Abstract
Faithful propagation of transcription programs through cell division underlies cell-identity maintenance. Transcriptional regulators selectively bound on mitotic chromatin are emerging critical elements for mitotic transcriptional memory; however, mechanisms governing their site-selective binding remain elusive. By studying how protein-protein interactions impact mitotic chromatin binding of RBPJ, the major downstream effector of the Notch signaling pathway, we found that histone modifying enzymes HDAC1 and KDM5A play critical, regulatory roles in this process. We found that HDAC1 knockdown or inactivation leads to increased RBPJ occupancy on mitotic chromatin in a site-specific manner, with a concomitant increase of KDM5A occupancy at these sites. Strikingly, the presence of KDM5A is essential for increased RBPJ occupancy. Our results uncover a regulatory mechanism in which HDAC1 negatively regulates RBPJ binding on mitotic chromatin in a KDM5A-dependent manner. We propose that relative chromatin affinity of a minimal regulatory complex, reflecting a specific transcription program, renders selective RBPJ binding on mitotic chromatin.
INTRODUCTION
Cell identity maintenance requires specific transcription programs to be transmitted through mitosis with high fidelity. Upon the onset of mitosis, the chromosomes condense, the nuclear membrane breaks down, transcription essentially ceases, and a large fraction of the transcriptional machinery, including RNA polymerases and transcription factors, dissociate from mitotic chromatin (1,2). Nonetheless, most transcription programs are faithfully propagated through cell division. Covalent modifications on DNA and histones play critical roles to maintain cell type-specific transcription programs through cell division (3–5). More recently, sequence-specific transcription factors that are selectively bound on mitotic chromatin, termed ‘mitotic bookmarking factors’, have emerged as potential players in the maintenance of transcriptional memory through cell division (6–15).
Mitotic bookmarking factors identified so far have demonstrated site-selective binding on mitotic chromatin (4,5,16–21); however, it remains unclear how this selectivity is accomplished. GATA1 is one of the better characterized mitotic bookmarking factors (17). Although meticulously examined, the critical determinants that differentiate genomic sites bound by GATA1 in interphase as compared to mitosis were not identified: co-factors that interact with GATA1 are absent on mitotic chromatin, GATA1 binding does not correlate with DNA hypersensitive sites, and specific histone modifications do not correlate with GATA1 binding sites (17).
By biochemical fractionation of mitotic chromatin followed by mass spectrometry, we discovered that the sequence-specific transcription factor RBPJ binds to the mitotic chromatin of F9 cells (18,22). F9 cells are derived from a mouse embryonal carcinoma and have many characteristic features of embryonic stem (ES) cells (23). RBPJ, also known as CSL, CBF1, Su(H), Lag-1, is an evolutionarily conserved protein that binds to the motif CGTGGGAA (24,25). The default activity of RBPJ is often considered to be transcriptional repression; RBPJ accomplishes this task, in part, by tethering a histone deacetylase (HDAC) corepressor complex to the promoter of target genes (26,27). The corepressor proteins SMRT, CIR, SAP30, HDAC1 and HDAC2 have been shown to be components of this complex (26,28,29). Histone demethylases, such as KDM5A or LSD1, are additional components of repressor complexes (30–32). Indeed, multiple repressive complexes have been identified, but how these complexes interact with RBPJ to repress transcription remains incompletely understood.
On the other hand, RBPJ is the major downstream effector of the Notch signaling pathway (33,34). Notch signaling is one of the critical pathways in cell-fate determination and is also frequently employed by tumor cells for their development and progression (35). Upon Notch activation, RBPJ complexes with the Notch intracellular domain as well as acetyltransferase p300, in addition to other transcriptional co-activators, and binds to its DNA consensus sequence to activate target genes in a cell-type specific manner (22,36,37). Furthermore, work from Drosophila indicates that RBPJ occupancy is enhanced at its targets after Notch activation (38).
RBPJ associates with mitotic chromatin through its interaction with specific DNA bases, as well as the phosphodiester backbone (18). While nucleosomes restrict DNA access of a large fraction of sequence-specific transcription factors, RBPJ binds directly to nucleosomal DNA with a preference for sites close to the entry/exit positions of the nucleosomal DNA (18). Genome-wide analysis in F9 cells revealed that roughly 60% of the sites occupied by RBPJ in asynchronous cells are also bound in mitotic cells (18). How RBPJ selectively binds to these specific sites during mitosis has remained unknown.
In this study, we found that the histone-modifying enzymes HDAC1 and KDM5A are present at RBPJ-occupied sites on mitotic chromatin, and a fraction of their site-specific association with mitotic chromatin is RBPJ-dependent. Altering transcription programs of F9 cells through HDAC1 knockdown or histone deacetylation inhibition with trichostatin A (TSA), alters RBPJ occupancy on mitotic chromatin. These observations support the notion that RBPJ may function as a mitotic bookmarking factor to maintain specific transcription programs through cell division. We further identified KDM5A as a critical determinant for mitotic chromatin bookmarking by RBPJ. Functionally, mitotic chromatin occupancy of RBPJ is sensitive to changes in HDAC1 protein levels or TSA treatment, which results in altered kinetics of transcription reactivation of RBPJ target genes upon mitotic exit.
MATERIALS AND METHODS
Cell culture and synchronization
Murine embryonal carcinoma F9 cells were cultured on gelatin-coated plates in DMEM supplemented with 10% FBS. To enrich for mitotic cells, cells were treated with 1 μg/ml nocodazole (Sigma) for 4 h. Mitotic cells were then collected by gently washing the loosely adherent mitotic cells off the culture dishes with PBS containing 1 μg/ml nocodazole, resulting in a mitotic index greater than 98% (18).
Nocodazole washout and cell cycle analysis
F9 cells arrested in mitosis were allowed to enter the cell cycle by nocodazole removal. The cells were then grown in suspension, using uncoated plates, and cell aliquots were removed at different time points. To calculate the percentage of cells remaining in the mitosis, cells were fixed with formaldehyde, stained with DAPI, and then transferred to a microscope slide using a cytocentrifuge (Wescor Inc.) with the following parameters: medium acceleration, 1000 rpm, 3 min cycle. Cells were mounted using ProLong™ Gold Antifade Mountant (ThermoFisher) and imaged with a Zeiss AxioObserver microscope equipped with Hamamatsu Flash4.0 sCMOS monochrome camera. At least 5 random fields of each slide were examined for each time point and treatment. Mitotic indices were calculated as the ratio of cells in mitosis to total number of cells.
Generation of RBPJ knockout F9 cell lines by CRISPR-Cas9
pX330-U6-Chimeric_BB-CBh-hSpCas9 was a gift from Feng Zhang (Addgene plasmid # 42230) (39). The engineered pX330-based plasmids target the introns upstream of Exon 6 and downstream of Exon 7 of mouse RBPJ and were constructed using the following primers: de7mRBPJF (5′ CACCGGTTAGAGCAGACGTAGCTCCAGG 3′), de7mRBPJR (5′ AAACCCTGGAGCTACGTCTGCTCTAACC 3′), ue6mRBPJF2 (5′ CACCGGGGGATCCTATTTAGATGCGGGG 3′), and ue6mRBPJR2 (5′ AAACCCCCGCATCTAAATAGGATCCCCC 3′) (39). The resulting two pX330-based plasmids were transfected along with pCR2.1-puro (ThermoFisher) using Lipofectamine 3000 (ThermoFisher) at a molar ratio of 10:10:1. Following transfection, F9 cells were treated with puromycin (2 μg/ml) for 16 h. Single colonies were isolated and deletions in the RBPJ locus were verified using PCR (Supplementary Table S1). The presence of deletions and whether or not a clone was homozygous or heterozygous was determined by PCR using primers surrounding the two guide RNAs (Supplementary Table S1). The deletions were further confirmed by sequencing the PCR products covering the genomic regions containing the deletion in each candidate line. Two RBPJ knockout F9 cell lines, RBPJKO17 and RBPJKO43, were obtained. By sequencing the genomic loci covering the deleted RBPJ regions, we found that the RBPJKO17 line is missing 1044 bps (53 649 339–53 650 382, GRCm38/mm10 assembly), and the RBPJKO43 line is missing 1065 bps (53 649 328–53 650 392, GRCm38/mm10 assembly). These deletions are predicted to remove 84 amino acids encoding part of the DNA-binding region (Supplementary Figure S1). The candidate lines were further examined by Southern blot and western blot analyses. A polyclonal anti-RBPJ antibody was raised in rabbits against a GST-RBPJ fusion protein containing full-length RBPJ (18). As shown in Supplementary Figure S1C, full-length RBPJ in F9 cells has a relative molecular mass of ∼58 KDa. Importantly, we did not detect any protein species between 25 and 80 KDa in the RBPJKO17 and RBPJKO43 cell lines, using the polyclonal anti-RBPJ antibody. Although we cannot exclude the formal possibility that a truncated RBPJ protein, which is not detected by our polyclonal antibody, is still expressed, such a protein would, nevertheless, be unable to bind DNA, as it would be missing 84 amino acids of the DNA-binding domain (18).
TSA and CPI-455 treatment
F9 cells were plated on a 100 mm tissue culture dishes coated with 0.1% gelatin at a density of ∼4.5 × 103 cells/cm2. Twenty four hours after seeding, F9 cell were treated with 30 nM trichostatin A (TSA) and/or 50 mM CPI-455 for 24 h. Control cells were treated with an equal volume of DMSO.
Lentiviral shRNA-mediated knockdowns
Mission non-targeting shRNA control (SHC002), shRNA targeting HDAC1 (TRCN0000229438, Sigma), or shRNA targeting KDM5A (TRCN0000113532, Sigma) were used in this study. Virus was produced as previously described (40).
