Abstract
Posttranscriptional RNA modifications occur in all domains of life. Modifications of anticodon bases are of particular importance for ribosomal decoding and proteome homeostasis. The Elongator complex modifies uridines in the wobble position and is highly conserved in eukaryotes. Despite recent insights into Elongator's architecture, the structure and function of its regulatory factor Kti12 have remained elusive. Here, we present the crystal structure of Kti12′s nucleotide hydrolase domain trapped in a transition state of ATP hydrolysis. The structure reveals striking similarities to an O-phosphoseryl-tRNA kinase involved in the selenocysteine pathway. Both proteins employ similar mechanisms of tRNA binding and show tRNASec-dependent ATPase activity. In addition, we demonstrate that Kti12 binds directly to Elongator and that ATP hydrolysis is crucial for Elongator to maintain proper tRNA anticodon modification levels in vivo. In summary, our data reveal a hitherto uncharacterized link between two translational control pathways that regulate selenocysteine incorporation and affect ribosomal tRNA selection via specific tRNA modifications.
INTRODUCTION
The eukaryotic Elongator complex (Elp1-Elp6) was originally isolated from yeast in association with actively transcribing RNA polymerase II (1) and hence implicated in transcription elongation (2). However, to date a robust body of scientific evidence indicates that the major, if not genuine, role of Elongator lies with its tRNA modification activity (3–5). Accordingly, Elongator binds tRNAs with different subunits (6–8) and constitutes the central element of a conserved tRNA modification pathway that is functionally exchangeable between eukaryotic organisms (4,5,9). In detail, the Elongator complex attaches carboxymethyl (cm5) groups to uridine bases located in the wobble position (U34) of tRNA anticodons. This pivotal substitution can be further converted to 5-carbamoyl-methyl (ncm5), 5-methoxy-carbamoyl-methyl (mcm5) or 5-methoxy-carbamoyl-methyl-2-thio (mcm5s2) uridines by other modification cascades (4,10).
Physiologically, the modified wobble uridines ensure efficient tRNA selection in the ribosomal A-site (11,12), promote proper anticodon–codon interaction and guarantee reading frame maintenance during mRNA translation (13–15). Elongator inactivation by mutations or subunit deletion lead to pleiotropic phenotypes (16) due to improper translation (17), altered co-translational folding dynamics (18) and disturbed proteome homeostasis (11,19). Furthermore, altered U34 modification levels are found in animal disease models (20–22) as well as in patients suffering from certain forms of cancer (23–26) and different neurodegenerative diseases (27–29), thus reinforcing the importance of Elongator for cell survival and proliferation control.
Hence, one might expect that Elongator is constitutively active to ensure proper protein synthesis rates at all times. Nevertheless, recent studies in yeast show that U34 modification levels do in fact oscillate during the cell cycle and in response to varying environmental conditions (30–34). Moreover, genetic screens (35,36) helped identifying accessory proteins, which transiently interact with Elongator and are as important for U34 modifications as the core Elongator complex itself (37–39). In particular, an isoform of the casein kinase 1 (also known as Hrr25 or Kti14), the Sit4 protein phosphatase as well as Kluyveromyces lactis Toxin Insensitive 12 (Kti12), were shown to affect the phosphorylation pattern of Elongator (40–42). Specific phosphorylation sites in Elp1 are critical for proper tRNA modification activity and the recruitment of Hrr25 and Kti12 to Elongator (25,40). Thus, the activity of Elongator appears to be dynamically regulated by a complicated equilibrium of phosphorylation and de-phosphorylation events.
Sequence similarities suggest that Kti12 shares a common ancestor with O-phosphoseryl-tRNA kinase (PSTK), which in archaea and many eukaryotes is required for synthesis of tRNASec and incorporation of selenocysteine (Sec) into selenoproteins (43,44). Sec, the 21st naturally occurring amino acid, lacks its own specific tRNA synthetase and is not directly encoded by the standard genetic code. Instead, the translating ribosome specifically incorporates Sec after recognizing ‘selenocysteine insertion sequences’ in the mRNA using tRNASec at recoded UGA stop codons. The respective tRNASec is initially charged with serine (tRNA[Ser]Sec) by seryl-tRNA synthetase (SerRS), phosphorylated by PSTK (tRNA[Sep]Sec) and finally converted to selenocysteine tRNA (tRNA[Sec]Sec) by SecS synthetase (45–47). PSTK specifically phosphorylates tRNA[Ser]Sec due to its unique structure (48,49) and does not accept serine attached to canonical tRNASer (44,45).
Here, we present the crystal structure of the N-terminal domain of Kti12 from Chaetomium thermophilium (CtKti12NTD) in complex with ADP·AlF3 and demonstrate that Kti12 binds to tRNAs in a PSTK-like fashion. Strikingly, the ATPase activity of Kti12 is tRNA dependent, specifically induced by tRNASec and requires the 3′CCA maturation signal. Structure-guided mutations within the catalytic cavity and tRNA interaction surfaces of Kti12 validate our comprehensive model of Kti12 and reveal that its ATPase activity is essential for anticodon modification by the Elongator complex. In summary, we identify yet uncharacterized similarities between two translational control pathways that regulate the synthesis of selenoproteins and translation elongation via cm5U modifications in the wobble base position.
MATERIALS AND METHODS
Protein expression and purification
Genes encoding Kti12 proteins from Saccharomyces cerevisiae (ScKti12) and C. thermophilum (CtKti12) were cloned into pETM24d vector, which adds a C-terminal 6x-His tag to the protein of interest. Proteins were expressed in the Escherichia coli pRARE strain at 18°C using overnight induction with 1 mM IPTG. Bacterial cells expressing Kti12 protein were suspended in an ice-cold 50 mM Tris–HCl pH 7.5 buffer containing 300 mM NaCl, 10 mM imidazole, 2 mM DTT, 2 mM MgCl2 and a cocktail of protease inhibitors. After addition of lysozyme and DNAse the suspension was sonicated to homogeneity and centrifuged for 30 min at 87 000 g. The supernatant obtained was next subjected to sequential Kti12 purification at 4°C on NiNTA (Qiagen), HiTrap Q (GE Healthcare) and Superdex 200 10/300 GL (GE Healthcare) columns. In all cases, the purification protocol yielded protein samples close to homogeneity. The ScKti12 fusion construct (ScKti12-5x[GS]–first WD40-6xHis) was purified similarly to ScKti12 WT protein. Use of a HiTrap Heparin column (GE Healthcare) after the HiTrap Q step yielded a homogenous protein sample that was subsequently refined on a Superdex 200 column. Full length Elp1 protein and its domains were expressed in Escherichia coli pRARE strain from a pETM30 expression vector that introduces an N-terminal 6xHis-GST tag followed by a TEV cleavage site. The bacterial pellet was resuspended in an identical buffer without imidazole and lysed using a High Pressure Homogenizer Emulsiflex C3 (Avestin). After centrifugation supernatant was circled overnight on a GSTPrep FF 16/10 (GE Healthcare) column. The column was washed with lysis buffer and protein was eluted using a buffer that additionally contained 18 mM glutathione. Overnight dialysis in the presence of GST-TEV protease resulted in tag cleavage. GST tag and GST-TEV were removed from the solution during subsequent rebinding to the GSTPrep column. Both tagged and tag-free versions of Elp1 were further purified on Superdex 200 10/300 GL column (GE Healthcare).
Crystallization and data collection
Homogeneous samples of CtKti12NTD were concentrated and mixed with 3 mM ADP, 6 mM AlCl3 and 60 mM NaF to a final protein concentration of 45 mg/ml. The crystals were obtained after 3 days using the sitting-drop vapor diffusion method. Mother liquor contained 100 mM MMT buffer (DL-malic acid, MES monohydrate, TRIS at pH 5.5) and 25% (w/v) PEG1500. Harvested crystals were cryo-protected in an identical solution containing 30% (v/v) glycerol. For experimental phase determination, a JBS Magic Triangle® (50) (Jena Bioscience) compound was added to the cryo-protectant. X-ray diffraction data were collected at MX-14-2 at BESSY II synchrotron source in Berlin, Germany.
Structure determination and refinement
The crystal structure of CtKti12NTD•ADP/AlF3/Mg2+ was solved by the SAD method using crystals soaked with JBS Magic Triangle. Native and phasing datasets were processed with XDS package (51). An anomalous difference map revealed the positions of six iodine atoms in the vicinity of protein. Shelx, Resolve and Phaser software (52–54) were used during initial phase determination and initial density modification steps. The Coot program (55) was used for manual building of a model that suits the obtained electron density map. Preliminary structures were refined against diffraction data using PHENIX software (54).
