Abstract
The HIV-1 protein Rev is essential for virus replication and ensures the expression of partially spliced and unspliced transcripts. We identified a ULM (UHM ligand motif) motif in the Arginine-Rich Motif (ARM) of the Rev protein. ULMs (UHM ligand motif) mediate protein interactions during spliceosome assembly by binding to UHM (U2AF homology motifs) domains. Using NMR, biophysical methods and crystallography we show that the Rev ULM binds to the UHMs of U2AF65 and SPF45. The highly conserved Trp45 in the Rev ULM is crucial for UHM binding in vitro, for Rev co-precipitation with U2AF65 in human cells and for proper processing of HIV transcripts. Thus, Rev-ULM interactions with UHM splicing factors contribute to the regulation of HIV-1 transcript processing, also at the splicing level. The Rev ULM is an example of viral mimicry of host short linear motifs that enables the virus to interfere with the host molecular machinery.
INTRODUCTION
Eukaryotic cells use the process of splicing to excise introns from precursor messenger RNAs (pre-mRNAs) and ligate exons in order to create mature mRNAs. The completion of splicing is a prerequisite for mRNA export and successful translation in the cytoplasm (1). Splicing contributes to proteome diversity through alternative splicing, a process where alternative joining of 5′ and 3′ splice sites allows for exon extension, shortening or skipping, as well as intron inclusion (2–4). The regulation of constitutive and alternative splicing relies on a complex network of weak interactions involving cis-regulatory elements in the pre-mRNA and trans-acting splicing factors, thus allowing regulatory triggers to direct spliceosome assembly at splice sites depending on cellular needs (5–7).
RNA processing relies on both protein-RNA and protein–protein interactions. The RNA recognition motif (RRM) is the most abundant RNA-binding domain, and often present in splicing factors. The RRM fold is composed of two α-helices and four antiparallel β-strands (βαββαβ). RNA binding takes place on the β-strand face of the domain via ribonucleoprotein motifs (RNP) 1 and 2 (8). Interestingly, atypical RRM domains can also mediate protein–protein interactions. This type of domain was first identified in the heterodimeric U2 snRNP auxiliary factor (U2AF) and was therefore called U2AF homology motif (UHM). In contrast to canonical RRM domains, UHM have degenerate RNP1 and RNP2 motifs, harbor an Arg-Xxx-Phe (RXF) sequence between helix α2 and strand β4 and acidic residues in helix α1. Binding of the protein ligand occurs on the α–helical side of the domain (9–12).
Biochemical, functional and structural studies have identified basic, tryptophan-containing linear peptide motifs, called UHM Ligand Motifs (ULM) that are specifically recognized by UHM domains. ULM motifs are characterized by a conserved tryptophan residue that is preceded by a stretch of positively charged residues (Arg/Lys) and followed by a negatively charged amino acid (Asp/Glu). So far, structural biology and functional analysis of ULMs in U2AF65, SF1, SF3b155 and Ataxin-1 have been reported (9,10,12–16). UHM–ULM interactions are involved in constitutive splicing (U2AF35-U2AF65, U2AF65-SF1, U2AF65-SF3b155) and also play important roles in alternative splicing regulation (e.g. SPF45-SF1, SPF45- SF3b155) (Figure 1A). Recently, it has been shown that ULM-derived cyclic peptides can be used to inhibit UHM domains and thereby modulate splicing by stalling spliceosome assembly at early stages (17).
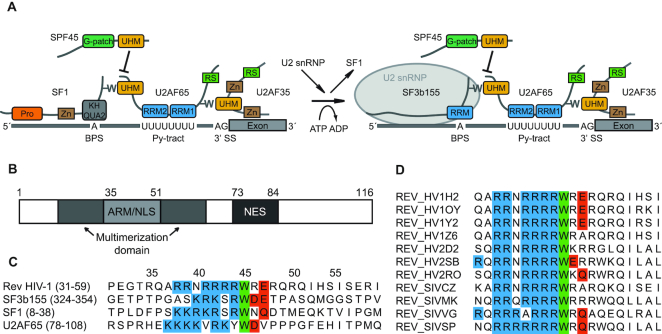
Figure 1.
HIV-1 Rev protein has a conserved ULM resembling motif. (A). Schematic representation of ULM–UHM interactions required for constitutive and alternative splicing. Pro – proline-rich region; Zn – zinc-finger domain; BPS – branch point sequence; Py – polypyrimidine; RS – arginine–serine-rich domain. (B). Domain organization of full-length Rev. (C). Alignment of ULM sequences of HIV-1 Rev, SF3b155 (ULM5), SF1 and U2AF65. Conserved residues are marked: blue – basic residues preceding the conserved tryptophan; green – conserved tryptophan; red – acidic and N/Q-type residues following the conserved tryptophan. (D). Alignment of the ULM motif in selected HIV-1, HIV-2 and SIV strains. With the exception of single HIV-2 and SIV strains, the tryptophan is strictly conserved in all aligned isolates (a total of 2000 HIV-1, HIV-2 and SIV sequences was used for the alignment).
Splicing of viral transcripts is a crucial step for human immunodeficiency virus type 1 (HIV-1) replication. HIV-1 is a lentivirus implicated in the pathogenesis of AIDS. During its replication cycle, the virus makes use of the host splicing machinery to produce >30 different spliced and partially spliced mRNAs. This process depends on a combination of suboptimal splice sites and intronic and exonic regulatory sequences. Viral gene expression begins with the production of fully spliced mRNAs encoding Rev, Tat and Nef. The splicing pattern is thereafter shifted towards production of partially and unspliced mRNAs. Partially spliced transcripts encode for Env, Vif, Vpr and Vpu proteins, while unspliced mRNAs encode for Gag and Pol and serve as viral genomes (18–20).
The shift from early to late stage of gene expression is attributed to the Rev protein (regulator of expression of viral proteins). Rev is a small 116 amino acid protein that is absolutely essential for HIV-1 replication. It has an arginine-rich motif (ARM) serving as an RNA binding domain and nuclear localization signal (NLS). The ARM is flanked by residues required for oligomerization and the Rev C-terminus harbors a leucine-rich effector domain containing the nuclear export signal (NES) (Figure 1B). Rev recognizes RNA sequences within the Rev Response Element (RRE) localized in the second (tat/rev) intron. The RNA interaction is initialized by Rev binding to stem-loop IIB (SLIIB); subsequently Rev forms multimers on the entire RRE, involving both Rev–RNA and Rev–Rev interactions (21–26).
An important function of Rev is to enable the export of partially spliced and unspliced viral mRNAs to the cytoplasm. Without Rev those mRNAs species are accumulated and degraded in the nucleus (27). Moreover, Rev has been shown to stabilize RRE-containing RNAs and to be involved in viral mRNA splicing, translation and packaging of viral particles (28–33). An inhibitory effect on splicing has been one of Rev's earliest assigned functions (34). It was established that Rev interferes with the spliceosome by inhibiting tri-snRNP binding (35). Rev was further reported to interact with a number of splicing-related proteins: the ASF/SF2-associated protein p32, DEAD/H proteins and hnRNPs (36–38). Nonetheless, mechanistic details of how Rev interferes with splicing remain elusive. Structural studies in recent years have provided insight into structural features involving Rev RNA binding and oligomerization. However, a molecular rationale for interactions and modulation of splicing factors is elusive.
Here, we report that Rev has a functional ULM that is able to interact with UHMs of host splicing factors. Our high-resolution crystal structure, NMR data and biochemical results show that, similarly to other UHM–ULM interactions, Rev ULM recognition by UHMs depends on tryptophan 45, a fully conserved residue across all HIV strains. Our results suggest that this interaction is involved in the processing of HIV-1 transcripts by contributing to splicing inhibition during the transition from early to late stage of viral gene expression.
