoplasmic localization of DXO1 strongly endorses its functions in these compartments, perhaps by providing a metabolic coupling to chloroplasts. Additional activities of DXO1 are possibly related to 5′QC and/or deNADding, but they do not contribute to the severe dxo1 morphology.
Catalytic activities of DXO1 are inhibited by PAP, which is a component of the FRY1-dependent chloroplast retrograde signaling pathway and an inhibitor of mammalian DXO and 5′-3′ exoribonucleases from the Xrn family (25,26). Although the outcome of DXO1 inhibition by PAP in vivo is unknown, it provides another link between DXO1 and chloroplast function that in turn affects nuclear gene expression. FRY1 may to some extent control enzymatic capacity of DXO1 by PAP-mediated inhibition of its deNADding and 5′-3′ exoribonuclease activities, but our data show that these are rather not relevant for chloroplast-associated phenotypes of dxo1 plants. Alternatively, DXO1 may be a component of an additional, independent retrograde signaling pathway. An attractive possibility, which requires further verification, is the modulation of nuclear gene expression as a result of DXO1 involvement in retrograde chloroplast-to-nucleus signaling. In this case, dxo1 phenotypes not related to DXO1 enzymatic activity, including mRNA stabilization and rqc-siRNA accumulation, may be due to deregulation of nuclear-encoded transcripts in response to altered chloroplast functioning and retrograde signaling.
Altogether, we show that Arabidopsis DXO1 retains certain features of its mammalian and fungal counterparts, such as deNADding and weak 5′-3′ exoRNase activities. However, DXO1 has also divergently evolved to play its most striking plant-specific functions that are associated with chloroplasts and rely on the N-terminal extension that stabilizes RNA binding and endows plant DXO1 with unique enzymatic features. Our study adds another layer of diversity to the complex, but yet not fully explored DXO protein family in eukaryotic cells.
DATA AVAILABILITY
RNA-seq data were deposited in the Gene Expression Omnibus Database (https://www.ncbi.nlm.nih.gov/geo/) under accession codes GSE95473 (total RNA-seq for Col-0 plants) and GSE99600 (total RNA-seq for dxo1-2 and small RNA-seq for Col-0 and dxo1-2 plants). Coordinates and structure factors were deposited in the Protein Data Bank (https://www.rcsb.org/) under accession code PDB 6DKN. Molecular modeling data were deposited in the Model Archive with the DOI: 10.5452/ma-hnvfv (https://www.modelarchive.org/doi/10.5452/ma-hnvfv).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Megerditch Kiledjian (Rutgers University, USA) for yeast strains and helpful discussions; Neil Whalen, Stuart Myers and Howard Robinson for access to the X29A beamline at the NSLS; Marcin Ziemniak (University of Warsaw) for the human Dcp2 enzyme; Aleksander Chlebowski (IBB PAS, Warsaw) for the assistance with confocal microscopy; Radosław Mazur (University of Warsaw) for antibodies against PsbO and RbcL and advice on chloroplast isolation; Hervé Vaucheret (INRA Centre de Versailles-Grignon) for rdr6 plants.
Author Contributions: A.K. performed the experiments unless otherwise indicated. V.Y.-F.W. and L.T. carried out the structural analysis and interpretations. M.K. performed all bioinformatics analyses, NAD capture and contributed to RT-qPCR analyses. A.G. provided recombinant proteins and performed experiments with FAM-labeled RNA substrates. M.Z.P. performed cordycepin experiment and obtained dxo1-1 and dxo1-2 lines. K.S. participated in biochemical studies. J.P. performed fluorescence polarization analysis, structural modeling and data interpretation. A.K., M.K., L.T. and J.K. wrote the manuscript.
Notes
Present address: Vivien Ya-Fan Wang, Faculty of Health Sciences, University of Macau, Avenida da Universidade, Taipa, Macau SAR, China.
Present address: Michal Krzyszton, Institute of Biochemistry and Biophysics Polish Academy of Sciences, Pawinskiego 5a, 02-106 Warsaw, Poland.