Visualization of protein structure and bioinformatics
Sequence alignments were performed using Jalview software (56). Molecules were visualized using PyMol software (57). Protein conservation was analyzed using ConSurf server (58). Electrostatic potential of the protein surface was estimated using APBS (59).
Mutagenesis and expression of Kti12 mutants
The QuickChange approach was used to perform an alanine screening of several positions within CtKti12, namely K14, T15, T16, D77, R84, K106/R109, D129Y, Y133/I134, K135, Y139, R171, Y202, E203, R213, R222/W223, W232. Corresponding residues in ScKti12 were identified using multiple sequence alignment and homolog-based modelling information, namely K14, T15, T16, D45, H52, R62/R65, D85, Y89/I90, K91, Y95, K125, W132, E133, R143, R152/W153 and W123 respectively. All CtKti12 protein mutants were purified using standard purification protocols described above for the wild type (WT) protein.
In vitro transcription of tRNA
Single tRNA species used herein were transcribed by T7 RNA polymerase from a linearized pUC19 vector harboring the corresponding tRNA genes under T7 promoter control. In vitro transcribed tRNA species were subsequently purified on HiTrap DEAE FF columns (GE Healthcare) and precipitated overnight in isopropanol. After removal of supernatant, tRNAs were annealed by gradual cooling from 80°C to 40°C at a speed of 0.5°C/min in the presence of annealing buffer composed of 200 mM HEPES pH 7.5, 500 mM NaCl and 500 mM KCl. Next, the mixtures were enriched with 1 mM MgCl2 and 1 mM DTT and tRNA species were additionally purified by size-exclusion chromatography on Superdex 75 10/300 GL column.
Measurement of Kti12 nucleotide binding affinity
Parameters of nucleotide binding by CtKti12 were dissected using MANT-labeled (2′/3′-O-(N-Methyl-anthraniloyl), Jena Bioscience) ATP or its non-hydrolysable analog, AMPPNP (adenosine-5′-[(β,γ)-imido] triphosphate). 4 μM protein was titrated with two-fold serial dilutions of MANT-labeled nucleotide. MANT concentrations ranged from 100 μM to 50 nM and the final volume of the mixture was 100 μl. Affinity to the ATP and AMPPNP in the presence of tRNASec was determined using only 1 μM CtKti12 protein and 1 μM tRNASec. In experiments involving tRNASec and CtKti12NTD/CTD three-fold serial dilutions covered a range between 15 μM and 61 nM. Protein was incubated with a nucleotide and incubated for 30 min at 37°C, then the mixture was transferred to a 96-well, black, flat-bottom, chimney-well microplate and incubated for additional 2 min inside of the preheated to 37°C SpectraMAX GeminiEM plate reader (Molecular Devices). FRET was induced using λex: 280 nm incident light, MANT fluorescence was measured at λem: 448 nm. Data for protein (λex: 280 nm; λem: 340 nm) and fluorophore (λex: 340 nm; λem: 448 nm) fluorescence were also collected as a control of protein and fluorophore amount. FRET (λex: 280 nm; λem: 448 nm) values after subtraction of respective protein-free controls are shown, all experiments were conducted in triplicates.
Measurement of Kti12 enzymatic activity
Nucleotide hydrolysis capacity of Kti12 proteins was investigated using the Malachite Green Phosphate Assay Kit (Sigma Aldrich). Reactions were performed in a buffer composed of 20 mM Tris–HCl, 150 mM NaCl, 2 mM DTT, 2 mM MgCl2 and 900 μM nucleotide. Final protein and tRNA concentrations were adjusted to 1 μM. In case of reactions conducted in the presence of Elongator subunit Elp1, the protein was added to the reaction mixture at indicated molar ratios. Assembled reactions were incubated for 15 hours at 37°C. After incubation, the reaction mixtures were diluted 10 times in water and the phosphate concentrations were measured according to a standard absorption protocol at λ = 620 nm. The data are presented as the mean ± standard deviation of three independent experiments with at least 2 technical replicas each.
Thermal shift assay
The thermostability of Kti12 proteins was assessed using thermal shift assays in the presence or absence of nucleotides. In each case, 7 μg of protein was incubated in 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT, 2 mM MgCl2 with the addition of 1 mM nucleotide. Temperature gradients spanned from 4°C to 98°C. Samples were heated at a speed of 0.6°C/min in presence of SYPRO Orange (Sigma Aldrich) hydrophobic fluorescent dye. Presented data derive from three independent experiments with at least 2 technical replicas each.
Yeast genetic manipulations
S. cerevisiae strains containing mutated kti12 alleles were generated and are listed in Supplementary Table S1. The parent strain for generation and expression of HA-tagged Kti12 point mutants was YAH76, which has the KTI12 coding sequence replaced by the marker gene KlURA3 (kti12Δ::KlURA3) (60) and allows for expression of c-myc-tagged Elongator subunit Elp1 [ELP1-(c-myc)3]. YAH76 is congenic with UMY2893 (4), which contains the SUP4 tRNA suppressor allele and the SUP4 suppressible ade2–1 and can1–100 reporters. Mutant strains mentioned in Supplementary Table S1 were generated by site-directed mutagenesis (61) on genomic DNA from strain YAH3 (KTI12-(HA)6::KlTRP1) to enable simultaneous HA-tagging. The kti12-mutated PCR fragments were transformed into YAH76 (62), and transformants were selected for tryptophan prototrophy and loss of the KlURA3 marker on the basis of 5-fluoroorotic acid resistance. Correct replacement of the KlURA3-knockout cassette by the mutagenized kti12 alleles was diagnosed by PCR, and all mutations were verified by DNA sequencing.
Bulk yeast tRNA isolation
Saccharomyces cerevisiae strains were grown in YPD to an OD600 ∼1, harvested and washed once with water, then directly subjected to tRNA isolation using RotiZOL reagent (Carl Roth, Germany) according to the manufacturer's instruction. Large RNAs were precipitated for 3 h at –20°C with 1/3 vol. of 8 M LiCl. Small RNAs in the supernatant were precipitated with 1/10 volume of 3 M sodium acetate pH 5.2 and 2.5 volumes 100% ethanol for 60 min at –80°C. The pellet was washed twice with 75% ethanol, then air dried and resuspended in 10 mM sodium acetate pH 5.2, to prevent deacylation of aminoacylated tRNAs as described by Köhrer and Rajbhandary (63).
Measurement of CtKti12 binding affinity to tRNA by EMSA
Binding of CtKti12 to S. cerevisiae WT bulk yeast tRNA (ytRNA) was analysed by electrophoretic mobility shift assays (EMSA). The binding reactions were performed in 4 μl binding buffer (20 mM Tris–HCl pH 7.5 at RT, 150 mM NaCl, 2 mM DTT, 1 mM MgCl2), containing 25 ng ytRNA and 1.25, 2.5 or 5 μM CtKti12 (WT or annotated mutant CtKti12 full length (FL), NTD or CTD fragments). For dissociation constant (KD) estimation of WT CtKti12-FL to ytRNA, 4 μl binding buffer containing 6.25 ng or 12.5 ng ytRNA (∼0.055 or 0.11 μM, respectively) and 0.06, 0.125, 0.25, 0.5, 0.75, 1.0, 1.25, 1.5, 2.0, 2.5 3.0, 4.0, 5.0 and 7.5 μM ChKti12-FL were mixed. After 30 min incubation at RT, 1 μl 50% sucrose (v/v) was added to the reactions and they were loaded on a 5% polyacrylamide gel containing TB-Buffer (45 mM Tris–HCl pH 7.0 at 4°C, 45 mM boric acid) and 10% sucrose. Electrophoresis was performed in TB buffer for 90 min and 60 V at 4°C. The gel was stained with SYBR Gold (Thermo Fisher Scientific) for 20 min and visualized on an UV table at 306 nm. Analysis of bound and free tRNA bands by densitometry was carried out using ImageJ2 (64) from three replicate experiments. The tRNAbound/tRNAtotal ratios were calculated, plotted against log10 [CtKti12-FL] and used to estimate the KD for the interaction by sigmoidal dose response fitting (GraphPad Prism version 6.01 for Windows, GraphPad Software, La Jolla, CA, USA).