MATERIALS AND METHODS
Protein and peptide sample preparation
Recombinant SPF45 UHM (301–401) and U2AF65 UHM (370–475) were expressed from modified pET-24d vectors with tobacco etch virus protease (TEV) cleavable tags: pET Z2-1a (N-terminal His6 and Z2 tag) and pETM11 (N-terminal His tag), respectively (http://www.helmholtz-muenchen.de/en/pepf/materials/vector-database/bacterial-expression-vectors/index.html). Unlabelled proteins were expressed in Escherichia coli BL21(DE3) in LB medium. The proteins were purified with Ni-NTA agarose (Qiagen) under standard conditions. Tags were cleaved with TEV protease and removed with a second Ni-NTA purification step. Subsequently, the proteins were purified by size-exclusion chromatography on a SuperdexTM 75 16/60 prep grade column. 15N-labelled proteins were expressed in minimal (M9) medium supplemented with 15NH4Cl.
Synthetic Rev peptides 38–50, 38–51, 41–49 and W45A 38–51 were purchased from Peptide Specialty Laboratory, Heidelberg, and used without further purification.
NMR titration experiments
For NMR, protein samples were concentrated to 250 μM in 20 mM sodium phosphate buffer (pH 6.8), 150 mM NaCl and 5 mM β-mercaptoethanol. UHM domains were titrated with increasing amounts of Rev peptide 38–50. 1H,15N-HSQC spectra were collected for the free UHMs and for the following UHM:Rev molar ratios: 1:0.3; 1:0.6; 1:1.0; 1:1.3; 1:1.6; 1:2.0. NMR spectra were recorded at 300 K on a Bruker DRX500 spectrometer, processed with NMRPipe (39) and analyzed with CCPN analysis software.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) measurements were carried out at 25°C using a MicroCal™ ITC200 calorimeter (GE Healthcare). Before calorimetry, both interaction partners were dialyzed against 50 mM sodium phosphate buffer (pH 7.0), 150 mM NaCl and 2 mM β-mercaptoethanol. Rev peptide 38–51 or Rev peptide W45A 38–51 at a concentration of 500–900 μM were injected into the cell containing SPF45 UHM 301–401 or U2AF65 UHM 370–475 at a concentration of 50–90 μM. After correction for heat of dilution, the data were fitted to a one-site binding model using the Microcal Origin 7.0 software. ITC experiments were repeated 2–5 times for each combination of peptide and UHM domain.
Crystallization, data collection, structure determination and refinement
For crystallization, SPF45-UHM in 20 mM Tris (pH 6.7), 150 mM NaCl and 5 mM DTT at 24 mg ml–1 (2 mM) was mixed at 1:10 molar ratio with Rev peptide (41–49). Crystals of the complex were grown at room temperature by vapor diffusion in sitting drops composed of equal volumes (100 nl each) of protein solution and crystallization buffer (22.5% (w/v) PEG 3350, 0.1 M HEPES pH 6.0, 0.2 M sodium acetate). They were cryoprotected by serial transfer into reservoir solution containing 20% (v/v) glycerol. Cryogenic data at 1.2-A° resolution were recorded at beamline ID14-1 of the European Synchrotron Radiation Facility (for data collection details see Table 1). The structure of SPF45 in complex with Rev (41–49) was determined with PHASER using the refined model of free SPF45-UHM. The solution comprises two SPF45 UHM molecules in the asymmetric unit and shows clear difference density for one peptide moiety. The structure was refined in alternating cycles of model correction using COOT and REFMAC5 refinement. Structural quality was checked with PROCHECK. Structural visualization was done with PyMOL (http://pymol.sourceforge.net/). In the final model, 94,7%, 5,3%, 0% and 0% of residues are in the most-favored, favored, generously-allowed and disallowed regions of the Ramachandran plot, respectively (for structure statistics see Table 1).
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P21 21 21 |
| Cell dimensions | |
| a, b, c (Å) | 48.21 63.68 67.63 |
| α, β, γ (°) | 90 90 90 |
| Resolution (Å) | 50–1.20 (1.27–1.20) |
| R merge (%) | 12.9 (58.9) |
| I / σI | 8.18 (2.89) |
| Completeness (%) | 98.5 (91.3) |
| Redundancy | 7.82 (3.98) |
| Observed reflexions | 507 235 (36 921) |
| Unique reflexions | 64 821 (9278) |
| Refinement | |
| Resolution (Å) | 18.49–1.20 |
| No. reflections | 60832 |
| R work/Rfree | 0.1489 / 0.1750 |
| No. atoms | |
| Protein | 1743 |
| Ligand/ion | 119 |
| Water | 323 |
| B-factors (Å) | |
| Protein | 9.003 |
| Ligand | 9.253 |
| Water | 28.287 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.015 |
| Bond angles (°) | 1.800 |
| Ramachandran plot (%) | |
| Most favored | 94.7 |
| Allowed | 5.3 |
| Outlier | 0.0 |
Cell culture
HeLa cells (obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen) were grown and propagated in low glucose (1 g/l) Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (P/S, Gibco) and 10% fetal calf serum (FCS, Gibco), at 37°C and 5% CO2.
Plasmids and mutagenesis
pDM128 (RRE CAT reporter based on pSV9B (49)), pRSV Rev (wt)-FLAG, pRSV Rev (W45A)-FLAG, pRLSV40 (Renilla luciferase reporter) and pBSIISK+ (as plasmid DNA for dilution) were a kind gift of Thomas Guettler (Harvard Medical School). pCsrevsg143 expressing GFP-tagged Rev and GFP-expressing pFred143 are described in (40). W45A mutation in pCsrevsg143 was introduced using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer's protocol.
Transfection with plasmid DNA
HeLa cells were transfected with plasmids of choice using FuGENE HD (Roche), following the manufacturer's guidelines. In short, plasmid DNA and FuGENE were diluted in serum-free medium and incubated for 15 min at room temperature. The FuGENE:DNA complexes were added drop-wise to 50–60% confluent cells plated 24 h prior to transfection. The analysis was performed 48 h after transfection. The amounts of FuGENE/plasmid DNA used in transfection were: 1.65 μl/0.6 μg for 12-well plate and 30 μl/12 μg for 14 cm plate. For RNA analysis, cells were transfected with pRSV Rev (wt)-FLAG/pRSV Rev (W45A)-FLAG/pRSV-FLAG + pDM128 + pRLSV40 (Renilla luciferase expression plasmid used to control for transfection efficiency) + pBSIISK+ (as plasmid DNA for dilution). For Co-IP, cells were transfected with pCsrevsg143-WT/pCsrevsg143-W45A (GFP-tagged Rev)/pFred143 + pBSIISK+ (as plasmid DNA for dilution). HeLafB cells (48) were seeded in 6-well plates at a density of 2 × 105 cells per well prior transfection and cultured 48 h after transfection. Transfection of 200 ng Rev WT or REV W45A was performed with FuGENE HD Transfection Reagent (Roche Diagnostic).
RNA isolation and RT-qPCR (pDM128-transfected HeLa cells)
Total RNA was isolated using NucleoSpin® RNA Kit (MN), according to manufacturer's instructions. Removal of potential DNA contamination was conducted using DNA-free™ DNA Removal Kit (Life Technologies). Isolated RNA was reverse transcribed using the SuperScript III kit (Invitrogen) with an oligo(dT)18 reverse primer. The cDNA was subjected to quantitative real-time PCR (Q-PCR). Q-PCRs were conducted using Absolute qPCR SYBR Green mix (Thermo Scientific) and Stratagene MX3005P instrument. Primer concentration ranged between 30 and 600 nM based on prior optimization (aimed at obtaining primer combinations amplifying the DNA template with and efficiency of 100 ± 3%). The PCR program was the following: 15 min at 95°C (enzyme activation); 35 cycles of 30 s at 95°C, 1 min at 55°C and 30 s at 72°C. To calculate fold changes in mRNA levels between control and experimental samples, the following equation was used: 2ΔCt(target)/2ΔCt(norm), where ΔCt = Ct (control sample) – Ct (experimental sample). Ct stands for threshold cycle value, target – gene of interest and norm – normalizer mRNA. ‘No reverse transcriptase’ RNA samples were always included to control for potential DNA contamination.