Chromatin immunoprecipitation (ChIP)
ChIP was carried out as previously described (18). Briefly, ChIP assays were performed using anti-RBPJ, anti-HDAC1, anti-p300, anti-KDM5A antibodies, and antibodies specific for histone H3 acetylated at lysine 9, 14, 27 or differentially methylated at lysine 4. Antibodies source and dilutions used for ChIP are listed in Supplementary Table S2. The protein-DNA complexes were captured by incubation with protein A+G agarose beads (Millipore) for 4 h at 4°C. Eluates were reversed-crosslinked at 65 °C for 16 h. DNA was purified using the SpinSmart nucleic acid purification columns (Denville Scientific).
RNA isolation, cDNA synthesis, and qPCR
Total RNA was extracted from cell pellets using TRIzol reagent (Ambion). cDNA was synthesized by reverse transcription of total RNA (1 μg) using a SensiFAST cDNA synthesis kit (Bioline) following the manufacturer's protocol. Reactions without reverse transcriptase were used as controls. Real-time PCR reactions were performed using SensiFAST SYBR Hi-ROX Master Mix (Bioline) in a 384-well format with a QuantStudio 5 real-time PCR system (Applied Biosystems). Expression levels were normalized to β-actin. All primers used for the assaying of nascent transcript levels and ChIP are listed in Supplementary Table S3.
Protein isolation and western blotting
Whole cell extracts for western blot analysis were prepared using 1× SDS sample buffer without DTT or bromophenol blue (125 mM Tris-base (pH 6.8), 20% glycerol and 2.5% SDS). Proteins were quantified using BCA protein assay kit (Thermo Fisher) according to the manufacturer's instructions. Samples were resolved on NuPAGE 4–12% Bis–Tris gels in MOPS-SDS buffer (Thermo Fisher) at 30 μg/lane. The sources and dilutions of the antibodies used for western blot analyses are listed in Supplementary Table S2. HRP-conjugated secondary antibodies (Epigentek) were used at 1:10,000. Immunoblots were developed using SuperSignal West Pico or Dura chemiluminescent substrate (ThermoFisher), and imaged using a Kodak Processor M35A. The imaged films were scanned and quantified using ImageStudio 5.2 software (Li-Cor).
RESULTS
HDAC1 and KDM5A, but not p300, complex with RBPJ in F9 cells
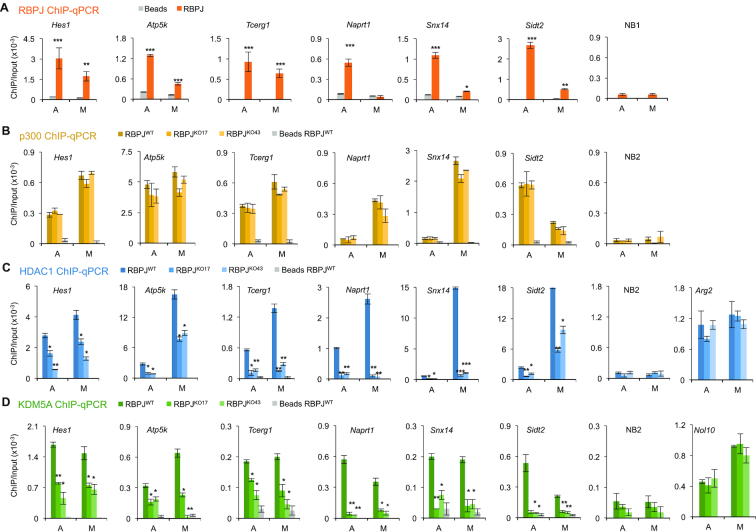
Using mass spectrometric analysis of biochemically purified mitotic chromatin, we found that, in addition to RBPJ, the histone modifying enzymes, HDAC1, p300 and KDM5A, were also retained on F9 cell mitotic chromatin (Lake RJ and Fan HY unpublished observations). Given that RBPJ has been found to interact with HDAC1, p300 and KDM5A-containing complexes and that RBPJ interacts directly with KDM5A (32), we sought to determine if HDAC1, p300 and KDM5A contribute to the selective mitotic chromatin binding of RBPJ. We first determined if these proteins also occupy RBPJ-binding sites and whether they bind to these sites in an RBPJ-dependent manner. We examined six loci from the most pronounced RBPJ occupied sites identified in our previous RBPJ ChIP-seq study (18). The promoters of Hes1, Atp5k and Tcerg1 genes (Figure 1A) represent RBPJ occupancy sites common to both asynchronous and mitotic cells, as enrichment of the RBPJ protein at these loci is within a four-fold difference between asynchronous and mitotic cells (18). On the other hand, the promoters of the Naprt1, Snx14 and Sidt2 genes are preferentially occupied by RBPJ in asynchronous cells, as the RBPJ enrichment at these sites is at least four times greater in interphase cells than in mitotic cells, as previously defined (Figure 1A) (18).
Figure 1.
HDAC1 and KDM5A complex with RBPJ in F9 cells. (A) Anti-RBPJ ChIP-qPCR showing RBPJ enrichment at different loci in asynchronous (A) or mitotic (M) F9 cells. Paired t-tests were used to compare RBPJ enrichment relative to beads only. NB1 represents an RBPJ nonbinding region. (B) ChIP-qPCR analyses were used to determine the site-specific enrichment of p300 in wild-type (RBPJWT) and two RBPJ knockout (RBPJKO17 and RBPJKO43) F9 cell lines. NB2 represents a p300 nonbinding region. (C) Results obtained from HDAC1 ChIP-qPCR. NB2 represents a HDAC1 nonbinding region. Arg2 represents a RBPJ-independent HDAC1 binding region. (D) Results obtained from KDM5a ChIP-qPCR. NB2 represents a KDM5A nonbinding region. Nol10 represent a RBPJ-independent KDM5A binding region. Shown are means ± SEM from two biological replicates. Paired t-tests were used to compare enrichment of the indicated proteins in RBPJ knockout cells relative to wild-type cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Using CRISPR-Cas9 technology, we generated RBPJ knock-out cell lines (RBPJKO17 and RBPJKO43), in which the genomic region that encodes part of the DNA-binding domain (amino acids 167–250 of variant 2) was deleted (Supplementary Figure S1A). Anti-RBPJ ChIP-qPCR was used to examine the six loci shown in Figure 1, and there is essentially no RBPJ enrichment at these sites in either of the RBPJ knockout lines, demonstrating the specificity of our anti-RBPJ antibody (Supplementary Figure S1d). Anti-p300 ChIP-qPCR in wildtype F9 cells revealed that p300 was significantly enriched at all RBPJ-binding sites in both asynchronous and mitotic cells (Figure 1B). Moreover, we found that p300 occupancy at these sites was not affected by loss of RBPJ, indicating that p300 occupies these sites in an RBPJ-independent manner. On the other hand, anti-HDAC1 ChIP-qPCR revealed that HDAC1 was significantly enriched at Hes1, Atp5k and Tcerg1 in both asynchronous and mitotic F9 cells (Figure 1C); however, HDAC1 occupancy at these sites was significantly decreased in the RBPJ knockout cell lines, suggesting that a fraction of HDAC1 bound to these sites in a RBPJ-dependent manner. Similar HDAC1 occupancy results were found at RBPJ-binding sites in the promoters of Naprt1, Snx14 and Sidt2 (Figure 1C), which are preferentially occupied by RBPJ in asynchronous cells. Interestingly, while RBPJ does not show significant occupancy at the Naprt1 promoter in mitotic cells (Figure 1A), there was, nonetheless, a substantial decrease in HDAC1 occupancy at this site in mitotic RBPJKO17 and RBPJKO43 cells, suggesting that RBPJ occupancy during interphase may contribute to the binding of HDAC1 on the Naprt1 promoter in mitotic F9 cells (Figure 1C).
Lastly, we examined KDM5A occupancy at these sites in both asynchronous and mitotic cells (Figure 1D). ChIP-qPCR analysis revealed that KDM5A was also significantly enriched at all RBPJ-binding sites and, similar to HDAC1, KDM5A occupancy at these sites was dependent upon the presence of RBPJ. However, HDAC1 displayed a generally higher occupancy in mitotic F9 cells as compared to asynchronous F9 cells, while KDM5A displayed relatively similar levels of occupancy in both asynchronous and mitotic F9 cells (Figure 1C and D).
Together, these results suggest that HDAC1 and KDM5A, but not p300, may complex with RBPJ in F9 cells, and these interactions may contribute to the selective binding of RBPJ to mitotic chromatin.