Yeast protein extraction, co-immunoprecipitation and Western blot analysis
Yeast cells from 100 ml liquid cultures were harvested at an OD600 ∼ 1 and resuspended in 800 μl ice-cold B60 buffer (50 mM HEPES–KOH pH 7.3, 60 mM KAc, 5 mM Mg(Ac)2, 0.1% Triton X-100, 10% glycerol, 1 mM NaF, 20 mM β-glycerophosphate, 1 mM DTT) supplemented with Complete protease inhibitor cocktail (Roche, Germany). Following addition of 600 μl glass beads, cell disruption was carried out at 4°C by vigorously shaking (5× for 1 min). Cell debris was removed by centrifugation at 15 000 rpm and 4°C for 5 min. An additional clearing step of the supernatant was performed at 15 000 rpm and 4°C for 20 min. Protein concentration of cleared lysates was determined using Protein Assay Dye reagent (Bio-Rad, USA). Co-immunoprecipitation was done on the cleared lysate, using either anti-c-myc (9E10, Santa Cruz Biotechnology, USA) or anti-HA (F-7, Santa Cruz Biotechnology, USA) antibodies coupled to magnetic Dynabeads (Invitrogen, USA) according to manufacturer's instructions. 2 mg lysate was incubated with 2 μg antibody-coupled Dynabeads for 60 min with end-over rotation at 4°C. Antibody bound fractions were collected using a magnetic rack and washed 3× with B60 buffer. Proteins were eluted from the beads by addition of 50 μl 1× Laemmli sample buffer (65) and incubating at 99°C for 10 min. Control samples prior to immunoprecipitation (Pre-IP) were taken from lysates before addition of antibody-coupled Dynabeads and boiled at 99°C for 10 min in 1× Laemmli sample buffer. Protein samples were separated by SDS-PAGE and analyzed by Western blotting using anti-c-myc (9E10, Thermo Fisher Scientific, USA), anti-HA (Ab-1, Dianova, Germany) and anti-Cdc19 serum (kindly provided by Dr J. Thorner, University of California, Berkley, CA, USA).
GST pull down assay
Equimolar amounts of ScKti12 and GST-ScElp1 were incubated on a rotating wheel in the presence of Glutathione Sepharose 4B (GE Healthcare) for 2 h at 4°C in a buffer composed of 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 2 mM DTT, 2 mM MgCl2, 0,5% (v/v) Tween 20. Pure GST protein was used as a specificity control. Glutathione beads were collected by gentle spinning (500 × g) and subsequently washed three times with a binding buffer. Bound proteins were denatured at 95°C in the presence of Laemmli sample buffer and analyzed on 12% SDS-PAGE.
Phenotypic analyses of yeast mutants using assays indicative for U34 modification defects
To analyze the function of mutated kti12 alleles in tRNA modification by Elongator, sensitivity of yeast strains towards growth inhibition by exogenous zymocin toxin complex or intracellular expression of zymocin's γ-toxin tRNase subunit was assayed as previously described (66). Zymocin was partially purified from supernatants of Kluyveromyces lactis killer strain AWJ137 (66) by ultrafiltration (67). Strains were spotted in ten-fold serial dilution on synthetic drop-out (SD) media plates (pH 7.0) or on rich media plates (YPD, 0.5% yeast extract, 2% peptone, 2% glucose and 2% agar) containing zymocin (2–10% v/v) and incubated for 2–3 days at 30°C. To test the effect of intracellular expression of zymocin's γ-toxin tRNase subunit on cell growth, strains were transformed with vector pLF16, which expresses the γ-toxin gene from the galactose-inducible GAL1 promoter (68). To monitor the response to γ-toxin induction, transformed strains were spotted in ten-fold serial dilution on synthetic drop-out media plates (SD) containing either 2% glucose (repressing conditions) or 2% galactose (inducing conditions) and incubated for 3 days at 30°C. Kti12 and Elongator dependent U34 modifications confer growth inhibition by the tRNase activity of zymocin whereas defects in Kti12 and Elongator cause resistance to the toxin complex (66). Analysis of the effect of kti12 mutations on ochre (UAA) stop-codon read-through by the Elongator-dependent suppressor tRNA SUP4 was essentially as described (4). Yeast strains based on UMY2893 (ade2–1 can1–100; Supplementary Table S1) were spotted on SD plates lacking adenine (–Ade) or medium without arginine but containing the toxic arginine analog canavanine. Plates were incubated for 4 d at 30°C. The SUP4 tRNA is non-functional in cells that lack Kti12 activity or Elongator dependent U34 modifications, and thus does not allow for growth in the absence of adenine but confers resistance to growth inhibition by canavanine (4).
Relative quantitation of tRNA wobble uridine modifications by LC–MS/MS analysis
Total tRNA was isolated from yeast cultures as described previously (5). Prior to LC–MS/MS analysis, 5 μg of each tRNA sample were digested into nucleosides according to the following protocol: samples were incubated in the presence of 5 mM ammonium acetate (pH 5.3), 0.3 U nuclease P1 (both Sigma Aldrich, Munich, Germany) and 0.1 U snake venom phosphodiesterase (Worthington, Lakewood, USA) at 37°C for 2 h. Next, 1/10 volume of 100 mM ammonium acetate (pH 8) and 1 U fast alkaline phosphatase (Thermo Scientific, Waltham, MA, USA) were added, and samples were incubated for additional 60 min at 37°C. The digested tRNA samples were analyzed on an Agilent 1260 HPLC series equipped with a diode array detector (DAD) and a triple quadrupole mass spectrometer (Agilent 6460). A Synergy Fusion RP column (4 μm particle size, 80 Å pore size, 250 mm length, 2 mm inner diameter) from Phenomenex (Aschaffenburg, Germany) was used at 35°C column temperature. The solvents consisted of 5 mM ammonium acetate buffer adjusted to pH 5.3 using acetic acid (solvent A) and pure acetonitrile (solvent B). The elution was performed at a flow rate of 0.35 ml/min using a linear gradient from 0% to 8% solvent B at 10 min, 40% solvent B at 20 min and 0% solvent B at 23 min. For an additional 7 min, the column was rinsed with 100% solvent A to restore the initial conditions. Prior to entering the mass spectrometer, the effluent from the column was measured photometrically at 254 nm by the DAD for the detection of the four canonical nucleosides. The triple quadruple mass spectrometer, equipped with an electrospray ion source (Agilent Jet Stream), was run at the following ESI parameters: gas (N2) temperature 350°C, gas (N2) flow 8 l/min, nebulizer pressure 50 psi, sheath gas (N2) temperature 350°C, sheath gas (N2) flow 12 l/min and capillary voltage 3000 V. The MS was operated in the positive ion mode using Agilent MassHunter software and modified nucleosides were monitored by multiple reaction monitoring (dynamic MRM mode). For identification of s2U and retention time determination, comparison to a synthetic standard (Berry & Associates, Dexter, USA) was used. Identification of ncm5U, mcm5U and mcm5s2U peaks were performed as described previously (69). All mass transitions and retention times used for identification of the modified nucleosides are listed in Supplementary Table S2. Peak areas were determined employing Agilent MassHunter Qualitative Analysis Software. In the case of the major nucleosides, peak areas were extracted from the recorded UV chromatograms in order to avoid saturation of the mass signals. For intersample comparability of the detected modifications, the peak areas of the modified nucleosides were normalized to the UV peak area of uridine.