Quantification of HIV-1 transcripts by RT-PCR (HeLafB cells)
RT-PCR was performed on cDNA generated from DNA-free RNA samples by reverse transcription using GoTaq® Green Master Mix (Promega). RNA was transcribed with Superscript II (Invitrogen) (1 U/mg RNA) according to the manufacturer's protocol, using random hexamers, with a final RNase H digestion step. The following primers were used for the amplification of spliced, single spliced and unspliced HIV-1 transcripts:
BSS-S: 5′-GGCTTGCTGAAGCGCGCACGGCAAGAG-3′;
GAG-A: 5′-AACTGCGAATCGTTCTAGCTCCCT-3′;
KPN-A: 5′-AGAGTGGTGGTTGCTTCCTTCCACACAG-3′
SJ4.7-A: 5′-TTGGGAGGTGGGTTGCTTTGATAGAG-3′.
Quantification of HIV-1 production
Gag production was quantified by HIV-1-p24-Antigen-ELISA as described in (41). Briefly, HeLafB cells were seeded in 6-well plates at a density of 2 × 105 cells per well. The next day, cells were transfected with 200 ng Rev WT or REV W45A using FuGENE HD according to the manufacturer's protocol. 48 hours after transfection supernatant was collected and quantification of Gag p24 was performed using the Coulter HIV-1-p24-Antigen-ELISA assay (Beckman Coulter) according to the manufacturer's protocol.
Protein extraction and Western blot analysis
HeLa cells were washed with PBS and lysed in RIPA buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP40, 0.5% Na deoxycholate, 5 mM EDTA, 0.05% SDS) supplemented with complete protease inhibitor cocktail (Roche) for 10 min on ice. The lysates were centrifuged for 10 min at 20 000g at 4°C. The supernatants were collected and the protein concentration was determined using Bradford assay. For western blot analysis, 10–40 μg of total protein extract was resolved on a SDS-PAGE gel. The following primary antibodies at 1:1000 dilutions were used: mouse mAb α-U2AF (MC3, kind gift of Karla Neugebauer, MPI-CBG), rabbit pAb α-GFP (Cell Signaling), rabbit pAb α-SPF45 (kind gift of Juan Valcarcel, CRG). As secondary antibodies HRP-conjugated α-rabbit and α-mouse IgGs (Cell Signaling) were used at 1:2000 dilutions.
Co-immunoprecipitation (Co-IP)
HeLa cells were washed with PBS, scraped, pelleted and lysed for 10 min on ice in NET-2 buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.05% Nonidet P-40) supplemented with complete protease inhibitor cocktail (Roche). The lysate was sonicated, centrifuged and treated with 100 μg/ml RNase A (Serva) for 30 min at 37°C. Subsequently, the lysates were incubated at 4°C on a rotary shaker with 10 μg of goat anti-GFP antibodies (kind gift of Karla Neugebauer, MPI-CBG) coupled to Dynabeads® Protein G (Life Technologies). The eluted material was analyzed by western blot as described.
RESULTS
A ULM motif in HIV-1 Rev binds to UHM domains of eukaryotic splicing factors in vitro
Since viral proteins commonly hijack cell systems using short linear motifs (42–44), we surveyed HIV protein sequences for novel candidates, using the linear motif patterns annotated in the ELM resource (45). The search identified a putative ULM sequence in the HIV-1 Rev protein (Figure 1C). The matching sequence is contained within the so-called ‘basic peptide’ in the Rev RNA binding ARM region. However, the tryptophan residue (Trp45) that is anticipated to be crucial for the UHM–ULM interaction does not contribute to the interaction with the SLIIB RNA, which is the principal Rev-binding site in the RRE (46,47), as RNA binding by the Rev ARM region (residues 33–55) or full-length Rev is not impaired by a W45A mutation (Supplementary Figure S1A and B, respectively). Interestingly, Trp45 is strictly conserved across all HIV-1 and HIV-2 and the vast majority of simian immunodeficiency virus (SIV) species (Figure 1D). These observations indicate that the conserved tryptophan might be important for a functional Rev ULM and prompted us to investigate whether host UHM proteins could bind to the viral Rev ULM and thereby modulate HIV-1 gene expression.
In order to characterize the potential binding between the Rev ULM and UHM proteins, we performed NMR titrations using 1H,15N correlation experiments. Increasing amounts of Rev ULM peptides were added to 15N-labeled UHM domains of splicing factors involved in constitutive (U2AF65) and alternative (SPF45) splicing (Figure 2A). Significant chemical shift perturbations (CSPs) confirm a specific interaction with both UHMs. When the Rev-induced CSPs are compared to those induced upon binding of SF1 and SF3b155 ULMs to U2AF65 and SPF45 UHMs, respectively, similar regions in the UHM domains are affected (Supplementary Figure S2B and C). As seen for previously reported UHM–ULM interactions (U2AF65-SF1 and SPF45-SF3b155), the Rev binding site maps to the dorsal helical surface of the UHMs, where the largest CSPs cluster in the major ULM binding pocket composed of the RXF motif and negatively charged residues from helix α1 (Figure 2B,C) (10,12). Thus, Rev utilizes overall the same conserved binding surface in binding to the U2AF65 and SPF45 UHM domains.
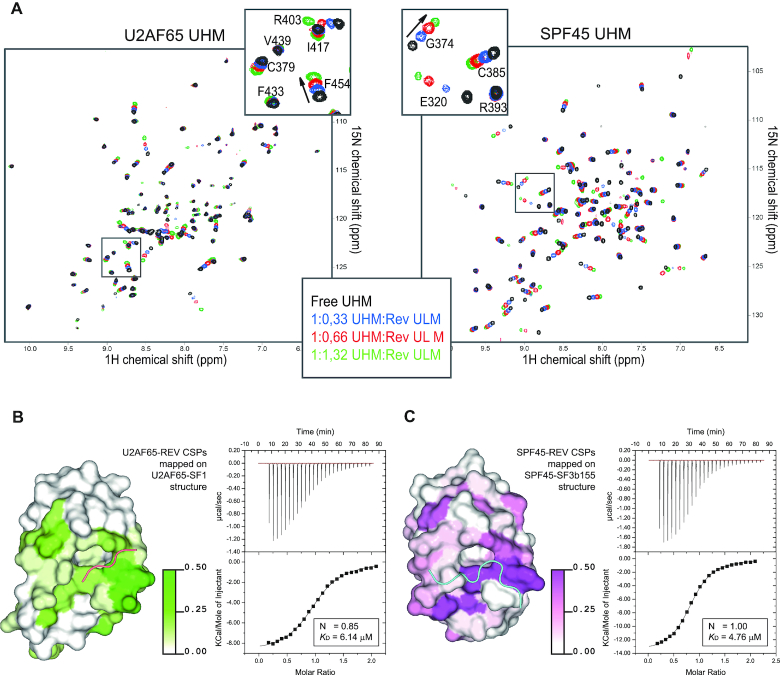
Figure 2.
Rev ULM binds U2AF65 and SPF45 UHMs. (A) 1H,15N-HSQC spectra of free 250 μM UHMs (black) titrated with increasing amounts of Rev ULM 38–50 (blue 1:0.3; red 1:0.6; green 1:1.3). Arrows in the close-up views indicate the direction of chemical shift perturbations. (B and C, left panels) Surface representation of U2AF65 and SPF45 UHMs in complex with SF1 and SF3b155 ULMs (PDB: 4FXW and 2PEH), respectively. The chemical shift perturbations induced by Rev ULM are mapped on the surface of the structures. Color gradients (white to green for U2AF65 and white to magenta for SPF45) indicate the extent of chemical shift perturbations. (B and C) right panels. Isothermal titration calorimetry of U2AF65/SPF45 UHMs and Rev ULM (38–51). 900 μM Rev peptide was titrated to 90 μM UHMs. The graphs are representative of five independent ITC measurements. Control titration of Rev peptide to the buffer was subtracted from experimental runs before KD calculations.