HDAC1 negatively regulates sequence-specific RBPJ binding on mitotic chromatin
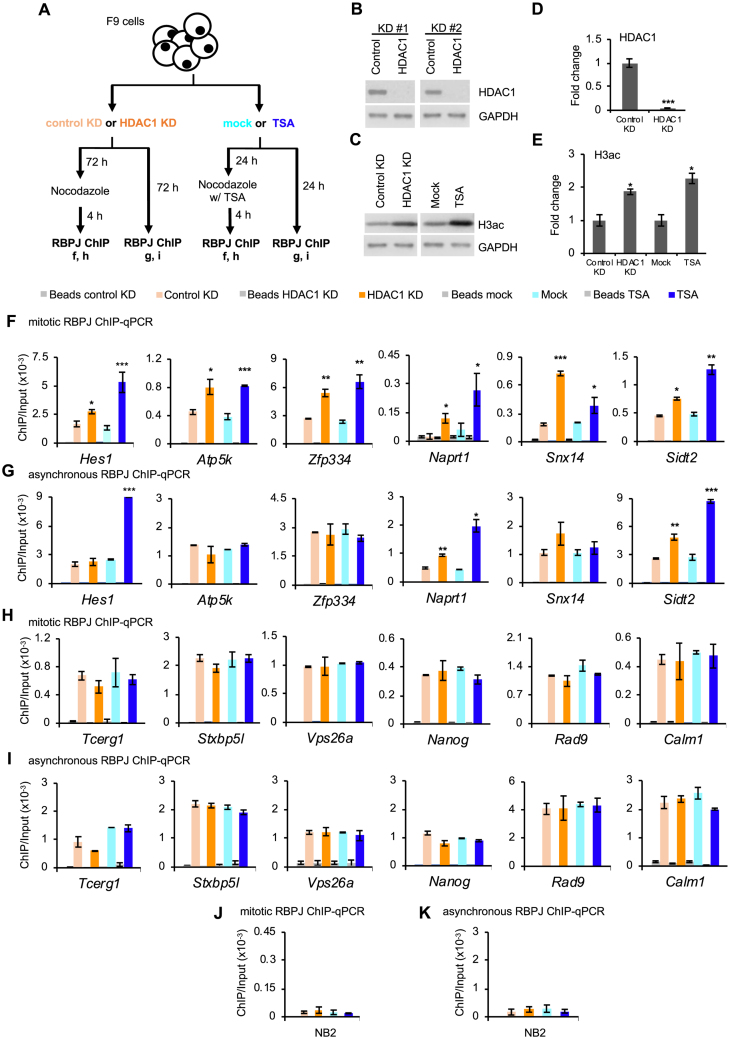
To test the hypothesis that HDAC1 contributes to the selective mitotic chromatin binding of RBPJ, we compared RBPJ occupancy in F9 cells treated with shRNA targeting HDAC1 or a control shRNA. Separately, we also treated F9 cells with trichostatin A (TSA), a reagent that inhibits class I and class II histone deacetylases (41) (Figure 2A). Infecting F9 cells with lentivirus expressing shRNA targeting HDAC1 for 72 h resulted in ∼95% decrease of the HDAC1 protein level (Figure 2B and C) and ∼2-fold increase in global histone H3 acetylation (Figure 2D-E), as compared to cells expressing the control shRNA (Figure 2A-E). Treating cells with TSA for 24 h also resulted in a ∼2-fold increase in histone H3 acetylation (Figure 2D and E). Using ChIP-qPCR, we analyzed RBPJ occupancy on mitotic chromatin at twelve genomic loci, which were among the top occupancy sites revealed from our RBPJ ChIP-seq study (18). Six of the RBPJ-binding sites contain the RBPJ-binding consensus sequence (Figure 2F-G), and the other six RBPJ-occupancy sites do not (Figure 2H and I) (18). As shown in Figure 2F, both HDAC1 knockdown and TSA treatment significantly increased RBPJ occupancy on mitotic chromatin of the Hes1, Atp5k, Zfp334, Naprt1, Snx14 and Sidt2 genes, which contain the RBPJ-binding consensus sequence, indicating that HDAC1 negatively regulates RBPJ occupancy on mitotic chromatin at these sites.
Figure 2.
RBPJ occupancy in mitotic cells is negatively regulated by HDAC1 at sites containing the RBPJ-binding consensus. (A) Experimental scheme. Both asynchronous and mitotic cells were collected under the indicated experimental conditions and subjected to RBPJ ChIP-qPCR analysis. (B–E) Representative immunoblots showing relative HDAC1 and histone H3 acetylation (H3ac) abundance (B, D) and immunoblot quantification (C, E). (F–G) RBPJ ChIP analyses at loci that contain the RBPJ-binding motif. (F) RBPJ ChIP-qPCR in mitotic cells. (G) RBPJ ChIP-qPCR in asynchronous cells. (H–I) RBPJ ChIP analyses at loci that do not contain the RBPJ-binding motif (H) RBPJ ChIP-qPCR in mitotic cells. (I) RBPJ ChIP-qPCR in asynchronous cells. (J) Control RBPJ ChIP assay from mitotic cells analysing RBPJ enrichment at an RBPJ nonbinding region (NB3). (K) Control RBPJ ChIP assay from asynchronous cells used to analyse RBPJ enrichment at an RBPJ nonbinding region (NB2). Shown are means ± SEM from two biological replicates. Paired t-tests were performed to compare RBPJ enrichment in treated relative to untreated cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
We also analyzed RBPJ occupancy at these six sites in asynchronous F9 cells treated with shRNA targeting HDAC1 or TSA (Figure 2G) and found a significant increase in RBPJ at the Naprt1 and Sidt2 promoters, but not the Hes1, Atp5k, Zfp334 and Snx14 promoters, indicating that mitotic chromatin RBPJ binding may occur by mechanisms that are distinct from its binding on interphase chromatin. Moreover, these results are consistent with previous findings that not all binding sites occupied by transcription factors in interphase cells remain bound by these factors in mitotic cells (18).
Of great interest, HDAC1 knockdown or TSA treatment did not alter RBPJ occupancy at the Tcerg1, Stxb5l, Vps26a, Nanog, Rad9 or Calm1 promoters, in either mitotic or interphase cells (Figure 2H and I). Given that these sites do not contain RBPJ-binding motifs, this result suggests that HDAC1 negatively regulates RBPJ occupancy on mitotic chromatin, only at sites containing an RBPJ-binding consensus sequence, where RBPJ binds to DNA in a sequence-specific manner.
It is possible that decreasing the HDAC1 protein level or TSA treatment might increase p300 occupancy at RBPJ-binding sites, and this could account for the increased RBPJ occupancy observed in mitotic cells. To test this hypothesis, we determined the levels of p300 enrichment at RBPJ-bound sites in mitotic cells using anti-p300 ChIP-qPCR (Supplementary Figure S2). We observed an increase in mitotic p300 enrichment at Hes1, Naprt1, Sidt2, Stxbp5l and Calm1 upon HDAC1 knockdown (Supplementary Figure S2); however, HDAC1 knockdown only increased the enrichment of RBPJ at Hes1, Naprt1, Sidt2, but not at Stxbp5l or Calm1 (Figure 2F and H). In addition, HDAC1 knockdown did not increase the enrichment of p300 at Atp5k, Zfp334, and Snx14 (Supplementary Figure S2B), but did increase mitotic RBPJ occupancy at these sites. These results, together, argue against the possibility that increased p300 occupancy alone underlies the HDAC1 knockdown-induced increase in mitotic RBPJ occupancy at sites containing the RBPJ-binding consensus sequence. Moreover, while TSA treatment prevents the occupancy of HDAC1 at these loci (Supplementary Figure S3), we did not observe any direct correlation between TSA treatment and site-specific p300 occupancy (Supplementary Figure S2). Taken together, these results support the hypothesis that HDAC1 negatively regulates sequence-specific RBPJ occupancy on mitotic chromatin in F9 cells in a manner independent of p300.
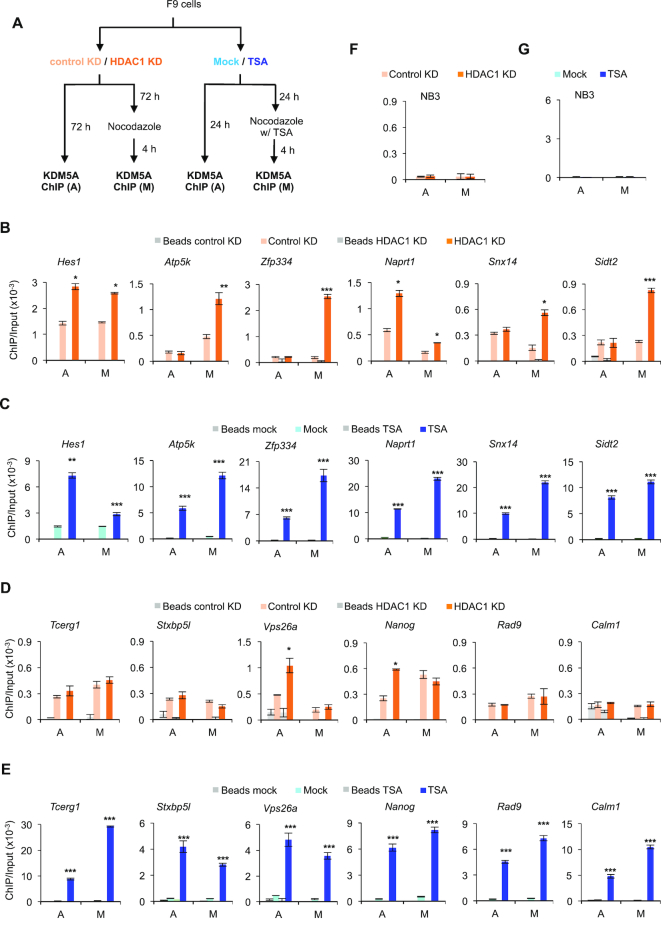
HDAC1 knockdown or TSA treatment increases KDM5A occupancy on mitotic chromatin at RBPJ occupancy sites containing the RBPJ-binding motif
We next determined the effect of HDAC1 knockdown or TSA treatment on mitotic chromatin occupancy of KDM5A at RBPJ-binding sites in F9 cells (Figure 3A). Remarkably, decreasing HDAC1 levels by shRNA also increased KDM5A occupancy at the Hes1, Atp5k, Zfp334, Naprt1, Snx14 and Sidt2 promoters in mitotic cells, indicating that HDAC1 negatively regulates KDM5A occupancy at these sites on mitotic chromatin (Figure 3B). However, HDAC1 knockdown did not have the same impact on KDM5A binding to all six sites on interphase chromatin (Figure 3B). Similarly, as expected, TSA treatment also increased mitotic chromatin KDM5A occupancy at these sites that contain the RBPJ-binding motif (Figure 3C). Together, these data support the notion that HDAC1 negatively regulates mitotic KDM5A occupancy at RBPJ-binding sites that contain an RBPJ-binding motif. In contrast to HDAC1 KD, however, TSA treatment increased KDM5A occupancy at all six sites examined in asynchronous cells (Figure 3C), suggesting that additional class I or II histone deacetylases contribute to the regulation of KDM5A occupancy at RBPJ-binding sites in asynchronous cells. Interestingly, HDAC1 knockdown had no effect on mitotic KMD5A occupancy at the Tcerg1, Stxb5l, Vps26a, Nanog, Rad9 or Calm1 promoters, sites without RBPJ-binding motifs (Figure 3D), arguing against the possibility that HDAC1 negatively regulates mitotic chromatin occupancy of KDM5A at sites lacking RBPJ-binding motifs. Although HDAC1 knockdown also increased KDM5A occupancy at the Vps26a and Nanog promoters in asynchronous cells, this pattern was not maintained during mitosis (Figure 3D).
Figure 3.