RESULTS
The structure of Kti12 resembles PSTK
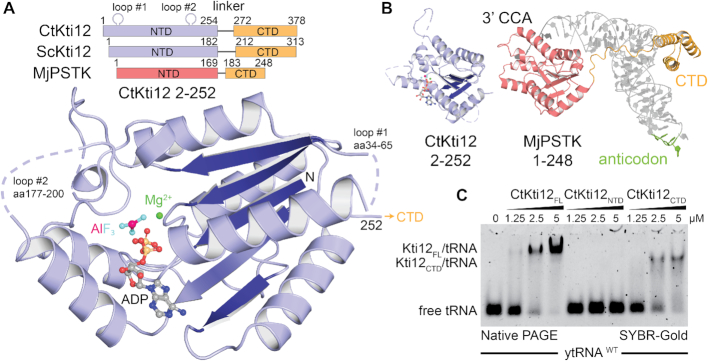
The highly conserved eukaryotic Kti12 proteins can be divided into two separate domains, which are connected by a flexible linker region. The N-terminal domain (NTD) harbors a typical ATPase signature, but the C-terminal domain (CTD) has no assigned enzymatic or functional motif (Figure 1A). Kti12 shares high sequence similarity and domain architecture with PSTK (43,70,71). We were able to express and produce large quantities of full length and truncated versions of Kti12 from Saccharomyces cerevisiae (ScKti12) and C. thermophilum (CtKti12; Supplementary Figure S1A). Purified Kti12 protein samples were used in co-crystallization experiments in the presence and absence of various ATP analogs. We obtained well diffracting crystals of CtKti12NTD (aa2–252) after addition of magnesium, ADP and aluminum fluoride (ADP·AlF3; (72)). The crystal structure of CtKti12NTD was solved in the P3121 space group using single wavelength anomalous diffraction (SAD) phasing (Figure 1A). Six individual iodine atoms could be located within the asymmetric unit belonging to two bound ‘magic triangle’ molecules that had been soaked into existing crystals (Supplementary Figure S1B). Using a native dataset at 2.4 Å, a final atomic model was built and refined to R/Rfree values of 23.9/27.5% (Table 1) obeying perfect stereochemistry.
Figure 1.
Kti12 resembles PSTK both structurally and functionally. (A) Structure of a CtKti12NTD locked in a transition state of ATP hydrolysis shown in a cartoon representation. Aluminum fluoride, a hydrolysis transition state mimetic (magenta and cyan) and magnesium (green sphere) are highlighted. The protein core consists of a single β-sheet composed of five parallel β-strands (dark blue). Domain architectures of CtKti12 and ScKti12 are similar to that of MjPSTK. ATPase motifs located on the N-terminal domain (NTD) of Kti12 (blue) and PSTK (red); C-terminal domain (CTD; yellow). (B) Structure of CtKti12NTD (blue) resembles the one of MjPSTKNTD bound to tRNASec (red; PDB ID 3ADB). The 3′ CCA of tRNASec is inserted into the PSTKNTD while the tRNA body is mainly held by PSTKCTD (yellow). The anticodon region (green) is not involved in this interaction. C. Binding affinity of CtKti12 with bulk yeast tRNA was examined by electrophoretic mobility shift assay (EMSA) using 5% native PAGE. Free tRNA and tRNA in complex with CtKti12-FL or CtKti12CTD were visualized using SYBR-Gold. Both full-length (FL) protein and CtKti12CTD bind tRNA in a PSTK-like fashion.
Table 1.
Data collection and refinement statistics
| Ct Kti12 1–254 | Ct Kti12 1–254 (phasing) | |
|---|---|---|
| PDB ID | 6QP0 | - |
| Data collection | ||
| Beam line | BESSY II, MX14–2 | BESSY II, MX14–2 |
| Wave length (Å) | 0.982 | 1.645 |
| Space group | P31 2 1 | P31 2 1 |
| Cell dimensions (Å) | 71.02 71.02 88.57 90 90 120 | 72.21 72.21 89.20 90 90 120 |
| Resolution (Å)a,b | 50–2.4 (2.48–2.4) | 50–2.1 (2.15–2.1) |
| Unique reflections | 10 334 (723) | 29 320 (2087) |
| Completeness (%) | 99.5 (95.0) | 96.1 (92.0) |
| Redundancy | 7.9 (4.8) | 20.7 (20.4) |
| Rsym | 0.15 (0.886) | 0.15 (3.06) |
| I/σ(I) | 12.3 (1.98) | 14.97 (1.02) |
| Iodine atoms per ASU | 0 | 6 |
| Structure refinement | ||
| Working set reflections | 10 300 | |
| Test set reflections | 517 | |
| Resolution (Å) | 35.5–2.4 | |
| Number of protein atoms | 1526 | |
| Number of ligand | 44 | |
| Number of solvent molecules | 46 | |
| R work/Rfree | 0.239/0.275 | |
| Average B factor (Å2) | ||
| Overall | 49.9 | |
| Protein | 50.1 | |
| Ligand | 55.3 | |
| Solvent | 36.7 | |
| RMSD values | ||
| Bond lengths (Å) | 0.0017 | |
| Bond angles (°) | 0.61 | |
| Ramachandran-plot analysis | ||
| Most favored regions (%) | 96.67 | |
| Allowed regions (%) | 3.33 | |
| Disallowed regions (%) | 0 | |
aValues in parentheses are for the highest-resolution shell.
bResolution limits according to an I/σ of 2 are 2.7 Å for the native ChKti12 dataset.
The structure shows a globular ATPase domain that consists of five parallel β-sheets and eight α-helices surrounding the hydrophobic core of the domain (Supplementary Figure S1C). Except for two flexible loop regions, that are not conserved and only exist in CtKti12, all residues are clearly visible and traceable in the density. As previously predicted (73), Kti12NTD indeed harbors a canonical P-loop motif, located between the first strand (β1) and the first helix (α1). The superimposition of CtKti12NTD with known structures of full length Methanocaldococcus jannaschii PSTK (MjPSTK; PDB IDs 3ADB and 3A4L) revealed very strong structural similarities (Figure 1B). Furthermore, likeness between Kti12 and PSTK spans beyond the NTD as both proteins display a short linker region followed by a conserved CTD that is composed of three predicted helices (Supplementary Figure S2A).
As the CTD of PSTK binds to tRNA[Ser]Sec (74), we tested whether Kti12 is able to bind to endogenous bulk tRNAs and uses a similar mechanism for tRNA recognition. CtKti12 indeed binds tRNAs mainly via its CTD in an almost identical fashion as PSTK (Figure 1C). The affinity constants (KD ∼1.5 μM) are typical for proteins binding transiently to tRNAs (6,75). As Kti12 shifted the entire population of yeast bulk tRNAs (Supplementary Figure S2B,C) and bound with similar affinities to different in vitro transcribed tRNAs (Supplementary Figure S2D) it does not seem to prefer or select any particular tRNA species at the initial binding step. Kti12 does not share any larger protein-protein interfaces with neighboring molecules in crystals and full length and truncated forms of CtKti12 or ScKti12 eluted as a single monomeric peak from the gel filtration columns (Supplementary Figure S1A). Therefore, we assume that unlike PSTK, which has been previously reported to form dimers (70,74,76), Kti12 stays monomeric in solution.
The active site of Kti12 resembles a canonical ATPase
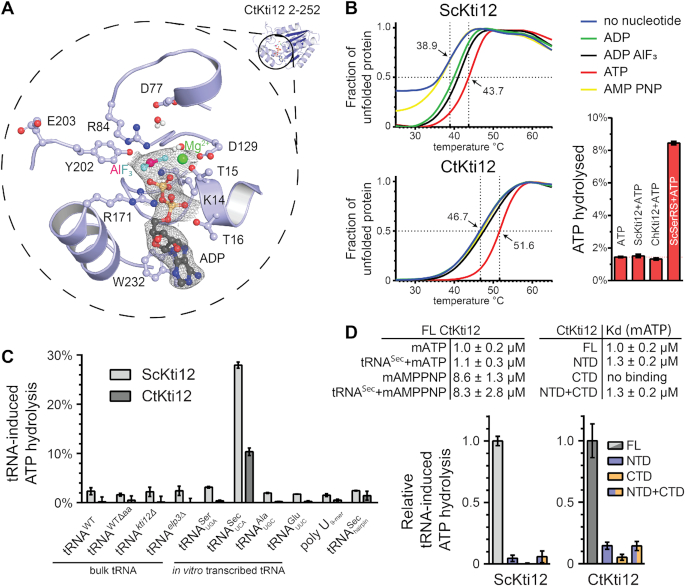
We solved the structure of CtKti12NTD in complex with ADP·AlF3 mimicking a bi-pyramidal transition state during ATP hydrolysis (77). The ADP·AlF3 molecule as well as the expected magnesium ion are clearly visible in the electron density. The nucleotide binding pocket of CtKti12 utilizes the evolutionarily conserved architecture typical of canonical P-loop ATPases (Figure 2A). In detail, several conserved residues, namely K14, T15, T16, D129 and R171 interact with the first two phosphate atoms (Pα and Pβ). D77, R84, Y202 are in close contact with the AlF3 molecule, which mimics the third phosphate (Pγ) and shows a typical distance for a transition state of 3.0 Å from the Pβ. The addition of ATP contributed to an increased thermostability of CtKti12 and ScKti12 in vitro. ADP, ADP·AlF3 and AMPPNP, a non-hydrolysable ATP analog, however had no significant effects on melting temperatures (Figure 2B). Thermal shifts after addition of ATP and in the absence of tRNA show a similar stabilization of both the full-length proteins and the truncated NTDs alone (Supplementary Figure S3A). Therefore, we speculate that the observed effects do not result from large conformation rearrangements of the two domains but rather represent local compaction of the defined ATP binding site. Despite intense co-crystallization trials, no analyzable crystals were obtained in the presence of ATP or other nucleotides.