The interaction between the Rev ULM and host UHM domains was further characterized by isothermal titration calorimetry (ITC). The dissociation constant (KD) of the interaction is found to be in the low micromolar range for both SPF45 and U2AF65 UHM (Figure 2B, C, right panel) and thus comparable to the majority of known UHM–ULM interactions (12,16). Importantly, mutation of Trp45 in the Rev ULM to alanine (W45A) decreases the binding affinity by >15-fold. (Supplementary Figure S3). Consistent with the calorimetric data, we confirmed that the full length Rev protein does bind to SPF45 and U2AF65 UHMs in a GST pulldown experiment (Supplementary Figure S4). In summary, our biochemical and biophysical data show that the HIV-1 Rev ULM interacts with the UHM domains of SPF45 and U2AF65 in vitro.
Structure of Rev ULM bound to SPF45 UHM
In order to reveal the structural details of Rev binding to UHM domains, we determined the crystal structure of the Rev ULM peptide in complex with the SPF45 UHM domain at 1.2 Å resolution (Figure 3A). Strong difference electron density is detectable for the bound Rev peptide (residues 41–49) for one of the two UHM molecules of SPF45 in the crystallographic asymmetric unit. The symmetry-related SPF45 molecule packs against a composite interface formed by the Rev peptide and the bound SPF45 molecule (Supplementary Figure S5A). There is a weak difference density detectable for a tryptophan ring next to this SPF45 molecule, but other residues of the peptide are not visible. A similar crystal packing has been observed for other UHM–ULM complexes (12,16). There are no significant structural differences between the two SPF45 copies (r.m.s.d. of 0.46 Å for 103 Cα atoms).
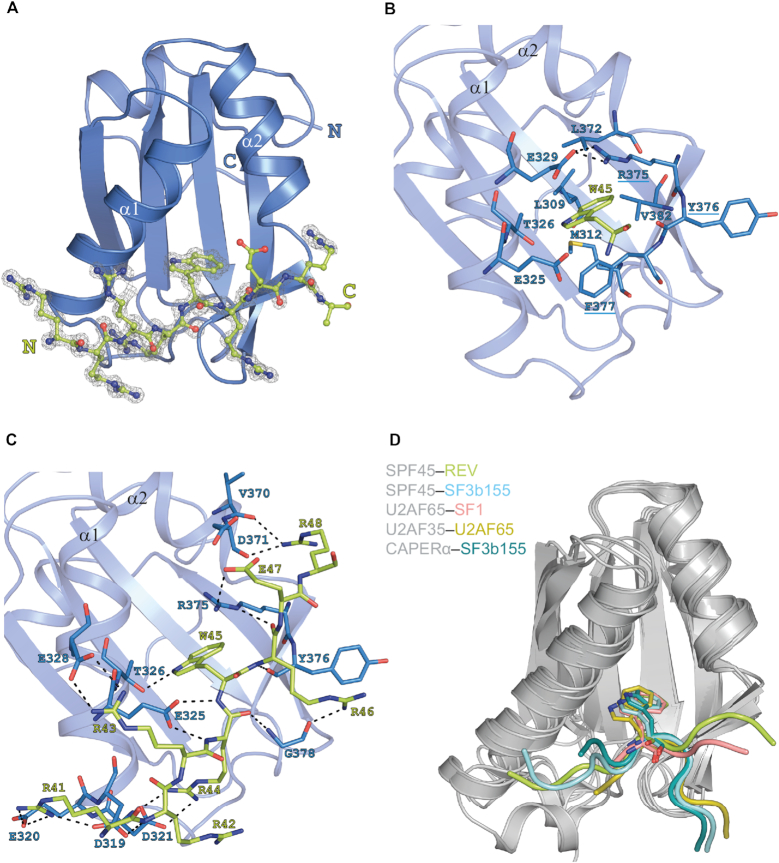
Figure 3.
Crystal structure of SPF45 UHM bound to Rev ULM. (A) Cartoon representation of SPF45 UHM with ball-and-stick representation of the bound Rev ULM surrounded by experimental omit-electron density map contoured at 1.5σ. Please note, that a number of alternative conformations of amino acid side chains are present in the structural model. In order to simplify the graphical representation of the structure, the conformations presented in this figure corresponds to the electron density with higher occupancy. (B) Recognition of the critical Rev tryptophan (W45) by the hydrophobic cleft of the SPF45 UHM-fold. Interacting residues are shown as sticks and numbered. SPF45 residues of the RXF motif are underlined. (C) Network of intermolecular hydrogen bonds that stabilize the UHM–ULM interaction. Residues are shown as sticks and numbered. (D) Overlay of SPF45, U2AF65, U2AF35 and CAPERα UHMs bound to ULM peptides (PDB: 6HIP, 2PEH, 4FXW, 1JMT, 4OZ1)
The binding mode of the SPF45–Rev complex displays the principal features of UHM–ULM interactions. The Rev peptide binds SPF45 with a total buried surface area of 816 Å2, of which one quarter (230 Å2) is contributed by the ULM tryptophan. The Rev Trp45 side chain inserts into a hydrophobic pocket between the α-helices of the SP45 UHM (Figure 3B). Additional interactions are mediated (i) by residues from the UHM β-sheet at one side of the cavity, including the characteristic RXF motif (R375-Y376-F377), which flanks helix α2, and (ii) acidic residues located on helix α1 in the UHM fold. The conservation of these crucial residues in UHM domains is evident from a multiple sequence alignment (Supplementary Figure S2A). As seen with other UHM–ULM structures, the RXF motif mediates critical interactions for the recognition of Rev Trp45. In the UHM domain, Arg375 (the first residue in the SPF45 RYF motif) forms an intramolecular salt bridge with Glu329 in the acidic helix α1, thus covering one face of the Trp45 side chain, whereas Phe377 (the third position in RYF) forms T-stacking interactions with the other face of the Trp45 ring (Figure 3B). Another feature preserved in the SPF45–Rev complex structure is a large network of intermolecular hydrogen bonds and polar contacts between basic ULM residues preceding the central tryptophan and residues in the acidic helix α1 of UHM (Figure 3C). The RYF motif and helix α1 are critical for ULM recognition and exhibit significant conformational changes upon ULM binding (Supplementary Figure S5B) (12,16).
The UHM fold and the ULM tryptophan in the SPF45–Rev complex, superimpose very well with other UHM–ULM complexes (root mean square deviations 0.7–2.2 Å for 75 UHM backbone atoms against the binary complexes shown in Figure 3D). However, the C-terminal segment of Rev-ULM peptide adopts a linear conformation that runs perpendicular to the UHM helices, passing over the RXF motif. In most other UHM–ULM complex structures the ULM ligand adopts a U-shape conformation around the central tryptophan and bends before an aromatic residue (Tyr376) in the middle position of the RXF motif (Figure 3D). The only exception is the U2AF65-SF1 complex, where the SF1 ULM also adopts an extended conformation, similar to the Rev ULM. Rev-ULM extends beyond the central tyrosine residue of the SPF45 UHM motif and forms additional hydrogen bonds and electrostatic interactions with SPF45 residues in helix α2. Given the fact that there is no appreciable structural perturbation in the SPF45-UHM domain when bound to Rev- or SF3B1-ULMs, the residues C-terminal to the conserved tryptophan (i.e. Arg46, Glu47 and Arg48) in the ULM must account for the divergent conformations of the SPF45-bound ULM ligands. Thus, ULM motifs bearing a negatively charged residue after the conserved Tryptophan (SF3b155 and U2AF65) appear to adopt a bent conformation, while ULM motifs that lack the negative charge (SF1 and Rev) have a linear shape. Although crystals of Rev ULM bound to U2AF65 UHM were not obtained, we can infer that the principal binding mode is highly similar to SPF45 UHM based on NMR titrations (same area of both UHMs affected by Rev ULM binding) and ITC experiments (highly similar binding affinity) (Figure 2).