HDAC1 knockdown or TSA treatment increases KDM5A occupancy on mitotic chromatin at sites containing the RBPJ-binding consensus sequence. (A) Experimental scheme. Asynchronous (A) and mitotic (M) cells were collected under the indicated experimental conditions and subjected to KDM5A ChIP. (B, C) KDM5A ChIP-qPCR analyses at RBPJ binding sites that contain the RBPJ-binding motif. (D, E) KDM5A ChIP-qPCR analyses at RBPJ binding sites that do not contain the RBPJ-binding motif. (F) Control KDM5A ChIP assays as in panels B and D analysing KDM5A enrichment at a KDM5A nonbinding region (NB3). (G) Control KDM5A ChIP assays as in panels C and E analysing KDM5A enrichment at a KDM5A nonbinding region (NB3). Shown are means ± SEM from two biological replicates. Paired t-tests were performed to compare enrichment in treated relative to untreated cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
We next examined KDM5A occupancy at RBPJ-bound sites that do not contain an RBPJ-binding motif in F9 cells treated with TSA. We found that TSA treatment increased KDM5A occupancy at all RBPJ-bound sites examined, regardless of cell cycle phase or whether or not the sites contained an RBPJ-binding motif (Figure 3E), supporting the notion that additional histone deacetylases regulate KDM5A occupancy at all RBPJ-binding sites. Importantly, these results indicate that HDAC1 plays a key role in regulating KDM5A occupancy, specifically, at RBPJ-binding sites containing the RBPJ consensus sequence on mitotic chromatin.
In sum, reducing HDAC1 protein levels or inhibiting HDAC1 activity by treating cells with TSA increased mitotic chromatin occupancy of both RBPJ and KDM5A at sites containing RBPJ-binding motifs (Figures 2 and 3). These observations raise a hypothesis that KDM5A positively regulates mitotic chromatin RBPJ binding at sites containing the RBPJ-binding motif.
KDM5A positively regulates RBPJ occupancy on mitotic chromatin in HDAC1 shRNA or TSA-treated F9 cells
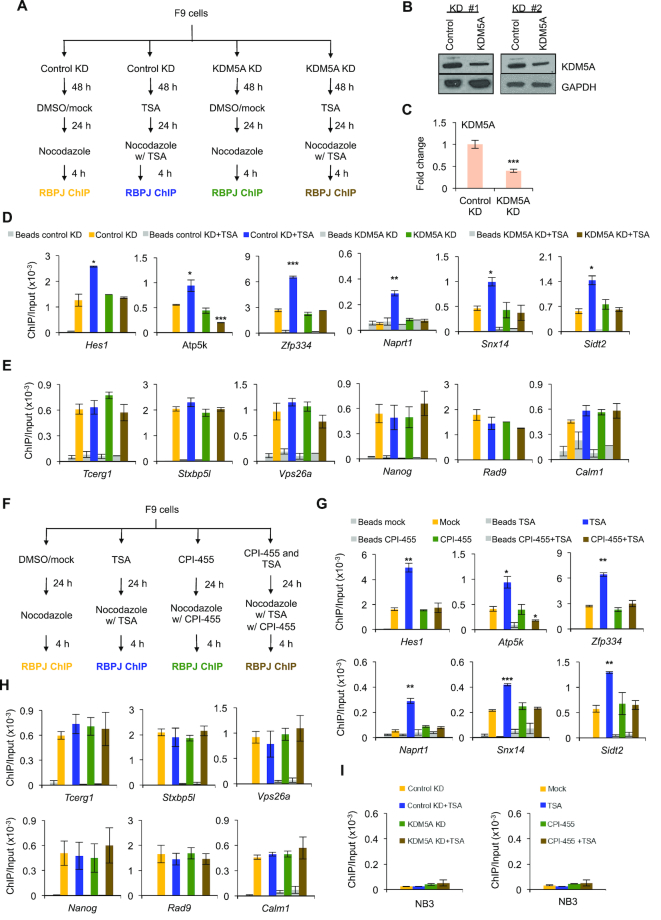
To determine if KDM5A positively regulates RBPJ occupancy on mitotic chromatin in F9 cells, we examined RBPJ occupancy using ChIP-qPCR in F9 cells treated with shRNA targeting KDM5A alone or with TSA co-treatment (Figure 4A). F9 cells expressing shRNA targeting KDM5A for 72 h had a >50% decrease in the KDM5A protein levels (Figure 4B and C). As shown in Figure 4D, TSA treatment increased mitotic RBPJ occupancy at the promoters of the Hes1, Atp5k, Zfp334, Naprt1, Snx14 and Sidt2 genes (blue versus yellow bars). Remarkably, KDM5A knockdown reversed the increase of RBPJ occupancy on mitotic chromatin induced by TSA treatment, but only at sites containing the RBPJ-binding motif (brown, Figure 4D and E). This result indicates that KDM5A positively regulates RBPJ-mitotic chromatin interactions at regions containing RBPJ-binding motifs in cells treated with TSA. On the other hand, KDM5A knockdown alone had no significant effect on mitotic RBPJ-binding, regardless of whether the sites contain RBPJ-binding motifs or not (Figure 4D and E, compare green to yellow).
Figure 4.
The KDM5A knockdown or CPI-455 treatment reverses the effect of TSA on mitotic chromatin RBPJ occupancy. (A) Experimental scheme for D–E. F9 cells were subjected to TSA treatment, shRNA-mediated KDM5A knockdown, or a combination of these treatments. Mitotic cells were collected under the indicated experimental conditions and subjected to RBPJ ChIP-qPCR analysis. (B) Immunoblots showing the extent of KDM5A knockdown from two independent experiments. (C) Quantification of the data in b. (D) Enrichment of RBPJ at loci that contain the RBPJ-binding motif. (E) Enrichment of RBPJ at loci that do not contain the RBPJ-binding motif (F) Experimental scheme for G–H. F9 cells were subjected to TSA treatment, treatment with the inhibitor of H3K4 demethylation CPI-455, or a combination of these drugs. (G) Enrichment of RBPJ at loci that contain the RBPJ-binding motif. (H) Enrichment of RBPJ at loci that do not contain the RBPJ-binding motif. (I) Control ChIP assays analysing RBPJ enrichment at an RBPJ nonbinding region (NB3). Shown are means ± SEM from two biological replicates. Paired t-tests were performed to compare enrichment relative to untreated cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
As a complementary approach, we used CPI-455, an inhibitor of KDM5 family of histone demethylases (Figure 4F–H). As shown in Figure 4G, treating cells with CPI-455 also reversed the effect of TSA on mitotic RBPJ occupancy at sites containing RBPJ motifs. Similar to KDM5A knockdown, CPI-455 treatment alone had no detectable effect on mitotic chromatin RBPJ occupancy at all sites examined (Figure 4G and H, compare green to yellow).
CPI-455 might reverse the TSA-induced mitotic chromatin-RBPJ interaction by directly inhibiting the KDM5A enzymatic activity or by interfering with the interaction of KDM5A with chromatin. To distinguish between these possibilities, we determined whether CPI-455 treatment impacts the interaction between KDM5A and mitotic chromatin, using anti-KDM5A ChIP-qPCR. As shown in Supplementary Figure S4, treating cells with CPI-455 reduced KDM5A-chromatin interactions at all loci tested. Together, our results raise an intriguing hypothesis that KDM5A enhances RBPJ-mitotic chromatin interaction by binding to RBPJ in the absence of HDAC1.
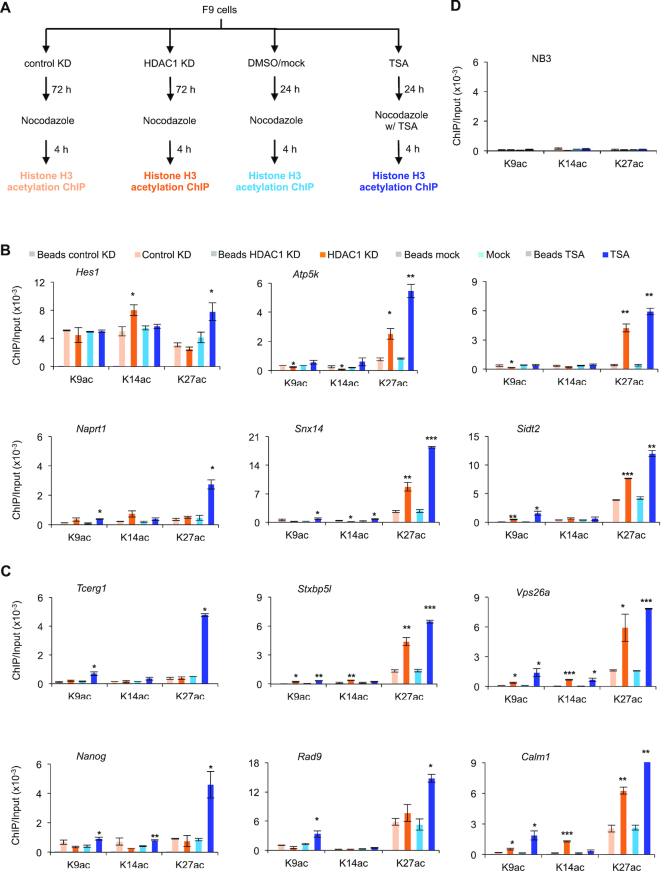
Changes in histone H3 acetylation at lysine 9, 14 and 27 do not correlate with changes in RBPJ occupancy on mitotic chromatin
We next determined if histone modifications contribute to the increased RBPJ occupancy on mitotic chromatin in HDAC1 shRNA expressing cells or in cells treated with TSA. Given that p300 can associate with RBPJ at promoters when the Notch receptor is activated, we examined the status of three p300-mediated histone acetylations (K9, K14 and K27) at RBPJ-occupied sites by ChIP-qPCR (Figure 5 and Supplementary Figure S5). Generally speaking, as compared to HDAC1 knockdown, TSA-treatment consistently increased H3K27Ac at all RBPJ-binding sites tested, and K9Ac or K14Ac at only a subset of loci examined, regardless of whether mitotic chromatin RBPJ occupancy was altered or not. These data argue against the notion that the acetylation status of histone H3 predominantly underlies the increased RBPJ binding on mitotic chromatin induced by TSA treatment (Figure 5B and C) and, therefore, suggests that the histone deacetylation activity of HDAC1 is likely not critical for selective mitotic chromatin binding of RBPJ. Furthermore, it is important to note that the acetylation status of histone H3 in interphase cells is not necessarily faithfully maintained in mitotic cells (compare Figure 5B and C to Supplementary Figure S5).