Figure 2.
Structural and functional characterization of ATP hydrolysis by Kti12. (A) A structural close-up of the ATP-binding pocket with important residues shown in ball-and-stick representation. Isomesh represents an OMIT map at σ = 1.0 around the ligand. Aluminum fluoride (magenta and cyan) and magnesium (green) are shown and the residues involved are labeled. (B) Thermal shift assays with ScKti12 (top) and CtKti12 (bottom) in the presence/absence of nucleotides. ATP hydrolysis (bottom right) was investigated using the malachite green method (right). Yeast seryl-tRNA synthetase was used as a positive control for basal ATP hydrolysis relative to ATP alone. (C) ATP hydrolysis of ScKti12 and CtKti12 in response to bulk or in vitro transcribed tRNAs (see labeling). All in vitro transcribed tRNAs possess 3′ CCA. Deamino-acylated (Δaa) tRNA was obtained by deamino-acylation of bulk tRNA obtained from WT yeast cells. A poly U (nine bases) and a tRNASec anticodon arm hairpin were used as a specificity control. Reactions contained tRNA and protein mixed at 1:1 stoichiometry and were incubated for 15 hours at 37°C. Results are presented as the percent of ATP hydrolyzed and calculated standard deviations are shown (n = 3). (D) Similarly, to (C), tRNASec was used to induce ATP hydrolysis in FL proteins, NTDs, CTDs or 1:1 stoichiometric mixture of NTDs and CTDs and calculated standard deviations are shown (n = 3).
Enzymatic activity of Kti12 depends on tRNA binding
Surprisingly, none of the purified Kti12 proteins displayed any basal ATPase activity in vitro (Figure 2B). To understand the mechanism of Kti12 action, we asked whether Kti12 might require a cofactor to activate its ATPase function. Therefore, Kti12 proteins were incubated with ATP and magnesium in the presence of bulk tRNAs from various yeast strains or different purified in vitro transcribed tRNA species containing CCA tails (e.g. tRNAGlu, tRNASer and tRNASec). In analogy to PSTK, both ScKti12 and CtKti12 showed a significant upregulation of ATPase activity upon treatment with in vitro transcribed tRNASec (Figure 2C and Supplementary Figure S3B,C,D). The observed ATP hydrolysis rate of Kti12 in the presence of tRNASec (kcat = 1.59 × 10−3 s−1) is ∼100× faster than the activation rate of archaeal MjPSTK (70). As neither any of the other tRNA samples, nor polyU RNA or the anticodon stem loop of tRNASec induce the ATPase activity of Kti12, the underlying mechanism seems highly specific (Figure 2C). Furthermore, we used MANT-labeled nucleotides to determine the binding constants of ATP (KD ∼ 1.0 μM) and AMPPNP (KD ∼ 8.6 μM) to ChKti12 (Figure 2D and Supplementary Figure S3E). We did not observe any changes in affinity for the nucleotides in the presence of tRNASec, showing that tRNA does not boost the activity by increasing the rate of ATP binding (Figure 2D).
A comparison between full length protein, NTD, CTD and a stoichiometric mixture of NTD and CTD revealed that both domains need to be connected within a single polypeptide chain to induce ATP hydrolysis efficiently and cannot be activated in trans (Figure 2D and Supplementary Figure S3E). Our data also clearly shows that nucleotides are bound only by the NTD and that the CTD does not influence the affinity of the NTD to ATP (Figure 2D). Notably, Kti12 does not require tRNA to be charged with an amino acid for ATPase activation, and fungi generally lack important components of the selenocysteine biosynthesis pathways (78). Although the rational for the observed specificity is not yet clear, we utilized human tRNASec to study the enzymatic properties of Kti12 in greater detail.
Identification of catalytic residues in Kti12
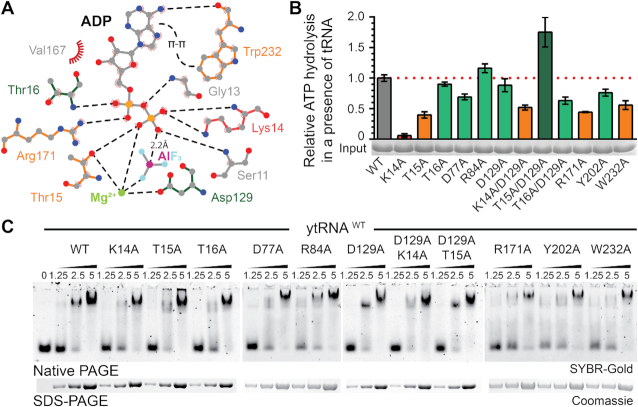
To dissect the specific contribution of active site residues for the catalytic reaction, we individually mutated all residues in the vicinity of the bound ADP·AlF3 molecule (Figure 3A). Next, we analyzed the ATP hydrolysis rates, thermostability profiles, ATP- and tRNA-binding capacities of all substituted variants. As the introduction of almost all equivalent mutations in ScKti12 led to insoluble proteins, our analyses in vitro focused on variants in CtKti12. Substituting the most central residue, K14A, resulted in complete loss of ATPase activity in vitro (Figure 3B). Other mutations, like T15A, R171A and W232A, strongly decreased Kti12’s ATP hydrolysis rates. D77A, Y202A and T16A/D129A show slightly decreased ATPase activity, whereas T16A, R84A and D129A behave like wild type Kti12. Certain mutants displayed decreased thermal stability profiles (e.g. T15A, D129A, T15A/D129A, T16A/D129A and T16A/D129A) partially explaining slightly decreased activities due to overall protein destabilization (Supplementary Figure S4A). Interestingly, the strong effect of K14A can be partially rescued by introducing D129A indicating a functional link between these two residues. As D129A also shows a stimulatory effect in combination with T15A (T15A/D129A) generating a hyperactive form of Kti12, D129 (D85 in ScKti12) might act as a negative regulator and molecular switch during the catalytic cycle. Strikingly, none of the ATPase mutants affects tRNA binding affinities, indicating that although tRNA binding is essential for ATPase activity, the rate of ATP hydrolysis does not affect tRNA binding by Kti12 in vitro (Figure 3C).
Figure 3.
Mutational analysis of the nucleotide binding pocket in CtKti12. (A) Two dimensional LigPlot (92) overview of the nucleotide binding pocket and residues targeted for mutations. Mutants with abolished (K14A, red), decreased (orange) or normal (green) ATPase activity are highlighted. Potential hydrogen bonds are shown as black dashed lines. Additionally, π-π stacking between adenine and W232 and a distance between the oxygen of phosphate beta and aluminum is shown. (B) ATP hydrolysis by wild type and mutant forms of CtKti12 in response to tRNASec showing the calculated standard deviations (n = 3). Mutants are labeled as in (A) and the upregulated combined mutant is shown in dark green. (C) Analysis of tRNA binding properties in selected nucleotide binding pocket mutants by EMSA. Increasing concentrations of protein (1.25, 2.5 and 5 μM) were incubated with 0.22 μM yeast bulk tRNA and resolved on a 5% native PAGE. tRNA was stained using SYBR-Gold. As a loading control, samples were also analyzed by denaturing SDS-PAGE and stained with Coomassie (lower panel).
In CtKti12, the adenine base of ATP is mainly coordinated by π-stacking interactions with W232 (Figure 2A), whereas archaeal PSTK employs arginine residues to coordinate the nucleotide base. W232 is not conserved, but in structural homology models of other Kti12 sequences, a highly conserved tryptophan residue (e.g. ScKti12 W123) located in the same position as the arginine in PSTK (helix α4) could substitute (74). Notably, Kti12 shows a clear preference towards ATP and only very weakly hydrolyzes UTP, GTP or CTP (Supplementary Figure S4B). As PSTK shows broad nucleotide hydrolysis activity (70), we speculate that the conserved tryptophan residue in Kti12 confers selectivity for ATP. Notably, substitution of W232 with alanine boosted the thermostability of CtKti12 (from 47°C to 57°C), but resulted in a lack of ATP-dependent stabilization (Supplementary Figure S4C, D). Thus, we hypothesize that the thermal shifts observed after addition of ATP partially originate from reduced degrees of freedom for this rather bulky residue. Foremost, all tested active site mutants lead to a detectable decrease in affinity to ATP and reduced ATP-dependent protein stabilization (Supplementary Figure S4C, D).