It is important to note that the extended conformation of the Rev ULM when bound to the eukaryotic UHM protein (Supplementary Figure S6A) is completely distinct from the helical conformation adopted when Rev is bound to the RRE RNA stem-loop (Supplementary Figure S6B). This indicates that a concurrent binding of the Rev ULM to a UHM and to the RRE RNA is not possible. EMSA experiments indeed confirm that stem-loop IIB-bound Rev (which has nanomolar affinity) is not able to bind SPF45 UHM (micromolar affinity) (Supplementary Figure S6C). In summary, the structural and biochemical data show that Rev harbors a functional bona fide ULM that is able to bind eukaryotic UHM domains.
Requirement of Trp45 in Rev ULM for proper biogenesis of viral mRNAs
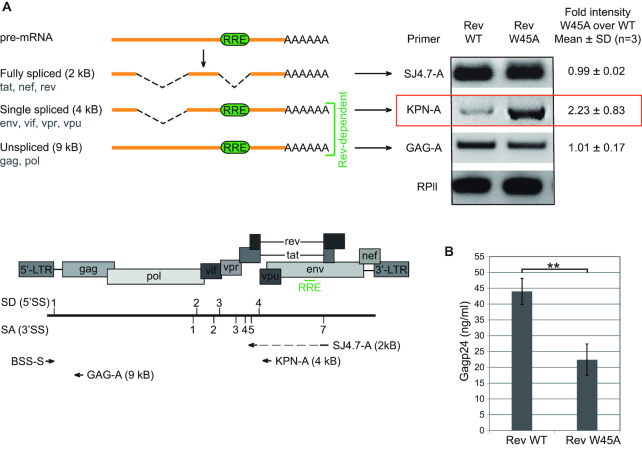
Next, we tested if the Rev ULM has a function in the HIV-1 replication cycle in human cells. We therefore used HeLafB cells that harbor a defective proviral HXB2c genome with an insertion of four nucleotides that creates a frameshift in the open reading frames of rev and env (48). In these cells, complementation of Rev by transfection with a Rev-expressing plasmid induces the production of HIV Gag proteins. Using this system, we compared the effect of Rev WT and Rev W45A on the levels of HIV-1 mRNA species. The mutated tryptophan residue lies within the nuclear localization signal. It was therefore important to verify that the Rev W45A intracellular localization is not altered, as this would have an effect on Rev function. HeLa cells were transfected with equal amounts of GFP-tagged Rev WT and W45A (Supplementary Figure S7A) and the ratio of cytoplasmic and nuclear GFP fluorescence was determined (Supplementary Figure S7B). Replacing the tryptophan residue with alanine had no effect on Rev cellular distribution. Total cellular protein levels of Rev WT and W45A after transfection also proved to be identical (Supplementary Figure S7C). We therefore proceeded with transfecting HeLafB cells with expression plasmids for Rev WT or Rev W45A and analyzed the levels of Rev-dependent (singly-spliced and unspliced) and Rev-independent (fully spliced) transcripts using RT-PCR. Figure 4A shows that the expression of Rev with the W45A mutation increased the levels of singly-spliced Rev-dependent transcripts (i.e. 4 kB transcript species) when compared to Rev WT expressing cells. In contrast, expression of Rev W45A did not affect expression levels of Rev-independent transcripts. Expression of Rev W45A also led to lower levels of Gag protein in the cell culture medium, indicating impaired viral biogenesis (Figure 4B). These experiments show that the ULM-tryptophan of Rev is necessary for proper processing of HIV-1 mRNAs, as complementation of HeLafB with Rev W45A but not Rev WT specifically increased levels of singly-spliced transcripts.
Figure 4.
Tryptophan at position 45 is required for proper biogenesis of HIV-1 mRNA species and viral production. (A) Left. Three different primer pairs were used to amplify alternatively spliced mRNAs produced during HIV-1 life cycle: fully spliced, single spliced and unspliced transcripts. Right. HeLafB cells expressing the Rev W45A mutant show increased levels of single spliced mRNAs compared to cells expressing Rev WT, as measured by RT-PCR. (B) Supernatant of HeLafB cells expressing the Rev W45A mutant show reduced p24 levels compared to cells expressing Rev WT, as measured by p24 ELISA.
The above experiments with full-length viral genome clearly point to defective mRNA processing upon mutation in Rev ULM. Nevertheless, the underlying molecular mechanisms could not be inferred from these experiments. The observed effects may be due to altered stability, splicing or export of viral mRNAs. We therefore decided to specifically monitor the splicing of the RRE-containing intron in a simplified system. We used a reporter plasmid pDM128 containing a single intron derived from the HIV-1 env gene flanked by D4 and A7 splice sites and tat exonic sequences (Figure 5A) (49). In pDM128 the 5′ end of env ORF (near 5′ SS) is replaced by a chloramphenicol acetyl transferase (cat) sequence for straightforward analysis of RRE dependent Rev activity. In our experimental approach, we do not utilize this feature of the pDM128, but instead focus on a precise analysis of the levels of spliced and unspliced mRNAs originating from pDM128. For this, we used quantitative real time PCR (qPCR) with primers specific for either spliced or unspliced mRNAs (Figure 5A). It was verified that the PCR amplification yielded single products whose sequence was further confirmed by sequencing (data not shown). Expression of Rev led to an expected increase in the ratio of unspliced versus spliced mRNA (Figure 5B), likely as a result of Rev-mediated export and stabilization of unspliced RRE-containing mRNAs. The Rev W45A mutant was less efficient than Rev wildtype (WT) in increasing the ratio of unspliced versus spliced mRNA. In order to look at the levels of spliced and unspliced mRNAs separately, the same qPCR data were plotted comparing changes in the levels of spliced and unspliced mRNA species in cells transfected with Rev W45A versus Rev WT (Figure 5C). This representation of the data clearly indicates that, compared with Rev WT, Rev W45A not only decreases the levels of unspliced mRNA but also increases the amount of spliced mRNA. The opposite effect on the two RNA species (spliced and unspliced) could result from distinct effects on splicing by Rev WT and Rev W45A, although additional effects of the W45A mutation on stability and export of unspliced mRNA might also exist.
Figure 5.
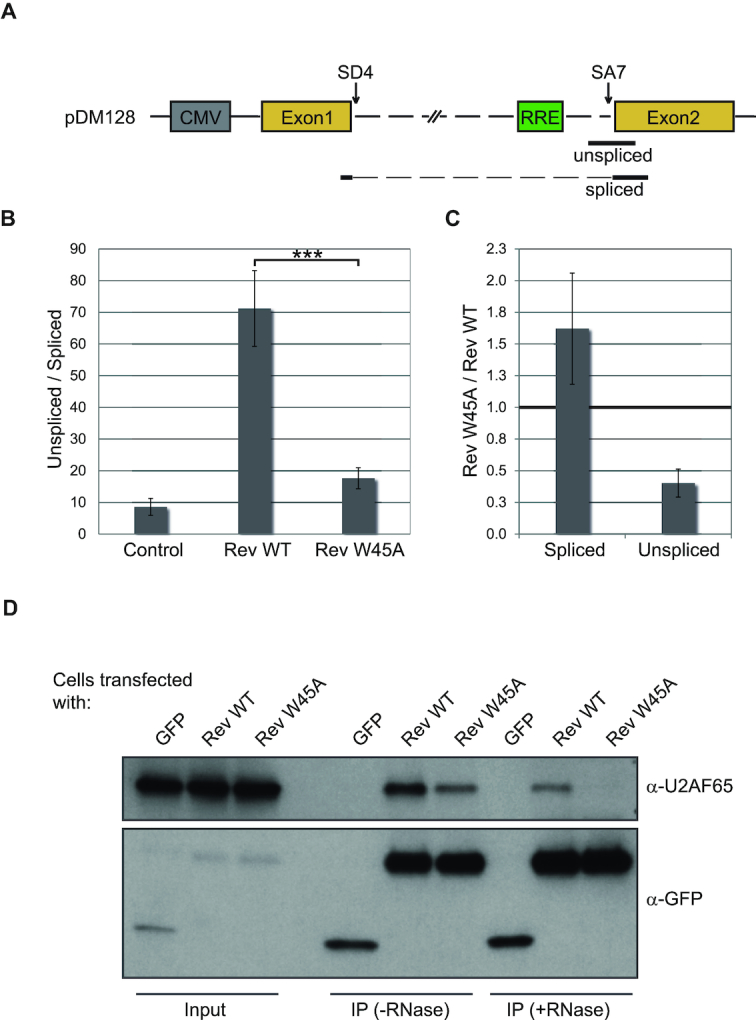
W45-dependent effect of Rev on the biogenesis of RRE-containing mRNA in HeLa cells. (A) Schematic representation of pDM128. The intronic sequence is represented as a dashed line, SD4 – splice donor 4, SA7 – spliced acceptor 7. The location of qPCR amplicons detecting either spliced or unspliced mRNA is shown. (B, C) HeLa cells were transiently transfected with pDM128 and Rev WT or W45A (+ pRLSV40 to control for transfection efficiency and pBSIISK+ as plasmid DNA for dilution). Total RNA was isolated 48 hours after transfection, reverse-transcribed with an oligo(dT) primer. (B) The relative abundance of spliced to unspliced poly(A)+ RNA was assessed by qPCR (left graph). (C) In order to look at the levels of spliced and unspliced mRNAs separately instead of as a ratio, the same qPCR data were plotted comparing changes in the levels of spliced and unspliced mRNA species in cells transfected with RevW45A versus Rev WT (right graph). (D) HeLa cells were transiently transfected with GFP-tagged Rev WT or W45A and assayed 48 hours after transfection. Extracts from HeLa cells were incubated with or without RNase A and subjected to immunoprecipitation with α-GFP antibody. 1% of input and 50% of the immunoprecipitate was analyzed by Western blot, using α-GFP and α-U2AF65 antibodies. The experiments were performed three times, a representative gel is shown.