Figure 5.
Effect of HDAC1 knockdown or TSA treatment on the acetylation status of histone H3K9, 14 and 27 at RBPJ-binding sites in mitotic F9 cells. (A) Experimental scheme. Mitotic cells were collected under the indicated experimental conditions and subjected to ChIP-qPCR analysis using antibodies specific to acetylated histone H3 at K9, K14 or K27. (B) Enrichment of H3K9ac, H3K14ac and H3K27ac at RBPJ binding sites that contain the RBPJ-binding motif. (C) Enrichment of H3K9ac, H3K14ac and H3K27ac at RBPJ binding sites that do not contain the RBPJ-binding motif. (D) Control ChIP assays analysing H3K9ac, H3K14ac and H3K27ac enrichment at a histone H3 nonbinding region (NB3). Shown are means ± SEM from two biological replicates. Paired t-tests were performed to compare enrichment relative to untreated cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
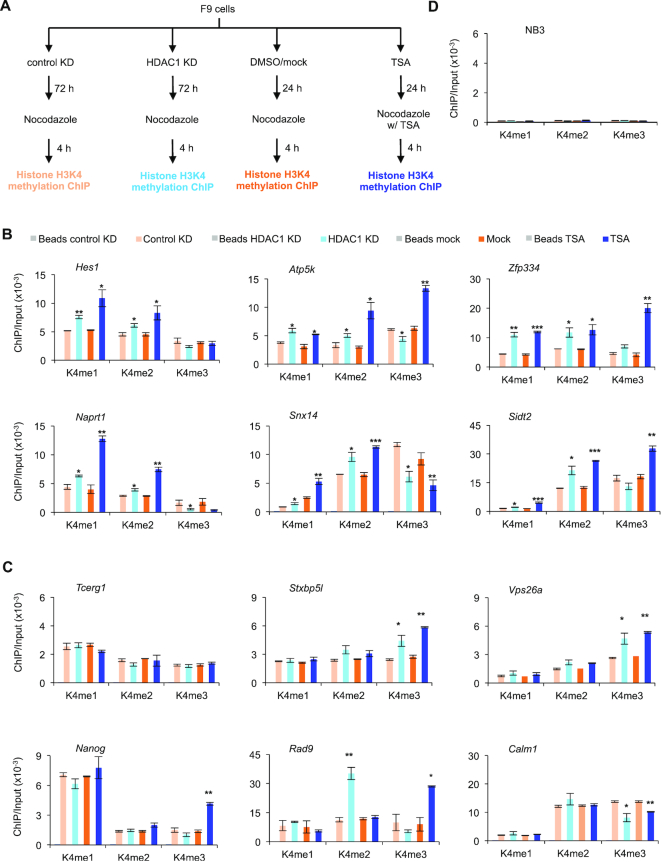
Changes in histone H3K4 methylation correlate with changes in RBPJ occupancy on mitotic chromatin in cells treated with HDAC1 shRNA or TSA
The positive regulation of RBPJ occupancy on mitotic chromatin by KDM5A in HDAC1 knockdown or TSA-treated F9 cells suggests that the status of histone methylation may play a role in the selective binding of RBPJ on mitotic chromatin. The primary function of KDM5A is to demethylate tri- and dimethylated histone H3 at lysine 4. Additionally, treating cells with, TSA can lead to changes in the levels of H3K4 methylation (42–45). Therefore, we examined the status of H3K4 methylation on RBPJ-binding sites using ChIP-qPCR in F9 cells treated with shRNA targeting HDAC1 or TSA (Figure 6 and Supplementary Figure S6).
Figure 6.
HDAC1 knockdown or TSA treatment changes the methylation status of histone H3 lysine 4 at sites containing the RBPJ-binding consensus in mitotic F9 cells. (A) Experimental scheme. ChIP-qPCR was performed to analyse the enrichment of histone H3 differentially methylated at lysine 4 in mitotic cells after HDAC1 knockdown or TSA treatment. (B) H3K4 methylation enrichment at RBPJ-binding sites that contain the RBPJ-binding motif. (C) H3K4 methylation enrichment at RBPJ-binding sites that do not contain the RBPJ-binding motif. (D) Control ChIP assays analysing methylated H3K4 enrichment at a histone H3 nonbinding region (NB3). Shown are means ± SEM from two biological replicates. Paired t-tests were used to compare enrichment relative to control cells. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Upon treatment, we found increased H3K4me1 and H3K4me2 at RBPJ-binding sites in the promoters of the Hes1, Atp5k, Zfp334, Naprt1, Snx14 and Sidt2 genes during mitosis, which contain RBPJ-binding sites (Figure 6A and B). In contrast, we observed no change in H3K4me1 or H3K4me2 status in the promoters of Tcerg1, Stxbp5l, Vps26a, Nanog, Rad9 or Calm1, which do not contain RBPJ-binding motifs (Figure 6C). The increase in H3K4me1 and/or H3K4me2 correlated well with increased RBPJ and KDM5A occupancy at these sites on mitotic chromatin in HDAC1 knockdown or TSA-treated F9 cells (Figures 2-3). While we also observed a decrease in H3K4me3 at RBPJ-binding sites in the promoters of Atp5k, Naprt1 and Snx14, we did not find changes in H3K4me3 levels in Hes1, Zfp334, and Sidt2 (Figure 6B and C). Moreover, we did not observe any consistent change in the methylation levels of H3K4 at RBPJ-binding sites in the Tcerg1, Stxbp5l, Vsp26a, Nanog, Rad9 or Calm1 promoters during mitosis (Figure 6C), where we also did not observe any increase in mitotic chromatin RBPJ occupancy in HDAC1 knockdown or TSA-treated cells (Figure 2H). Together, these results indicate that increased mitotic RBPJ occupancy at sites containing RBPJ-binding motifs is associated with increased histone H3K4 mono and dimethylation.
Increased mitotic chromatin RBPJ occupancy induced by HDAC1 knockdown or TSA treatment increases the rate of transcription reactivation upon mitotic exit
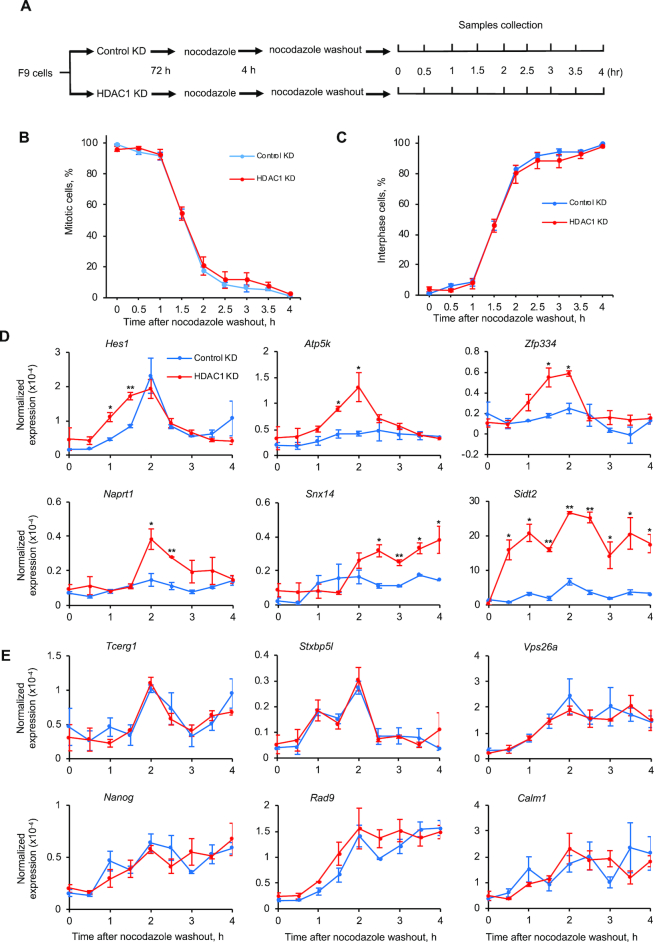
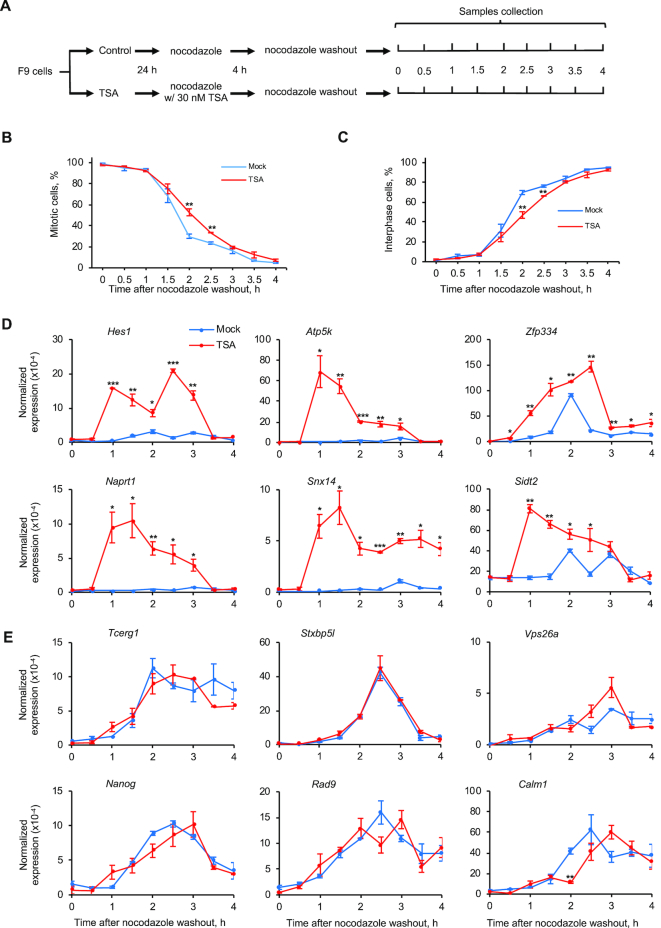
HDAC1 knockdown and TSA treatment selectively increase RBPJ occupancy on mitotic chromatin (Figure 2). To determine how the altered mitotic chromatin binding of RBPJ impacts transcription reactivation upon mitotic exit, we collected RNA from F9 cells expressing HDAC1 shRNA or treated with TSA at different time points after release of nocodazole-arrested cells (Figures 7 and 8).