The NTD of Kti12 couples tRNA recognition with ATP binding and hydrolysis
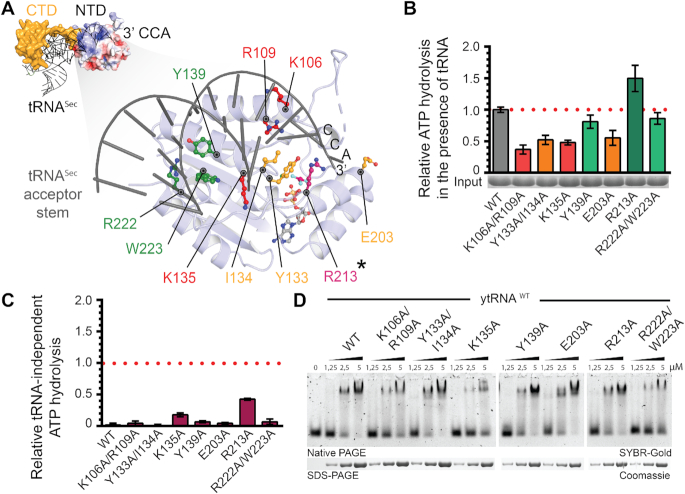
Analyses of sequence conservation and charge distribution within Kti12NTD revealed an evolutionary conserved and positively charged surface area (Supplementary Figure S5A–C). Structural modeling of tRNA-bound Kti12 complex based on the known MjPSTK structure in complex with tRNASec (PDB ID: 3ADB) specifically positioned the acceptor stem of the bound tRNA molecule next to the aforementioned basic surface patch (Figure 4A). The obtained structural model places the 3′CCA sequence potentially carrying the charged amino acid in close proximity to a channel leading directly to the ATPase site (Figure 4A). Therefore, our model may be indicative for a Kti12 reaction scheme that would be remarkably similar to that of PSTK. To verify our hypothesis, we checked the ATPase activity and tRNA binding properties of a number of Kti12 variants containing amino acid substitutions replacements located in the anticipated Kti12-tRNA interface of the NTD (Figure 4B).
Figure 4.
Analysis of tRNASec interaction with CtKti12NTD. (A) (Top left): Structural model of tRNA-bound CtKti12NTD based on superimposition with MjPSTKNTD (3ADB), r.m.s.d. = 3.8 Å2. tRNASec is shown in grey and MjPSTKCTD in yellow. The 3′ end of the superimposed tRNASec reaches to the active site of CtKti12NTD. Surface charge distribution of CtKti12NTD is shown in blue and red. Positively charged regions (blue) coincide with a modelled contact surface with tRNASec. Color coding and scale are identical to Figure S5. (Bottom right): The close up of the model shows the proposed tRNA interaction surface at the NTD, which is shown in light blue, acceptor stem of tRNASec is gray. Mutants with significantly downregulated ATPase activity are shown in red, mutants with moderate ATPase properties in orange. Hydrolytic activity after mutating residues shown in green does not differ significantly from WT protein. The gatekeeping R213 residue (asterisk) is shown in violet. (B) Analyses of hydrolytic properties of tRNA interaction surface mutants using the malachite green assay. Calculated standard deviations are shown (n = 3). Color coding as in (A). (C) tRNA-independent ATP hydrolysis of mutants within tRNA-interaction interface on the NTD using the malachite green assay. The results were normalized to the tRNASec-induced hydrolysis of WT protein and calculated standard deviations are shown (n = 3). (D) Binding affinity of tRNA-interface mutants to the bulk yeast tRNA. Samples were resolved on a 5% native PAGE. As a loading control, samples were also analyzed by denaturing SDS-PAGE stained with Coomassie. tRNA was stained using SYBR-Gold.
In case of K106A/R109A, Y133A/I134A, K135A and E203A, we observed a substantial decrease in tRNA-induced ATPase activity. This strongly suggests the involvement of these residues in tRNA binding and shows that the predicted tRNA interaction surface area is mechanistically coupled to the spatially separated ATPase site (Figure 4B). Strikingly, R213A not only has enhanced tRNA-induced ATPase activity but also shows increased basal hydrolysis activity in the absence of tRNAs (Figure 4C). As the location of R213 overlaps with the modeled 3′CCA sequence, we suspect it may act as a gatekeeper that has to be displaced by the incoming tRNA for hydrolysis to occur. We also observed weak basal hydrolysis rates in case of K135A, suggesting an involvement in correct positioning of the neighboring R213 residue. The most remote substitutions, Y139A and R222A/W223A, did not affect tRNA-induced ATPase activity (Figure 4B) or thermostability (Supplementary Figure S5D). The CTD of Kti12 is the major tRNA binding site and if at all, the NTD by itself has only very weak affinity for tRNAs (Figure 1C). Therefore, we suspected that NTD mutations in the full-length Kti12 protein (containing the CTD) may not necessarily affect tRNA binding. Nevertheless, certain mutations, namely K106A/R109A, K135A and R222A/W223A, resulted in a small but noticeable decrease in ATP- and tRNA-affinity (Figure 4D and Supplementary Figure S5D), further supporting the mechanistic role that the basic region identified in Kti12NTD plays in tRNA binding and recognition.
Kti12 ATPase activity is crucial for Elongator's tRNA modification activity in vivo
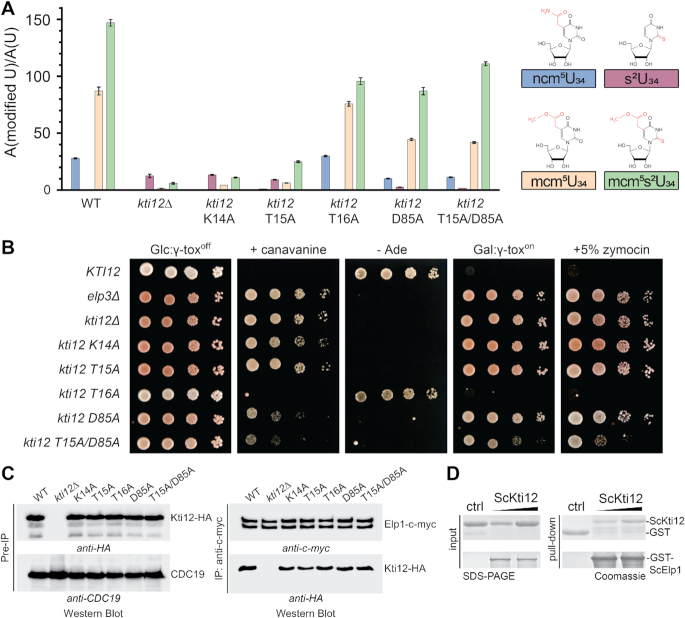
In order to better understand the in vivo role of Kti12 and its ATPase activity, we analyzed yeast strains carrying mutations in the nucleotide binding pocket of ScKti12 equivalent to CtKti12 (Figure 3A). Firstly, we isolated bulk tRNAs from these strains and used mass spectrometry (LC–MS/MS) to profile Elongator dependent tRNA modifications quantitatively. Strongly decreased levels of the U34 modification types ncm5U, mcm5U and mcm5s2U were observed in kti12Δ cells and in kti12 mutants expressing catalytically defective Kti12 proteins (Figure 3B), namely K14A and T15A (Figure 5A). In addition, these mutants accumulate s2U, an abnormal thio-modification not detected in tRNAs from the KTI12 wild-type strain or the T16A mutant (Figure 5A) and typical of Elongator mutants (4,79) when full U34 modifications cannot be accomplished. In case of T16A, D85A (analogous to D129A in CtKti12) and D85A/T15A, with intact ATPase activity (Figure 3B), the ncm5U, mcm5U and mcm5s2U modifications were detectable albeit at moderately lowered levels compared to the wild-type (Figure 5A).
Figure 5.