HIV-1 Rev co-precipitates with U2AF65 in human cells
The experiments described above show that Rev can interact with eukaryotic UHM containing splicing factors in vitro and that the tryptophan residue enabling this interaction affects Rev function in human cells. Binding of Rev to several splicing-related factors has been reported, however none of them exhibits a UHM domain. We therefore tested if cellular UHM-containing splicing factors can be precipitated with GFP-tagged Rev expressed in HeLa cells. In the case of the alternative splicing factor, SPF45, no protein could be unambiguously immunoprecipitated with Rev (Supplementary Figure S8). We conclude that Rev does not interact with SPF45 in HeLa cells and discard SPF45 as a significant mediator of the Rev effect on mRNA splicing in vivo. On the other hand, U2AF65 is clearly present in complexes with Rev in human cells (Figure 5D). Moreover, mutation of the ULM tryptophan weakened this interaction. As both U2AF65 and Rev are RNA-binding proteins, we wished to exclude the possibility that the observed Rev-U2AF65 interaction could be mediated by RNA. We therefore introduced RNase treatment of the lysates before the immunoprecipitation step. RNA depletion indeed partially decreased the amount of U2AF65 co-precipitated with Rev. Nevertheless, the interaction was not entirely disrupted and remained clearly dependent on Trp45. We conclude that Rev can bind to U2AF65 via an ULM–UHM interaction.
DISCUSSION
Rev is critical for HIV replication cycle. It is a small, yet multifunctional protein. Its best-characterized role is the export of unspliced viral mRNAs to the cytoplasm. Our knowledge of other functions often lacks mechanistic details. The identification of a putative ULM within the Rev sequence led us to investigate if this motif is functional and whether it could be related to the reported splicing inhibition triggered by Rev (34). Our data prove that Rev has a functional ULM that interacts in vitro and in HeLa cells with host UHM-containing proteins. The binding mode revealed by our crystal structure and confirmed by NMR and ITC titration experiments is highly similar to previously reported interactions between UHMs and ULMs of eukaryotic proteins involved in splicing regulation. Key features of the UHM–ULM recognition are conserved in the Rev ULM, indicating that Rev can mediate interactions with host UHM proteins. Moreover, the low micromolar affinity of Rev ULM to host UHMs is in the same range as most of the cellular ULM–UHM interactions (9,10,12–16,50–52). Rev ULM has therefore the capability to compete with host ULMs for the binding to UHM-containing splicing factors. Such mimicry of host linear motifs is a widespread strategy of viruses (and pathogens). It enables interacting with the host cell molecular machinery, thus expanding viral functionality without encoding additional viral proteins (44,53).
Notably, the tryptophan residue (Trp45) that is crucial for the ULM–UHM interaction is strictly conserved among HIV-1 strains and mutation of W45 interferes with Rev function and leads to altered HIV-1 mRNA processing in human cells (Figures 1D, 4A, 5B). Thus, the conservation and importance of W45 may also reflect its role in UHM binding. The Rev ULM overlaps with two other functional sites that mediate binding to importin-β and RNA, respectively. Moreover, the dimerization domain is also in close proximity, surrounding the ULM/RNA binding site (Figure 1B). This raises the question of whether mutating the Trp45 residue to disrupt ULM function could interfere with Rev dimerization, RNA binding or nuclear import. Several lines of evidence indicate that this is not the case. First, both published work and our own experiments indicate that Trp45 does not affect the oligomerization and binding to the main Rev binding site, the SLIIB, in vitro (22,46,47,54) (Supplementary Figure S1). Although a decreased affinity of Rev W45A towards full length RRE in vitro was reported (55), this result could not be explained from a mechanistic point of view as Trp45 is neither involved nor required for the binding of stem IIB (the main binding site) or stem IA (an additional binding site). Moreover, if RNA binding in cells was disturbed by W45A, the mutation would be expected to similarly affect the levels of both partially spliced and unspliced Rev-dependent mRNA species due to their decreased stability or export. The fact that only the levels of singly spliced mRNAs are changed by W45A implies a specific phenotype that is not linked to Rev–RNA binding capacity (Figure 4A). Based on an analysis of the Rev dimer structure and the buried surface area, Trp45 contributes to the stabilization of the helical hairpin within the Rev monomer (22,54). More recently, mutation of W45 to leucine was found to affect the formation of Rev filaments in vitro (23). However, the functional role of these observations in human cells remain unclear. Finally, we also show that the nucleocytoplasmic distribution is not disturbed by the W45A mutation, which indicates that W45 is not required for Rev import (Supplementary Figure S7A). We therefore infer that introducing the W45A mutation most significantly affects its ULM function. The Rev basic region (33–51) represents a multifunctional part of Rev protein conferring diverse and independent functions such as binding of RNA and nuclear import, as well as protein binding. It is interesting to note that a similar functional complexity of the ULM-comprising region is also observed in the case of Ataxin-1; where the ULM overlaps with the NLS and the 14-3-3 binding motif resulting in a 3-way molecular switch (15).
The structure of Rev ULM bound to a eukaryotic UHM and accompanying biochemical experiments provide evidence for direct protein–protein interaction of HIV-1 Rev with UHM-containing eukaryotic splicing factors. Both the binding mode and affinity are comparable to reported eukaryotic UHM–ULM interactions (Figures 2 and 3). Our Co-IP experiments in HeLa cells indicate a clear interaction of Rev with U2AF65, while we did not find evidence for an interaction of Rev with SPF45 in cells (Figure 5 and Supplementary Figure S8). Thus, U2AF65 appears as a sensible Rev-interacting factor and mediator of its function. It is involved in constitutive splicing, associates with the transcription site early during mRNA biogenesis through its binding to the RNA Pol II CTD and is an abundant cellular protein (56,57). In contrast, SPF45 is ten times less abundant and has a more specialized function in alternative splicing (12,58). Our results suggest that SPF45 is not the main cellular target of Rev but do not rule out that the two proteins could be involved in weak, transient or cell-type specific interaction. It should be considered that apart from U2AF35–U2AF65, UHM–ULM interactions are characterized by low affinity, specificity and selectivity (12,52). Initial weak and transient interactions between various UHM–ULM pairs allow for final stable multi-protein assemblies that permit splicing decisions fitting varying cellular requirements. Such combination of multiple low affinity interactions is a characteristic feature of other functionally important interactions within the spliceosome that ensures the plasticity required during both constitutive and alternative splicing (59,60). Finally the promiscuous character of UHM–ULM interactions do not rule out the possibility that Rev binds to other host UHM-containing proteins in vivo. There are many proteins harboring characterized or predicted UHM domains (11). In fact, such proteins, e.g. RBM39 and Matrin-3, were identified within the HIV-1 Rev interactome (37).