Figure 7.
Nascent transcript production in HDAC1 knockdown cells at different times after nocodazole release. (A) Experimental scheme. (B) Percentage of cells in mitosis at different times after nocodazole removal. A minimum of 500 DAPI-stained cells was examined for each time point. (C) Percentage of cells in interphase. (D, E) RT-qPCR analyses revealing nascent transcript expression levels relative to actin B. (D) Nascent transcript expression levels from genes that contain the RBPJ-binding motif in their promoters. (E) Nascent transcript expression levels from genes that do not contain the RBPJ-binding motif in their promoters. Shown are means ± SEM from two biological replicates. Paired t-tests were used to compare nascent transcripts level in cells with HDAC1 knockdown relative to control knockdown cells at the indicated time point. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Figure 8.
Nascent transcript production in TSA-treated cells at different times after nocodazole release. (A) Experimental scheme. (B) Percentage of cells in mitosis at different times after nocodazole removal. (C) Percentage of cells in interphase. A minimum of 500 DAPI-stained cells was examined for each time point. (D, E) RT-qPCR analyses revealing nascent transcript expression levels relative to actin B. (D) Nascent transcript expression levels from genes that contain the RBPJ-binding motif in their promoters. (E) Nascent transcript expression levels from genes that do not contain the RBPJ-binding motif in their promoters. Shown are means ± SEM from two biological replicates. Paired t-tests were used to compare nascent transcripts level in cells with TSA treatment relative to control cells at the indicated time point. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
As shown in Figure 7, we observed higher levels of primary transcripts originating from the Hes1 and Sidt2 genes in HDAC1 knockdown cells, as compared to control cells, as early as the first hour after nocodazole washout, when more than 90% of the cells were still in mitosis (Figure 7B). Additionally, we observed more primary transcript from the Apt5k and Zfp334 genes as early as 1.5 h after nocodazole washout, when ∼50% of the cells were still in mitosis. We also observed more nascent transcripts originating from the Naprt1 and Snx14 genes at 2 and 2.5 h, respectively, after nocodazole release, when more than 80% of the cells were in interphase (Figure 7C). On the other hand, we did not observe any change in the levels of nascent transcripts originating from the Tcerg1, Stxbp5l, Vps26a, Nanog, Rad9, Calm1 genes, where mitotic RBPJ occupancy did not change significantly in cells treated with HDAC1 shRNA (Figures 7E and 2H). These results are consistent with the notion that increased mitotic RBPJ occupancy leads to higher levels of RBPJ-dependent transcription upon mitotic exit.
Most importantly, we observed a significant increase in RBPJ-chromatin association during mitosis at the Hes1, Atp5k, Zfp334 and Snx14 genes, and this increased occupancy correlated with increased transcription reactivation kinetics of these genes in HDAC1 KD cells (Figures 2F, G and 7D). This, therefore, offers for the first time evidence that increased chromatin binding by RBPJ, specifically, on mitotic chromatin, and not on interphase chromatin, leads to faster transcription reactivation upon mitotic exit.
We next determined the effect of the increased, site-specific RBPJ occupancy on mitotic chromatin induced by TSA treatment on transcription reactivation upon mitotic exit. For these experiments, we removed TSA during nocodazole washout when releasing mitotic arrested cells into the cell cycle (Figure 8). By monitoring cells after nocodazole washout, we found that cells previously treated with TSA displayed slightly delayed kinetics exiting mitosis, specifically, at 2 and 2.5 h after nocodazole washout (Figure 8B-C); these cells had ∼25% and ∼15% more cells remaining in mitosis as compared to untreated cells. However, before 1.5 h or after 3 h post nocodazole washout, the fractions of cells that exited from mitosis were similar, regardless of TSA treatment.
Similar to the results obtained with HDAC1 knockdown (Figure 7), increased RBPJ occupancy induced by TSA treatment correlated with higher levels of nascent transcript production at early times upon mitotic exit (Figure 8D). This increase in the first run of RNA synthesis is specifically associated with increased RBPJ mitotic occupancy, as we observed no increase at the six genes in which the level of mitotic RBPJ occupancy was unchanged after TSA treatment (Figure 8E). Furthermore, TSA treatment had a more pronounced effect on nascent transcript production than HDAC1 knockdown. In fact, we detected a dramatic increase in RNA synthesis as early as 1 h at all six genes assayed, a time when more than 95% of the cells were still in mitosis. Together, these results indicate that accelerating the first runs of RNA synthesis upon mitotic exit may be a functional outcome of the increased site-specific RBPJ occupancy on mitotic chromatin.
DISCUSSION
In this study, we discovered a regulatory mechanism for the selective mitotic chromatin binding of the sequence-specific transcription factor RBPJ. Manipulating the levels of the RBPJ-interacting and histone-modifying enzymes HDAC1 and KDM5A, which are also present on the mitotic chromatin of F9 cells, strikingly alters sequence-specific RBPJ occupancy on mitotic-chromatin. Specifically, we found that HDAC1 knockdown or TSA treatment increases mitotic chromatin RBPJ binding at regions containing the RBPJ-binding consensus sequence. Furthermore, HDAC1 knockdown or TSA treatment also increased the mitotic chromatin occupancy of the histone H3 lysine 4 demethylase KDM5A, with a concomitant increase in the levels of histone H3K4 mono- and dimethylation at these RBPJ binding sites (Figures 3B-C and 6B). Intriguingly, site-specific KDM5A occupancy on mitotic chromatin was absolutely necessary for the increased RBPJ occupancy at these sites (Figure 4), indicating that KDM5A positively regulates mitotic chromatin binding of RBPJ in F9 cells under these growth conditions. Furthermore, the enhanced RBPJ occupancy on mitotic chromatin is unlikely mediated through the status of histone H3 acetylation at lysine's 9, 14 and 27, as evidenced by the absence of consistent changes in H3 acetylation at these lysines correlating with changes in mitotic chromatin occupancy of RBPJ. Our results are consistent with a model in which the site-specific, mitotic-chromatin occupancy of the HDAC1 protein, rather than histone acetylation status, negatively regulates mitotic chromatin occupancy of RBPJ and KDM5A, specifically, at regions containing RBPJ binding DNA consensus (Figures 1C, 2F, H and Supplementary Figure S2).
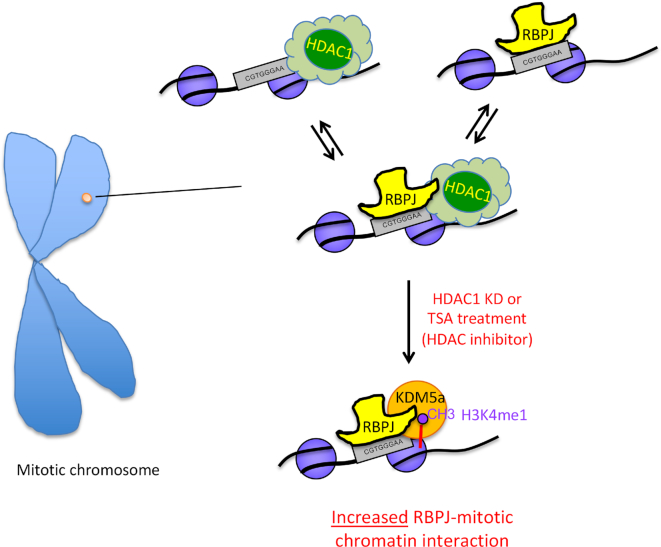
Together, our results uncover a mechanism by which selective mitotic chromatin binding of the sequence-specific transcription factor RBPJ is regulated (Figure 9). RBPJ can recognize its binding consensus located in both nucleosome-free regions or within nucleosomal DNA (18,46). We propose that RBPJ interacts with its binding motif on mitotic chromatin dynamically as in asynchronous cells (38,47). Accordingly, the RBPJ-binding site can exist in at least three states on mitotic chromatin, (i) only RBPJ bound to its binding consensus, (ii) RBPJ bound to its binding consensus in conjunction with an HDAC1-containing complex and (iii) the RBPJ binding site is vacant while an HDAC1-containing complex is bound (Figure 9). Changes in transcription programs resulting from cell-signaling events could lead to changes in HDAC1 levels, activity or induce HDAC1 redistribution. This may lead to enhanced RBPJ-KDM5A interactions. It is important to note that, although RBPJ has been shown to directly interact with KDM5A, the effects we observe here might involve an interaction between RBPJ and a KDM5A-containing complex. Once KDM5A alone or a KDM5A-containing complex binds to chromatin, it can catalyse the demethylation of trimethylated histone H3K4 to the di- and mono-methylated forms (32,48). Histone H3K4me1/me2 may further stabilize the chromatin association of KDM5A (49), and this may lead to lower rates of RBPJ dissociation from mitotic chromatin at sites containing the RBPJ-binding consensus sequence. Alteration in the affinity of RBPJ-containing complexes for mitotic chromatin through binding partner exchange may, therefore, be a mechanism to reinforce specific transcription programs through cell division.
Figure 9.