Effect of ScKti12 mutants on Elongator's U34 modification function. (A) Quantification of modified U34 nucleosides in tRNAs from yeast strains carrying kti12 nucleotide binding pocket mutations. For each mutant, the relative levels of ncm5U, s2U, mcm5U and mcm5s2U are shown. Modified nucleoside signals were normalized using the total uridine content A (modified U)/ A (U), to allow comparison between different samples. ncm5, s2, mcm5 and mcm5s2 are highlighted in red within their chemical structure (top). (B) Zymocin-/γ-toxin- and SUP4-based assays indicative for Elongator function and U34 modification in vivo. WT (KTI12) and Elongator deficient elp3Δ and kti12Δ strains served as growth control for loss-of-function mutants. Ten-fold serial cell dilutions of the strains expressing Kti12 variants with the indicated mutations in the nucleotide binding pocket mutants (see A) were spotted on SD glucose media with 60 μg/ml canavanine, or 5% (v/v) zymocin, lacking adenine (- Ade) or containing 2 % (w/v) galactose (Gal: γ-toxon) instead of 2% (w/v) glucose (Glc: γ-toxoff). For the γ-toxin assay, strains were transformed with vector pLF16 expressing the γ-toxin gene under the galactose inducible GAL1 promoter (68) and cultivated for 3–4 days at 30°C. (C) Kti12-Elongator interaction studies. Yeast strains co-expressing c-myc-tagged Elp1 and HA-tagged Kti12 variants were subjected to immunoprecipitation (IP) using anti-c-myc antibodies. IPs were examined by Western blotting with anti-c-myc (to detect Elp1) and anti-HA antibodies (to detect co-immunoprecipitated Kti12). Western blots of the extracts prior to the IPs (Pre-IP) controlled the input using anti-HA and anti-Cdc19 antibodies, to confirm similar levels of Kti12 and pyruvate kinase (Cdc19) in the different strains. (D) Purified ScKti12 and ScElp1 interact in vitro. Full length GST-ScElp1 successfully bound ScKti12–6xHis in a concentration-dependent manner. Free GST was used as a specificity control. For GST pull-down assay, gels were stained with Coomassie and respective proteins are labeled next to the gel.
We tested all kti12 mutants by previously established phenotypical surveys (zymocin, γ-toxin and SUP4 assays) diagnostic for Elongator's tRNA modification function in vivo (4,66). The zymocin and γ-toxin assays are based on a fungal anticodon nuclease which cleaves mcm5s2U34 modified tRNAs and thereby inhibits yeast growth in an Elongator dependent fashion (68,80). Thus, extracellular application of the zymocin complex (zymocin assay) or conditional expression of its tRNase γ-toxin subunit (γ-toxin assay) allow to monitor the U34 modification states of Elongator substrate tRNAs based on toxin sensitivity or resistance phenotypes (see method section) (81). As expected, elp3Δ and kti12Δ mutants were found to resist against zymocin and γ-toxin (Figure 5B) confirming the importance of Kti12 for Elongator function (66). The ATPase defective mutants K14A and T15A also conferred resistance indicating that nucleotide hydrolysis is vital for Kti12 to support Elongator's U34 modification function. Among the phenotypes of mutants with moderately reduced U34 modification levels (Figure 5A), resistance to zymocin and γ-toxin was pronounced in the D85A but weaker for the T15A/D85A mutant and entirely absent from T16A cells (Figure 5B). The phenotype of the latter is remarkable since the mcm5s2U levels measured by LC–MS/MS are similar to D85A or T15A/D85A mutants (Figure 5A). Of note, however, while the ncm5U and mcm5U modification levels are comparable between the T16A mutant and KTI12 wild-type cells, they are significantly reduced in the D85A and T15A/D85A mutants.
Therefore, we resorted to an alternative in vivo assay (SUP4) that is diagnostic specifically for the mcm5U modification. SUP4 encodes a tRNA suppressor that decodes ochre stop codons when its anticodon carries the Elongator dependent mcm5U modification (4,68). Hence ochre mutations in ade2–1 and can1–100 reporter genes are suppressed by SUP4 in an Elongator dependent fashion resulting in adenine prototrophy (growth on media lacking adenine) and canavanine sensitivity (due to uptake of the toxic arginine analog canavanine by restored expression of arginine permease Can1) (4,68). As a consequence, mcm5U modification defects typical of elp3Δ and kti12Δ mutants abolish SUP4 tRNA suppressor activity and cause adenine auxotrophy and canavanine resistance (Figure 5B). In the same SUP4 assay, the K14A and T15A mutants with Elongator defects show Kti12 loss-of-function phenotypes while the T16A mutant behaves like wild-type confirming the toxin assay data above and the importance of Kti12 ATPase activity for Elongator's U34 modification capacity (Figure 5B). In contrast, the D85A and T15A/D85A mutants displayed adenine auxotrophy but partial resistance toward canavanine. Intriguingly, the latter trait is intermediate between the loss-of-function (kti12Δ) and wild-type (Figure 5B). This indicates that the can1–100 reporter appears to be more sensitive than ade2–1 to variations in the mcm5U34 levels. The reduced mcm5U34 level in the D85A and T15A/D85A mutants is sufficient to give some protection against canavanine likely by reduced permease levels but insufficient to provide enough ADE2 gene product to support prototrophy. The wild-type growth behavior with both readouts of the T16A mutant (Figure 5B) is consistent with the wild-type like mcm5U34 modifications levels measured by LC–MS/MS (Figure 5A).
Thus, we explain the phenotypic variations between T16A on the one hand and the D85A and T15A/D85A mutants on the other, by differences in the measured ncm5U and mcm5U modification levels and by differential sensitivities between the ade2–1 and can1–100 reporters used to diagnose U34 modification defects with SUP4. Apparently, moderate reduction in mcm5s2U levels is not sufficient to provide γ-toxin/zymocin resistance in the T16A mutant in contrast to D85A and T15A/D85A. We therefore propose that the reduced levels of ncm5U and mcm5U modifications contribute to lower toxin sensitivity of the latter two mutants. The SUP4 and zymocin/γ-toxin assays show that the loss-of-function phenotype of the T15A mutant is partially rescued in the T15A/D85A double mutant. Genetic interaction observed between the residues T15 and D85 (analogous to D129 in CtKti12) in the double mutant (Figure 5B) is in agreement with our CtKti12 hydrolysis measurements (Figure 3B), where low ATPase activity caused by the T15A exchange was compensated in combination with the D129A substitution. In summary, our mass spectrometric and phenotypic data (Figure 5A and B) provide robust correlation between decreased ATPase activity, reduced tRNA modification levels and Elongator phenotypes and thus establish a direct link between the ATP hydrolysis activity of Kti12 and the U34 modification function of the Elongator complex
In addition, we have analyzed all structure guided mutants in CtKti12 (Supplementary Figure S6A) and confirmed the importance of the equivalent residues for the function of ScKti12 (Supplementary Figure S6B). Last but not least, we have performed unbiased randomized mutagenesis of Kti12 in yeast to identify additional functionally important residues. Here, phenotypic studies highlight the importance of the hydrophobic core of Kti12, its ATP binding pocket and the tRNA interacting interface on the NTD for proper functioning of Kti12 in vivo (Supplementary Figure S6C).