Present address: Karolina Stepniak, Nencki Institute of Experimental Biology, Polish Academy of Sciences, Pasteur 3, 02-093 Warsaw, Poland.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Centre [UMO-2014/15/B/NZ2/02302, UMO-2013/08/M/NZ1/00931 to J.K.]; NIH grants [R35GM118093 and S10OD012018 to L.T.]; Experiments were carried out with the use of CePT infrastructure financed by the European Union – the European Regional Development Fund [Innovative economy 2007–2013, Agreement POIG.02.02.00-14-024/08-00]. Funding for open access charge: Narodowe Centrum Nauki [UMO-2014/15/B/NZ2/02302].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jiao X., Xiang S., Oh C., Martin C.E., Tong L., Kiledjian M.. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature. 2010; 467:608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang J.H., Jiao X., Chiba K., Oh C., Martin C.E., Kiledjian M., Tong L. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat. Struct. Mol. Biol. 2012; 19:1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiao X., Chang J.H., Kilic T., Tong L., Kiledjian M. A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell. 2013; 50:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jiao X., Doamekpor S.K., Bird J.G., Nickels B.E., Tong L., Hart R.P., Kiledjian M. 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell. 2017; 168:1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kiledjian M. Eukaryotic RNA 5′-end NAD+ capping and deNADding. Trends Cell Biol. 2018; 28:454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xue Y., Bai X., Lee I., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W.. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell Biol. 2000; 20:4006–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang F., Phillips S., Butler J.S. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005; 11:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiang S., Cooper-Morgan A., Jiao X., Kiledjian M., Manley J.L., Tong L. Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009; 458:784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramanathan A., Robb G.B., Chan S.H. mRNA capping: Biological functions and applications. Nucleic Acids Res. 2016; 44:7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunn S., Cowling V.H. Myc and mRNA capping. Biochim. Biophys. Acta. 2015; 1849:501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Otsuka Y., Kedersha N.L., Schoenberg D.R. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol. Cell Biol. 2009; 29:2155–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schoenberg D.R., Maquat L.E. Re-capping the message. Trends Biochem. Sci. 2009; 34:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mukherjee C., Patil D.P., Kennedy B.A., Bakthavachalu B., Bundschuh R., Schoenberg D.R. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2012; 2:674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grudzien-Nogalska E., Kiledjian M.. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip. Rev. RNA. 2007; 8:e1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang V.Y.F., Jiao X., Kiledjian M., Tong L. Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes. Nucleic Acids Res. 2015; 43:6596–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bird J.G., Zhang Y., Tian Y., Panova N., Barvik I., Greene L., Liu M., Buckley B., Krasný L., Lee J.K. et al.. The mechanism of RNA 5′capping with NAD+, NADH and desphospho-CoA. Nature. 2016; 535:444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Julius C., Yuzenkova Y.. Bacterial RNA polymerase caps RNA with various cofactors and cell wall precursors. Nucleic Acids Res. 2017; 45:8282–8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Julius C., Yuzenkova Y.. Noncanonical RNA-capping: discovery, mechanism, and physiological role debate. Wiley Interdiscip Rev. RNA. 2018; e1512. [DOI] [PubMed] [Google Scholar]
- 19. Cahová H., Winz M.L., Höfer K., Nübel G., Jäschke A.. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature. 2015; 519:374–377. [DOI] [PubMed] [Google Scholar]
- 20. Höfer K., Li S., Abele F., Frindert J., Schlotthauer J., Grawenhoff J., Du J., Patel D.J., Jäschke A.. Structure and function of the bacterial decapping enzyme NudC. Nat. Chem. Biol. 2016; 12:730–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mlynarska-Cieslak A., Depaix A., Grudzien-Nogalska E., Sikorski P.J., Warminski M., Kiledjian M., Jemielity J., Kowalska J. Nicotinamide-containing di- and trinucleotides as chemical tools for studies of NAD-capped RNAs. Org. Lett. 2018; 20:7650–7655. [DOI] [PubMed] [Google Scholar]
- 22. Yoshimura K., Shigeoka S. Versatile physiological functions of the Nudix hydrolase family in Arabidopsis. Biosci. Biotechnol. Biochem. 2015; 79:354–366. [DOI] [PubMed] [Google Scholar]
- 23. Picard-Jean F., Brand C., Tremblay-Létourneau M., Allaire A., Beaudoin M.C., Boudreault S., Duval C., Rainville-Sirois J., Robert F., Pelletier J. et al.. 2′-O-methylation of the mRNA cap protects RNAs from decapping and degradation by DXO. PLoS One. 2018; 13:e0193804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reinholt S.J., Ozer A., Lis J.T., Craighead H.G.. Highly multiplexed RNA aptamer selection using a microplate-based microcolumn device. Sci. Rep. 2016; 6:29771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yun J.S., Yoon J.H., Choi Y.J., Son Y.J., Kim S., Tong L., Chang J.H.. Molecular mechanism for the inhibition of DXO by adenosine 3′,5′-bisphosphate. Biochem. Biophys. Res. Commun. 2018; 504:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dichtl B., Stevens A., Tollervey D.. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 1997; 16:7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pane A., Jiang P., Zhao D.Y., Singh M., Schüpbach T.. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011; 30:4601–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohn F., Sienski G., Handler D., Brennecke J.. The Rhino-Deadlock-Cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014; 157:1364–1379. [DOI] [PubMed] [Google Scholar]