Model depicting a regulatory mechanism for selective RBPJ binding on mitotic chromatin. RBPJ interacts dynamically with chromatin. The RBPJ-binding site can exist in at least three states: (i) with RBPJ bound, (ii) with RBPJ and an HDAC1-containing complex bound and (iii) with only an HDAC1-containing complex bound. Changes in transcription programs resulting from cell-signaling events could lead to changes in HDAC1 levels, activity or induce HDAC1 redistribution. This may lead to enhanced RBPJ–KDM5A interactions. Once KDM5A binds to RBPJ, it can catalyse the demethylation of trimethylated histone H3K4 to the di- and mono-methylated forms. Histone H3K4me1/me2 may further stabilize the chromatin association of KDM5A, and this may lead to lower rates of RBPJ dissociation from mitotic chromatin at sites containing the RBPJ-binding consensus sequence. Alteration in the affinity of RBPJ-containing complexes for mitotic chromatin through binding partner exchange may, therefore, be a mechanism to reinforce specific transcription programs through cell division.
Different RBPJ-containing complexes likely carry out different transcriptional functions on mitotic chromatin. For example, we previously found that RBPJ can occupy sites containing the CTCF-binding motif, that RBPJ and CTCF interact with high affinity (18), and that this association is necessary for RBPJ occupancy at CTCF-binding sites. Here, we found that HDAC1 impacts mitotic chromatin RBPJ occupancy, specifically, at sites that contain the RBPJ binding motif (Figure 2H). This indicates that RBPJ occupancy at sites which do not contain the RBPJ-binding consensus is not regulated by HDAC1 (Figure 2H). Therefore, different classes of RBPJ-containing complexes are anticipated to be selectively bound on mitotic chromatin through multiple regulatory mechanisms, which likely reflect specific transcription programs. It is important to note that we did not observe the same effect of HDAC1 knockdown on RBPJ and KDM5A occupancy in asynchronous cells as compared to mitotic cells (Figures 2F, G and 3B), indicating that, while HDAC1 has a critical effect on regulating RBPJ and KDM5A association with mitotic chromatin, additional factors are involved in controlling chromatin association of RBPJ and KDM5A in asynchronous cells.
Our results are consistent with a model in which stronger protein-protein and protein-histone interactions may underlie the selective binding of bookmarking factors on mitotic chromatin. In the case of murine embryonal carcinoma cell line F9, HDAC1 may play a critical role in maintaining specific RBPJ-dependent transcriptional program by regulating mitotic RBPJ occupancy at sites containing RBPJ binding DNA consensus. Upon HDAC1 knockdown, F9 cells may adopt an altered transcriptional state, and the enhanced mitotic chromatin occupancy of RBPJ and KDM5A may represent part of a mechanism by which an altered transcriptional program can be propagated through cell division. Indeed, HDAC1 is known to control the proliferation and differentiation of mouse embryonic stem cells (50,51).
By characterizing RBPJ knockout cells, we have found that RBPJ is critical to F9 cell identity. As shown in Supplementary Figure S7A, the RBPJ knockout cell line RBPJKO17 displays a slower growth rate (Supplementary Figure S7A) and altered differentiation kinetics upon retinoic acid (RA) treatment (Supplementary Figure S7B and C). Cells without RBPJ lose the stem cell marker OCT4 faster than the RBPJwt cells when treated with RA. Moreover, the knockout cells do not express the markers of endoderm differentiation, RUNX1 and GATA4, as robustly as RBPJwt cells after 5 days of RA treatment. Collectively, these results indicate that RBPJ is critical for growth and differentiation of F9 cells, and support a hypothesis that mitotic chromatin occupancy of RBPJ may contribute to the maintenance of mitotic transcriptional memory and, consequently, cell identity through cell division.
To assess the Notch signaling status in F9 cells, we profiled the expression of 10 Notch-induced target genes, as defined by Rao et al. (2009) (52,53). As shown in Supplementary Figure S8, expression of the signature Notch-dependent genes, Hes1, Hey1, Hey2, Dtx1, Nrarp and Myc is increased in RBPJ KO cells, suggesting that RBPJ functions as a transcriptional repressor. We next treated F9 cells with the γ-secretase inhibitor DAPT at 25, 50 and 100 μM for 72 h, and then assayed for the expression of ten Notch-target genes (Supplementary Figure S9). If Notch were active in F9 cells, treating cells with DAPT should result in inhibition of Notch signaling. As shown in Supplementary Figure S9a, treating cells with DAPT had no significant effect on the expression of these ten genes, supporting our conclusion that Notch signaling is inactive in F9 cells. To control for DAPT activity, we activated Notch by treating F9 cells with 4 mM EDTA for 1 h (yellow in Supplementary Figure S9b), as EDTA has been demonstrated to activate Notch signaling (38,54,55). This mode of receptor activation is believed to occur through EDTA-induced dissociation of the extracellular domain of the single-pass Notch receptor. Treating cells with EDTA did cause the upregulation of three known Notch target genes (yellow, Supplementary Figure S9b). Including DAPT with EDTA treatment inhibited the effect of EDTA (green, Supplementary Figure S9b) on the expression of these genes, demonstrating that DAPT was active. Together these results indicate Notch signaling is inactive in F9 cells under our culture condition.
Future experiments in which Notch signaling is activated and the status of RBPJ binding to mitotic chromatin is examined will be of great interest. Given that Naprt1 has not yet been described as a Notch target but is, nonetheless, regulated by RBPJ upon HDAC1 KD or TSA treatment (Figures 7 and 8), we do not expect all RBPJ bookmarked sites would be modulated by Notch signaling. Moreover, both HDAC1 and KDM5A may be components of different transcriptional repressive complexes. Therefore, it will be interesting to identify and characterize different RBPJ-containing complexes on mitotic chromatin, to provide additional insights into the mechanisms by which RBPJ contributes to the transmission of specific transcription programs through cell division.
The minimal function of a mitotic bookmarking factor is considered to be altering the kinetics of transcriptional reactivation upon mitotic exit (6–12,16,17,19). Altering mitotic chromatin RBPJ occupancy by manipulating the levels of HDAC1 (Figures 7 and 8) increased the binding of RBPJ at specific loci and increased the kinetics of transcription reactivation upon mitotic exit, supporting the notion that RBPJ functions as a bona fide bookmarking factor.
A key issue that arises from our study is: how does one define a bookmarked site and whether all the mitotic binding sites of a transcription factor are important or functionally equivalent? Our data suggest that it is not necessarily the relative amount of RBPJ bound between asynchronous and mitotic chromatin that is critical, but whether or not there is a direct functional consequence when the mitotic chromatin occupancy is altered. Upon HDAC1 knockdown, we detected faster transcription reactivation kinetics from both genes, where increased RBPJ occupancy was observed (Figure 7). The basic paradigm is that sites which display increased RBPJ enrichment on mitotic chromatin (Hes1, Atp5k, Zfp334, Naprt1, Snx14 and Sidt2) have faster transcription reactivation kinetics. In contrast, sites that do not show a change in RBPJ enrichment (Tcerg1, Stxbp5l, Vps26a, Nanog, Rad9 and Calm1), do not show a statistically significant difference in transcription reactivation kinetics upon mitotic exit. Therefore, we have established a direct functional correlation between the extent of mitotic chromatin occupancy and rates of transcription reactivation.
Importantly, our methods to manipulate the binding of a bookmarking factor make a significant advance from the other approaches available to date, evaluating the impact of mitotic bookmarking on transcription reactivation kinetics. To address this question rigorously, one needs to alter protein-DNA interactions specifically during mitosis and assay the impact of this alteration on transcription reactivation, which is extremely challenging technically. Before this work, the best method to address this question is to construct a fusion protein with a degron, which will specifically degrade a protein during mitosis. Meticulously designed studies by Kadauke et al. (17) found that mitosis-specific GATA1 degradation led to a delay in transcription re-activation, consistent with the hypothesis that mitotic bookmarking by GATA1 increases transcription reactivation kinetics. Nonetheless, there is one technical limitation associated with this methodology; that is the need to re-synthesize GATA1 in G1. Therefore, a delay in transcription reactivation is anticipated using this method. In strong contrast, we have developed a more rigorous approach that bypasses the need for protein re-synthesis in G1. As opposed to decreasing mitotic chromatin occupancy, we can enhance RBPJ binding to mitotic chromatin at select sites by decreasing HDAC1 levels (Figure 2F). Significantly we observe faster kinetics of transcription reactivation specifically from these sites (Figures 7 and 8), but not from sites where RBPJ occupancy does not change in response to HDAC1 knockdown (Figure 2H).
Formaldehyde cross-linking is known to dissociate a fraction of DNA binding proteins from mitotic chromatin; however, the fraction of proteins that survive cross-linking by formaldehyde likely represent more stable interactions (56). Here, we discovered that HDAC1 and KDM5A, two components of RBPJ-interacting complexes, both with critical functions in transcription regulation, modulate the selective mitotic chromatin binding of RBPJ. Therefore, our study revealed that enhanced RBPJ-mitotic chromatin binding can arise through selective protein-protein interactions and that interactions between transcriptional regulators on mitotic chromatin can govern transcriptional output upon mitotic exit.