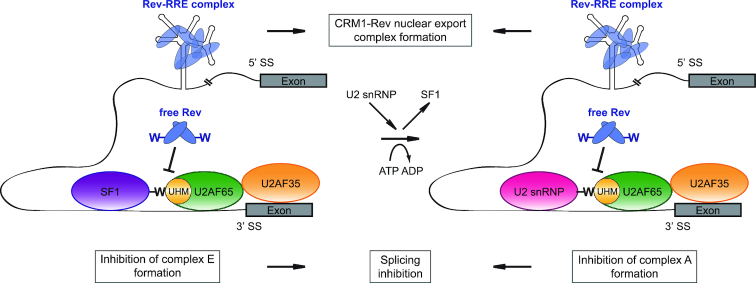
Our results suggest that Rev ULM–UHM interactions are involved in the HIV-1 replication cycle at the level of splicing regulation (Figures 4 and 5). Effects of Rev on splicing have been reported in earlier work by Kjems et al. (34,35). Also, the strength of HIV-1 3′ splice sites has been shown to affect Rev function (30). We speculate that binding of UHMs by Rev could compete with UHM–ULM interactions between constitutive splicing factors required for HIV-1 splicing. For example, Rev may bind endogenous U2AF65 impeding the interaction of this splicing factor with SF1-ULM or SF3b155-ULM during spliceosome assembly (Figure 6), as the binding affinities of all of those interactions are in a similar low-micromolar range (12). Indeed, the analysis of splicing complexes presented by Kjems and Sharp (35) shows that Rev has no effect on U1 snRNP assembly, but partially inhibits U2 snRNP binding and blocks the entry of U4/U6 and U5 snRNP. Interestingly, unlike in the case of the tri-snRNP, the Rev inhibitory effect on U2 snRNP is not dependent on the RRE.
Figure 6.
Model presenting possible mechanisms of Rev functions in HIV-1 mRNA splicing inhibition. Rev binds to the RRE in HIV-1 mRNA and interacts with the nuclear export factor CRM1 in order to transfer the transcript to the cytoplasm. Independently of its function in mRNA export, Rev can also inhibit the process of HIV-1 mRNA splicing by interfering with UHM–ULM interactions required for spliceosome assembly. We propose that Rev interactions with splicing factors may contribute to the protection of HIV-1 mRNA from splicing in order to promote rapid and efficient formation of CRM1-Rev nuclear export complex and subsequent transport of the mRNA to the cytoplasm. SS – splice site; -W – conserved tryptophan residue from the ULM motifs.
We propose that Rev-induced decrease of HIV-1 mRNA splicing efficiency would promote efficient Rev-mediated export of unspliced HIV-1 mRNA to the cytoplasm. Nuclear export requires binding of multiple copies of the Rev protein to HIV-1 mRNA, suggesting involvement of a larger proportion of Rev in interactions with the RNA than with splicing factors. Rev/RNA binding and formation of the nuclear export complex has been proposed to be a two-step process with checkpoints for higher-order Rev-RRE assembly (24). It is crucial that the HIV-1 mRNA remains unspliced during this process. HIV-1 mRNA is designed for inefficient splicing (20). As our data show that Rev can interact either with RRE RNA or with UHM domains in splicing factors it is conceivable that interaction of Rev with splicing factors may further protect the RNA from splicing during formation of the nuclear export complex by interfering with UHM–ULM interactions required for spliceosome assembly on HIV-1 mRNA (Figure 6). Considering that Rev originates from fully spliced transcripts it is plausible that initially translated Rev protein will be fully engaged in RRE RNA binding and thus not available to inhibit splicing. However, with increasing Rev production, excess of Rev protein may inhibit splicing by binding to UHM-containing splicing factors. Note, that in the context of Rev oligomers forming in vivo, higher order Rev assemblies could be simultaneously bound to the RRE and to UHM proteins, involving distinct Rev molecules and/or subunits, and thereby preferentially affecting splicing of RRE containing HIV transcripts. The resulting inhibition of splicing will reduce the amount of Rev protein synthesis, while at the same time permitting increased production of singly spliced and unspliced HIV transcripts. Thus, the Rev ULM may play a role in reducing/inhibiting HIV splicing during later stages of infection. Finally, we do rule out additional effects of the W45A mutation on stability and export of unspliced mRNA mediated through other yet uncharacterized Rev ULM – host UHM interactions.
To conclude, our study identifies a ULM sequence within the HIV-1 Rev protein that mimics a eukaryote-specific motif and follows the canonical ULM–UHM interaction mode. We propose that this motif mediates protein–protein interactions between HIV-1 Rev and host proteins that contribute to the complex viral mRNA processing required for viral replication cycle. Presumably, the mechanism involves interfering with host ULM–UHM interactions involved in splicing.
DATA AVAILABILITY
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data bank under accession number 6HIP.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Hothorn and Grzegorz Popowicz for help with crystal data acquisition and analysis. We are grateful to Karla Neugebauer and Juan Valcarcel for antibodies, Thomas Güttler for plasmid gifts. The authors acknowledge stimulating discussion with Cindy Will, Sophie Bonnal, Reinhard Lührmann, Thomas Güttler, Juan Valcarcel, Dirk Görlich, Marie-Louise Hammarskjold and David Rekosh.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB1035 and GRK1721 to M.S.]; Ministry of Education, Youth and Sports of Czech Republic under the project CEITEC 2020 [LQ1601 to K.T.]; Bavarian NMR Centre (BNMRZ) for NMR measurement time. Funding for open access charge: institutional funds/grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hocine S., Singer R.H., Grunwald D.. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010; 2:a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maniatis T., Tasic B.. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002; 418:236–243. [DOI] [PubMed] [Google Scholar]
- 3. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wahl M.C., Will C.L., Luhrmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 5. Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J.. Deciphering the splicing code. Nature. 2010; 465:53–59. [DOI] [PubMed] [Google Scholar]
- 6. Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H.. Function of alternative splicing. Gene. 2005; 344:1–20. [DOI] [PubMed] [Google Scholar]
- 7. Hertel K.J. Combinatorial control of exon recognition. J. Biol. Chem. 2008; 283:1211–1215. [DOI] [PubMed] [Google Scholar]
- 8. Maris C., Dominguez C., Allain F.H.. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005; 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- 9. Kielkopf C.L., Rodionova N.A., Green M.R., Burley S.K.. A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell. 2001; 106:595–605. [DOI] [PubMed] [Google Scholar]
- 10. Selenko P., Gregorovic G., Sprangers R., Stier G., Rhani Z., Krämer A., Sattler M.. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol. Cell. 2003; 11:965–976. [DOI] [PubMed] [Google Scholar]
- 11. Kielkopf C.L., Lucke S., Green M.R.. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004; 18:1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corsini L., Bonnal S., Basquin J., Hothorn M., Scheffzek K., Valcarcel J., Sattler M.. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat. Struct. Mol. Biol. 2007; 14:620–629. [DOI] [PubMed] [Google Scholar]
- 13. Thickman K.R., Swenson M.C., Kabogo J.M., Gryczynski Z., Kielkopf C.L.. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein–protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 2006; 356:664–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corsini L., Hothorn M., Stier G., Rybin V., Scheffzek K., Gibson T.J., Sattler M.. Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J. Biol. Chem. 2009; 284:630–639. [DOI] [PubMed] [Google Scholar]
- 15. de Chiara C., Menon R.P., Strom M., Gibson T.J., Pastore A.. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 2009; 4:e8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loerch S., Maucuer A., Manceau V., Green M.R., Kielkopf C.L.. Cancer-relevant splicing factor CAPERalpha engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem. 2014; 289:17325–17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jagtap P.K.A., Garg D., Kapp T.G., Will C.L., Demmer O., Luhrmann R., Kessler H., Sattler M.. Rational design of cyclic peptide inhibitors of U2AF Homology Motif (UHM) domains to modulate Pre-mRNA splicing. J. Med. Chem. 2016; 59:10190–10197. [DOI] [PubMed] [Google Scholar]
- 18. Frankel A.D., Young J.A.. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998; 67:1–25. [DOI] [PubMed] [Google Scholar]