- 29. Chen Y.C.A., Stuwe E., Luo Y., Ninova M., Le Thomas A., Rozhavskaya E., Li S., Vempati S., Laver J.D., Patel D.J. et al.. Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol. Cell. 2016; 63:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andersen P.R., Tirian L., Vunjak M., Brennecke J.. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature. 2017; 549:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herr A.J., Molnàr A., Jones A., Baulcombe D.C.. Defective RNA processing enhances RNA silencing and influences flowering of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2009; 103:14994–15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gy I., Gasciolli V., Lauressergues D., Morel J.-B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C.. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell. 2007; 19:3451–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De Alba A.E.M., Moreno A.B., Gabriel M., Mallory A.C., Christ A., Bounon R., Balzergue S., Aubourg S., Gautheret D., Crespi M.D. et al.. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015; 43:2902–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang X., Zhu Y., Liu X., Hong X., Xu Y., Zhu P., Shen Y., Wu H., Ji Y., Wen X. et al.. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science. 2015; 348:120–123. [DOI] [PubMed] [Google Scholar]
- 35. Zhao L., Kunst L. SUPERKILLER complex components are required for the RNA exosome-mediated control of cuticular wax biosynthesis in Arabidopsis inflorescence stems. Plant Physiol. 2016; 171:960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsuzuki M., Motomura K., Kumakura N., Takeda A. Interconnections between mRNA degradation and RDR-dependent siRNA production in mRNA turnover in plants. J. Plant Res. 2017; 130:211–226. [DOI] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W.. [20] Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 38. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al.. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 40. Murshudov G.N., Vagin A.A., Dodson E.J.. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997; 53:240–255. [DOI] [PubMed] [Google Scholar]
- 41. Emsley P., Cowtan K.. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 42. Kruszka K., Barneche F., Guyot R., Ailhas J., Meneau I., Schiffer S., Marchfelder A., Echeverría M.. Plant dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 2013; 22:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. Tape-Arabidopsis sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009; 5:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kubis S.E., Lilley K.S., Jarvis P. Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol. Biol. 2008; 425:171–186. [DOI] [PubMed] [Google Scholar]
- 45. Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., Gruissem W., Baginsky S.. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009; 150:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thran M., Link K., Sonnewald U. The Arabidopsis DCP2 gene is required for proper mRNA turnover and prevents transgene silencing in Arabidopsis. Plant J. 2012; 72:368–377. [DOI] [PubMed] [Google Scholar]
- 47. Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X.. Detection of pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015; 25:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gläßer C., Haberer G., Finkemeier I., Pfannschmidt T., Kleine T., Leister D., Dietz K.J., Häusler R.E., Grimm B., Mayer K.F.X.. Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol. Plant. 2014; 7:1167–1190. [DOI] [PubMed] [Google Scholar]
- 49. Estavillo G.M., Crisp P.A., Pornsiriwong W., Wirtz M., Collinge D., Carrie C., Giraud E., Whelan J., David P., Javot H. et al.. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011; 23:3992–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bruggeman Q., Mazubert C., Prunier F., Lugan R., Chan K.X., Phua S.Y., Pogson B.J., Krieger-Liszkay A., Delarue M., Benhamed M. et al.. Chloroplasts activity and PAP-signaling regulate programmed cell death in Arabidopsis. Plant Physiol. 2016; 170:1745–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ye J., Yang J., Sun Y., Zhao P., Gao S., Jung C., Qu J., Fang R., Chua N.H.. Geminivirus activates asymmetric levels 2 to accelerate cytoplasmic DCP2-mediated mRNA turnover and weakens RNA silencing in Arabidopsis. PLoS Pathog. 2015; 11:e1005196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Christie M., Croft L.J., Carroll B.J.. Intron splicing suppresses RNA silencing in Arabidopsis. Plant J. 2011; 68:159–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data were deposited in the Gene Expression Omnibus Database (https://www.ncbi.nlm.nih.gov/geo/) under accession codes GSE95473 (total RNA-seq for Col-0 plants) and GSE99600 (total RNA-seq for dxo1-2 and small RNA-seq for Col-0 and dxo1-2 plants). Coordinates and structure factors were deposited in the Protein Data Bank (https://www.rcsb.org/) under accession code PDB 6DKN. Molecular modeling data were deposited in the Model Archive with the DOI: 10.5452/ma-hnvfv (https://www.modelarchive.org/doi/10.5452/ma-hnvfv).