Supplementary Material
ACKNOWLEDGEMENTS
Support for microscopy imaging was provided by the University of New Mexico Cancer Center Fluorescence Microscopy Shared Resource, funded by NCI 2P30 CA118100 and NIGMS 5P50 GM085273. We thank Marisa Bartolomei, Joanne Thorvaldsen and Aimee Juan for their help in creating the RBPJ KO lines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Heart Association [17GRNT33400020]; Cancer Center Support Grant [P30CA118100 to H.Y.F., R.J.L., K.D.]. Funding for open access charge: Center for Strategic Scientific Initiatives, National Cancer Institute [P30CA118100].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ohta S., Bukowski-Wills J.C., Sanchez-Pulido L., Alves Fde L., Wood L., Chen Z.A., Platani M., Fischer L., Hudson D.F., Ponting C.P. et al.. The protein composition of mitotic chromosomes determined using multiclassifier combinatorial proteomics. Cell. 2010; 142:810–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Martinez-Balbas M.A., Dey A., Rabindran S.K., Ozato K., Wu C.. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell. 1995; 83:29–38. [DOI] [PubMed] [Google Scholar]
- 3. Wang F., Higgins J.M.. Histone modifications and mitosis: countermarks, landmarks, and bookmarks. Trends Cell Biol. 2013; 23:175–184. [DOI] [PubMed] [Google Scholar]
- 4. Liu Y., Pelham-Webb B., Di Giammartino D.C., Li J., Kim D., Kita K., Saiz N., Garg V., Doane A., Giannakakou P. et al.. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 2017; 19:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Javasky E., Shamir I., Gandhi S., Egri S., Sandler O., Rothbart S.B., Kaplan N., Jaffe J.D., Goren A., Simon I.. Study of mitotic chromatin supports a model of bookmarking by histone modifications and reveals nucleosome deposition patterns. Genome Res. 2018; 28:1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kadauke S., Blobel G.A.. Mitotic bookmarking by transcription factors. Epigenet. Chromatin. 2013; 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zaret K.S. Genome reactivation after the silence in mitosis: recapitulating mechanisms of development. Dev. Cell. 2014; 29:132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. John S., Workman J.L.. Bookmarking genes for activation in condensed mitotic chromosomes. Bioessays. 1998; 20:275–279. [DOI] [PubMed] [Google Scholar]
- 9. Xing H., Wilkerson D.C., Mayhew C.N., Lubert E.J., Skaggs H.S., Goodson M.L., Hong Y., Park-Sarge O.K., Sarge K.D.. Mechanism of hsp70i gene bookmarking. Science. 2005; 307:421–423. [DOI] [PubMed] [Google Scholar]
- 10. Zaidi S.K., Young D.W., Montecino M.A., Lian J.B., van Wijnen A.J., Stein J.L., Stein G.S.. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat. Rev. Genet. 2010; 11:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Young D.W., Hassan M.Q., Yang X.Q., Galindo M., Javed A., Zaidi S.K., Furcinitti P., Lapointe D., Montecino M., Lian J.B. et al.. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3189–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dey A., Nishiyama A., Karpova T., McNally J., Ozato K.. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell. 2009; 20:4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Festuccia N., Gonzalez I., Owens N., Navarro P.. Mitotic bookmarking in development and stem cells. Development. 2017; 144:3633–3645. [DOI] [PubMed] [Google Scholar]
- 14. Raccaud M., Suter D.M.. Transcription factor retention on mitotic chromosomes: regulatory mechanisms and impact on cell fate decisions. FEBS Lett. 2018; 592:878–887. [DOI] [PubMed] [Google Scholar]
- 15. Festuccia N., Owens N., Navarro P.. Esrrb, an estrogen-related receptor involved in early development, pluripotency, and reprogramming. FEBS Lett. 2018; 592:852–877. [DOI] [PubMed] [Google Scholar]
- 16. Caravaca J.M., Donahue G., Becker J.S., He X., Vinson C., Zaret K.S.. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013; 27:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kadauke S., Udugama M.I., Pawlicki J.M., Achtman J.C., Jain D.P., Cheng Y., Hardison R.C., Blobel G.A.. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012; 150:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lake R.J., Tsai P.F., Choi I., Won K.J., Fan H.Y.. RBPJ, the major transcriptional effector of Notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking. PLos Genet. 2014; 10:e1004204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teves S.S., An L., Bhargava-Shah A., Xie L., Darzacq X., Tjian R.. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife. 2018; 7:35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Festuccia N., Dubois A., Vandormael-Pournin S., Gallego Tejeda E., Mouren A., Bessonnard S., Mueller F., Proux C., Cohen-Tannoudji M., Navarro P.. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat. Cell Biol. 2016; 18:1139–1148. [DOI] [PubMed] [Google Scholar]
- 21. Deluz C., Friman E.T., Strebinger D., Benke A., Raccaud M., Callegari A., Leleu M., Manley S., Suter D.M.. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 2016; 30:2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fortini M.E., Artavanis-Tsakonas S.. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994; 79:273–282. [DOI] [PubMed] [Google Scholar]
- 23. Alonso A., Breuer B., Steuer B., Fischer J.. The F9-EC cell line as a model for the analysis of differentiation. Int. J. Dev. Biol. 1991; 35:389–397. [PubMed] [Google Scholar]
- 24. Tun T., Hamaguchi Y., Matsunami N., Furukawa T., Honjo T., Kawaichi M.. Recognition sequence of a highly conserved DNA binding protein RBP-J kappa. Nucleic Acids Res. 1994; 22:965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kovall R.A., Hendrickson W.A.. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004; 23:3441–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kao H.Y., Ordentlich P., Koyano-Nakagawa N., Tang Z., Downes M., Kintner C.R., Evans R.M., Kadesch T.. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998; 12:2269–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oswald F., Kostezka U., Astrahantseff K., Bourteele S., Dillinger K., Zechner U., Ludwig L., Wilda M., Hameister H., Knochel W. et al.. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 2002; 21:5417–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hsieh J.J., Zhou S., Chen L., Young D.B., Hayward S.D.. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mulligan P., Yang F., Di Stefano L., Ji J.Y., Ouyang J., Nishikawa J.L., Toiber D., Kulkarni M., Wang Q., Najafi-Shoushtari S.H. et al.. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell. 2011; 42:689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moshkin Y.M., Kan T.W., Goodfellow H., Bezstarosti K., Maeda R.K., Pilyugin M., Karch F., Bray S.J., Demmers J.A., Verrijzer C.P.. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol. Cell. 2009; 35:782–793. [DOI] [PubMed] [Google Scholar]
- 31. Di Stefano L., Walker J.A., Burgio G., Corona D.F., Mulligan P., Naar A.M., Dyson N.J.. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011; 25:17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liefke R., Oswald F., Alvarado C., Ferres-Marco D., Mittler G., Rodriguez P., Dominguez M., Borggrefe T.. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010; 24:590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Artavanis-Tsakonas S., Rand M.D., Lake R.J.. Notch signaling: cell fate control and signal integration in development. Science. 1999; 284:770–776. [DOI] [PubMed] [Google Scholar]
- 34. Bray S., Bernard F.. Notch targets and their regulation. Curr. Top. Dev. Biol. 2010; 92:253–275. [DOI] [PubMed] [Google Scholar]
- 35. Aster J.C., Pear W.S., Blacklow S.C.. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017; 12:245–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oswald F., Tauber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R.M.. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 2001; 21:7761–7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu L., Aster J.C., Blacklow S.C., Lake R., Artavanis-Tsakonas S., Griffin J.D.. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000; 26:484–489. [DOI] [PubMed] [Google Scholar]
- 38. Krejci A., Bray S.. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007; 21:1322–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lake R.J., Boetefuer E.L., Won K.J., Fan H.Y.. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 2016; 44:2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoshida M., Kijima M., Akita M., Beppu T.. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 1990; 265:17174–17179. [PubMed] [Google Scholar]
- 42. Henrique R., Oliveira A.I., Costa V.L., Baptista T., Martins A.T., Morais A., Oliveira J., Jeronimo C.. Epigenetic regulation of MDR1 gene through post-translational histone modifications in prostate cancer. BMC Genomics. 2013; 14:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pufahl L., Katryniok C., Schnur N., Sorg B.L., Metzner J., Grez M., Steinhilber D.. Trichostatin A induces 5-lipoxygenase promoter activity and mRNA expression via inhibition of histone deacetylase 2 and 3. J. Cell. Mol. Med. 2012; 16:1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu F.R., Liu Y., Shang M.B., Yang X.X., Ding B., Gao J.G., Wang R., Li W.Y.. Differences in H3K4 trimethylation in in vivo and in vitro fertilization mouse preimplantation embryos. Genet. Mol. Res.: GMR. 2012; 11:1099–1108. [DOI] [PubMed] [Google Scholar]
- 45. Marinova Z., Leng Y., Leeds P., Chuang D.M.. Histone deacetylase inhibition alters histone methylation associated with heat shock protein 70 promoter modifications in astrocytes and neurons. Neuropharmacology. 2011; 60:1109–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rozenberg J.M., Taylor J.M., Mack C.P.. RBPJ binds to consensus and methylated cis elements within phased nucleosomes and controls gene expression in human aortic smooth muscle cells in cooperation with SRF. Nucleic Acids Res. 2018; 46:8232–8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castel D., Mourikis P., Bartels S.J., Brinkman A.B., Tajbakhsh S., Stunnenberg H.G.. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013; 27:1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K.. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007; 128:1063–1076. [DOI] [PubMed] [Google Scholar]
- 49. Torres I.O., Kuchenbecker K.M., Nnadi C.I., Fletterick R.J., Kelly M.J., Fujimori D.G.. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 2015; 6:6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dovey O.M., Foster C.T., Cowley S.M.. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:8242–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lagger G., O’Carroll D., Rembold M., Khier H., Tischler J., Weitzer G., Schuettengruber B., Hauser C., Brunmeir R., Jenuwein T. et al.. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002; 21:2672–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rao S.S., O’Neil J., Liberator C.D., Hardwick J.S., Dai X., Zhang T., Tyminski E., Yuan J., Kohl N.E., Richon V.M. et al.. Inhibition of NOTCH signaling by gamma secretase inhibitor engages the RB pathway and elicits cell cycle exit in T-cell acute lymphoblastic leukemia cells. Cancer Res. 2009; 69:3060–3068. [DOI] [PubMed] [Google Scholar]
- 53. Grabher C., von Boehmer H., Look A.T.. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2006; 6:347–359. [DOI] [PubMed] [Google Scholar]
- 54. Rand M.D., Grimm L.M., Artavanis-Tsakonas S., Patriub V., Blacklow S.C., Sklar J., Aster J.C.. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol. 2000; 20:1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li J., Housden B.E., Bray S.J.. Notch signaling assays in Drosophila cultured cell lines. Methods Mol. Biol. 2014; 1187:131–141. [DOI] [PubMed] [Google Scholar]
- 56. Teves S.S., An L., Hansen A.S., Xie L., Darzacq X., Tjian R.. A dynamic mode of mitotic bookmarking by transcription factors. Elife. 2016; 5:e22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.