- 19. Karn J., Stoltzfus C.M.. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harbor Perspect. Med. 2012; 2:a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dlamini Z., Hull R.. Can the HIV-1 splicing machinery be targeted for drug discovery. HIV AIDS (Auckl.). 2017; 9:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pollard V.W., Malim M.H.. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998; 52:491–532. [DOI] [PubMed] [Google Scholar]
- 22. Daugherty M.D., Liu B., Frankel A.D.. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010; 17:1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DiMattia M.A., Watts N.R., Cheng N., Huang R., Heymann J.B., Grimes J.M., Wingfield P.T., Stuart D.I., Steven A.C.. The structure of HIV-1 rev filaments suggests a bilateral model for Rev-RRE assembly. Structure. 2016; 24:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bai Y., Tambe A., Zhou K., Doudna J.A.. RNA-guided assembly of Rev-RRE nuclear export complexes. eLife. 2014; 3:e03656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sherpa C., Rausch J.W., Le Grice S.F., Hammarskjold M.L., Rekosh D.. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res. 2015; 43:4676–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hammarskjold M.L., Heimer J., Hammarskjold B., Sangwan I., Albert L., Rekosh D.. Regulation of human immunodeficiency virus env expression by the rev gene product. J. Virol. 1989; 63:1959–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Malim M.H., Hauber J., Le S.Y., Maizel J.V., Cullen B.R.. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989; 338:254–257. [DOI] [PubMed] [Google Scholar]
- 28. Felber B.K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G.N.. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl Acad. Sci. U.S.A. 1989; 86:1495–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malim M.H., Cullen B.R.. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol. Cell Biol. 1993; 13:6180–6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kammler S., Otte M., Hauber I., Kjems J., Hauber J., Schaal H.. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology. 2006; 3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arrigo S.J., Chen I.S.. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991; 5:808–819. [DOI] [PubMed] [Google Scholar]
- 32. D’Agostino D.M., Felber B.K., Harrison J.E., Pavlakis G.N.. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell Biol. 1992; 12:1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Groom H.C., Anderson E.C., Lever A.M.. Rev: beyond nuclear export. J. Gen. Virol. 2009; 90:1303–1318. [DOI] [PubMed] [Google Scholar]
- 34. Kjems J., Frankel A.D., Sharp P.A.. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell. 1991; 67:169–178. [DOI] [PubMed] [Google Scholar]
- 35. Kjems J., Sharp P.A.. The basic domain of Rev from human immunodeficiency virus type 1 specifically blocks the entry of U4/U6.U5 small nuclear ribonucleoprotein in spliceosome assembly. J. Virol. 1993; 67:4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tange T.O., Jensen T.H., Kjems J.. In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem. 1996; 271:10066–10072. [DOI] [PubMed] [Google Scholar]
- 37. Naji S., Ambrus G., Cimermancic P., Reyes J.R., Johnson J.R., Filbrandt R., Huber M.D., Vesely P., Krogan N.J., Yates J.R. 3rd et al.. Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell. Proteomics: MCP. 2012; 11:M111 015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hadian K., Vincendeau M., Mausbacher N., Nagel D., Hauck S.M., Ueffing M., Loyter A., Werner T., Wolff H., Brack-Werner R.. Identification of a heterogeneous nuclear ribonucleoprotein-recognition region in the HIV Rev protein. J. Biol. Chem. 2009; 284:33384–33391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 40. Ludwig E., Silberstein F.C., van Empel J., Erfle V., Neumann M., Brack-Werner R.. Diminished rev-mediated stimulation of human immunodeficiency virus type 1 protein synthesis is a hallmark of human astrocytes. J. Virol. 1999; 73:8279–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vincendeau M., Kramer S., Hadian K., Rothenaigner I., Bell J., Hauck S.M., Bickel C., Nagel D., Kremmer E., Werner T. et al.. Control of HIV replication in astrocytes by a family of highly conserved host proteins with a common Rev-interacting domain (Risp). AIDS. 2010; 24:2433–2442. [DOI] [PubMed] [Google Scholar]
- 42. Davey N.E., Van Roey K., Weatheritt R.J., Toedt G., Uyar B., Altenberg B., Budd A., Diella F., Dinkel H., Gibson T.J.. Attributes of short linear motifs. Mol. Biosyst. 2012; 8:268–281. [DOI] [PubMed] [Google Scholar]
- 43. Chemes L.B., de Prat-Gay G., Sanchez I.E.. Convergent evolution and mimicry of protein linear motifs in host-pathogen interactions. Curr. Opin. Struct. Biol. 2015; 32:91–101. [DOI] [PubMed] [Google Scholar]
- 44. Hagai T., Azia A., Babu M.M., Andino R.. Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 2014; 7:1729–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dinkel H., Van Roey K., Michael S., Davey N.E., Weatheritt R.J., Born D., Speck T., Kruger D., Grebnev G., Kuban M. et al.. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014; 42:D259–D266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kjems J., Calnan B.J., Frankel A.D., Sharp P.A.. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 1992; 11:1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tan R., Chen L., Buettner J.A., Hudson D., Frankel A.D.. RNA recognition by an isolated alpha helix. Cell. 1993; 73:1031–1040. [DOI] [PubMed] [Google Scholar]
- 48. Hadzopoulou-Cladaras M., Felber B.K., Cladaras C., Athanassopoulos A., Tse A., Pavlakis G.N.. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 1989; 63:1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hope T.J., Huang X.J., McDonald D., Parslow T.G.. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl Acad. Sci. U.S.A. 1990; 87:7787–7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang W., Maucuer A., Gupta A., Manceau V., Thickman K.R., Bauer W.J., Kennedy S.D., Wedekind J.E., Green M.R., Kielkopf C.L.. Structure of phosphorylated SF1 bound to U2AF(6)(5) in an essential splicing factor complex. Structure. 2013; 21:197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Madl T., Bagdiul I., Kern T., Kang H.-S., Zou P., Maeusbacher N., Sieber S.A., Kraemer A., Sattler M.. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 2013; 41:1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stepanyuk G.A., Serrano P., Peralta E., Farr C.L., Axelrod H.L., Geralt M., Das D., Chiu H.J., Jaroszewski L., Deacon A.M. et al.. UHM–ULM interactions in the RBM39-U2AF65 splicing-factor complex. Acta Crystallogr. D Struct. Biol. 2016; 72:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davey N.E., Trave G., Gibson T.J.. How viruses hijack cell regulation. Trends Biochem. Sci. 2011; 36:159–169. [DOI] [PubMed] [Google Scholar]
- 54. DiMattia M.A., Watts N.R., Stahl S.J., Rader C., Wingfield P.T., Stuart D.I., Steven A.C., Grimes J.M.. Implications of the HIV-1 Rev dimer structure at 3.2 A resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:5810–5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Daugherty M.D., D’Orso I., Frankel A.D.. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell. 2008; 31:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M.. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011; 7:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. David C.J., Boyne A.R., Millhouse S.R., Manley J.L.. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011; 25:972–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lallena M.J., Chalmers K.J., Llamazares S., Lamond A.I., Valcarcel J.. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002; 109:285–296. [DOI] [PubMed] [Google Scholar]
- 59. Will C.L., Luhrmann R.. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011; 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tazi J., Bakkour N., Stamm S.. Alternative splicing and disease. Biochim. Biophys. Acta. 2009; 1792:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the reported crystal structure have been deposited with the Protein Data bank under accession number 6HIP.