Kti12 interacts directly with Elp1
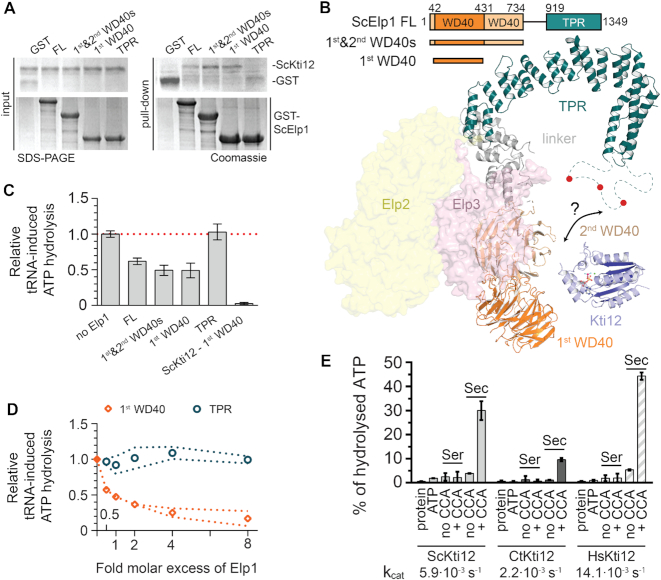
As Kti12 was shown to interact with Elongator in vivo (66,73,82), we were curious to see whether its ATPase activity is necessary for the temporary recruitment to this large macromolecular complex of ∼850 kDa (83). Strikingly, our co-immunoprecipitation analyses show that both wild-type and ATPase-deficient mutants of Kti12 interact with Elongator at comparable levels in vivo (Figure 5C). Hence, the ATPase activity of Kti12 neither constitutes a regulatory step for its interaction with Elongator nor for its binding to tRNA. These observations in yeast are fully consistent with direct protein-protein interaction observed between purified ScKti12 and full length ScElp1 in the absence of other co-factors in vitro (Figure 5D and Supplementary Figure S6D). In addition, we show that neither the presence of hydrolysable nucleotides nor non-hydrolysable analogs affects this interaction in vitro (Supplementary Figure S6D). We and others (83,84) have recently determined the architecture of the Elp123 subcomplex and the fully assembled yeast Elongator complex. Hence, we were curious to find out where Kti12 contacts Elongator and used individually purified domains of Elp1 to map the interaction site of Kti12 more precisely. Somewhat surprisingly, Kti12 specifically interacts with the first WD40 domain of Elp1 (Figure 6A), which seems spatially distant from the previously mapped phosphorylation sites in the CTD of Elp1 that are known to slightly affect Kti12 binding to Elongator in vivo (40). Nevertheless, our recent structural model of Elp1 shows that these two regions, which are roughly 1,000 residues apart from each other in the primary sequence, could in fact come into relatively close proximity in the assembled Elongator complex (Figure 6B). Hence, to fully understand the underlying regulatory circuitry it remains to be established where other regulatory factors (e.g. Hrr25 and Sit4) contact Elongator and how Kti12 could influence their interactions. Nevertheless, we tested whether the interaction of Kti12 with Elp1 would have any effect on Kti12’s ATPase activity. Notably, we observed a specific inhibition of its tRNA-induced ATPase activity in the presence of all Elp1 constructs that contain the first WD40 domain (Figure 6C). Furthermore, we show that increasing amounts of the interaction domain inhibit the ATPase activity in a strictly concentration dependent fashion (Figure 6D). In summary, we have identified a direct interaction between Elp1 and the Kti12 protein, which is independent of ATP hydrolysis and tRNA binding, but restricts the tRNA stimulated ATPase activity of Elongator-bound Kti12.
Figure 6.
Functional studies of ScKti12 binding to Elongator. (A) The interaction site on ScElp1 was mapped using domains of ScElp1, namely, first WD40 (aa 42–431), first and second WD40s (aa 1–734) and TRP (aa 919-1349). Interaction was observed only in case of constructs containing the first WD40 domain. Inputs before the pull down are shown on the left and GST-tagged Elp1 proteins are shown below. Gels were stained with Coomassie and respective proteins are labeled next to the gel. (B) A structural model of one lobe of the Elp123 subcomplex of Elongator built using integrative modelling approach. Elp1 consists of two N-terminal WD40 domains (shades of orange), flexible linker region (gray) and a tetratricopeptide motif (TPR; teal) used for dimerization of Elp1 molecules. Unstructured loop region (aa1175–1252) within Elp1 TRP is shown as dashed line. Red circles indicate presence of multiple phosphorylation sites within the loop region. Surfaces of Elp2 (yellow) and Elp3 (pink) proteins are also shown. Kti12NTD (blue) is shown in a vicinity of the identified interaction site. (C) Binding of ScElp1 to the ScKti12 inhibits tRNA-induced hydrolysis. 1 μM ScElp1 protein was added to the reaction mixture containing 1 μM ScKti12. ScKti12 – first WD40 indicates ATP hydrolysis by a fusion protein, in which the first WD40 was linked to the C-ter of ScKti12 using a flexible 5x(GS) linker. Results are normalized to the tRNASec-induced ATP hydrolysis in the absence of Elp1. Means and calculated standard deviations are shown (n = 3). (D) The first WD40 domain of Elp1 inhibits tRNA-dependent ATP hydrolysis in a dose dependent manner. Increasing amounts of first WD40 domain (orange squares) or TPR (tale circles) were added to the ATPase reaction, up to eight times greater molar concentration of Elp1. Results are normalized to the tRNASec-induced ATP hydrolysis in the absence of Elp1. Error corridors represent standard deviations (n = 3). (E) ATP hydrolysis of Kti12 is dependent on maturity of presented tRNA. Kti12 hydrolyses ATP in the presence of tRNASec with a 3′CCA (tRNASec, +CCA). Removal of 3′CCA from tRNASec (tRNASec, no CCA) resulted in almost complete diminishment of inducing properties of tRNASec. ATP hydrolysis was assessed with the malachite green assay. kcat values were calculated based on the ATP hydrolysis after 4 h (data not shown). Tested conditions resulted in following kcat values: kcat (ScKti12) = 5.85×10−3 ± 0.28×10−3 s−1; kcat (CtKti12) = 2.19×10-3 ± 0.05×10−3 s−1; kcat (HsKti12) = 14.05×10−3 ± 0.10·10−3 s−1. PSTK from hyperthermophylic archaea exhibited lower kcat values upon induction by tRNASec, kcat(MjPSTK) = 2.9×10−5 ± 0.3×10−5 s−1 (Sherrer et al., 2008b). Calculated means and standard deviations are shown (n = 3).
The 3′CCA tail of tRNA is essential for the activation of eukaryotic Kti12s
Last but not least, we were curious if Kti12 might be able to discriminate between different maturation stages of tRNASec. Therefore, we produced a version of human tRNASec lacking the 3′ CCA tail, which is added to tRNAs by a nucleotidyl transferase. Next, we compared its ATPase activation potential with tRNASer and mature tRNASec in the established assays. Strikingly, tRNASec(w/o CCA) is not able to induce the ATPase activity of neither ScKti12 nor CtKti12. This data not only further confirm our model of tRNA binding to the NTD (Figure 4A), but also raise the possibility that Kti12 helps the Elongator complex to discriminate between immature and mature tRNA species. Furthermore, we were able to express and purify human full length Kti12 and confirmed its identical behavior and specificity (Figure 6E), showing that the here proposed mechanism of activation is conserved between various eukaryotic Kti12s. Notably, Kti12s from three different species process ATP in the presence of non-aminoacylated tRNASec+CCA approximately one hundred times faster than MjPSTK (Figure 6E). This indicates that Kti12, in comparison to PSTK, might not require a coupled amino acid at the tRNA for its ATPase activation. Nevertheless, the activities of all Kti12 and PSTK proteins have been tested at 37°C (70) and the observed variations might partially result from differences in the optimal temperature ranges for the individual proteins, especially for proteins from thermophilic organisms like M. jannaschii or C. thermophilum.
DISCUSSION
Kti12 is the only known eukaryotic protein to share a sequence-wide similarity with PSTK (43,44). Our initial structural and functional analyses of Kti12 confirm this notion and show that both proteins indeed consist of two domains, an N-terminal ATPase domain and a C-terminal high affinity tRNA-tethering domain.
We performed an in-depth gene distribution analysis of confirmed Kti12 sequences, which revealed that Kti12 can be found almost exclusively in eukaryotes, including land plants and fungi that actually do not possess PSTK and other components of the selenocysteine pathway (Figure 7A). Although putative Kti12-like sequences could be identified in two closely related archaeal subclades, a search in all other archaeal species returned without any additional hits. Whereas the enzymatically active Elp3 subunit and the cm5U34 modification itself (85) are conserved in archaea, the remaining five Elongator subunits are only found in eukaryotes. After showing that the Elp1 subunit provides the necessary docking platform on the eukaryotic Elongator complex, we suspect that the appearance of Kti12 is coupled to the evolutionary occurrence and establishment of a multi-subunit Elongator complex in eukaryotes and more precisely Elp1. Strikingly, animals, including mammals and humans, possess both Kti12 and PSTK. According to publicly available tissue expression databases (86,87), human and murine Kti12 and PSTK mRNAs and proteins are ubiquitously expressed in different tissues, excluding the possibility of cell type specific production of only one of the proteins at a time. Here we demonstrate that the two proteins get activated by tRNASec and therefore it remains to be shown if their structural and functional similarities may lead to cross activation and/or functional redundancy in animals and specifically in humans.
Figure 7.
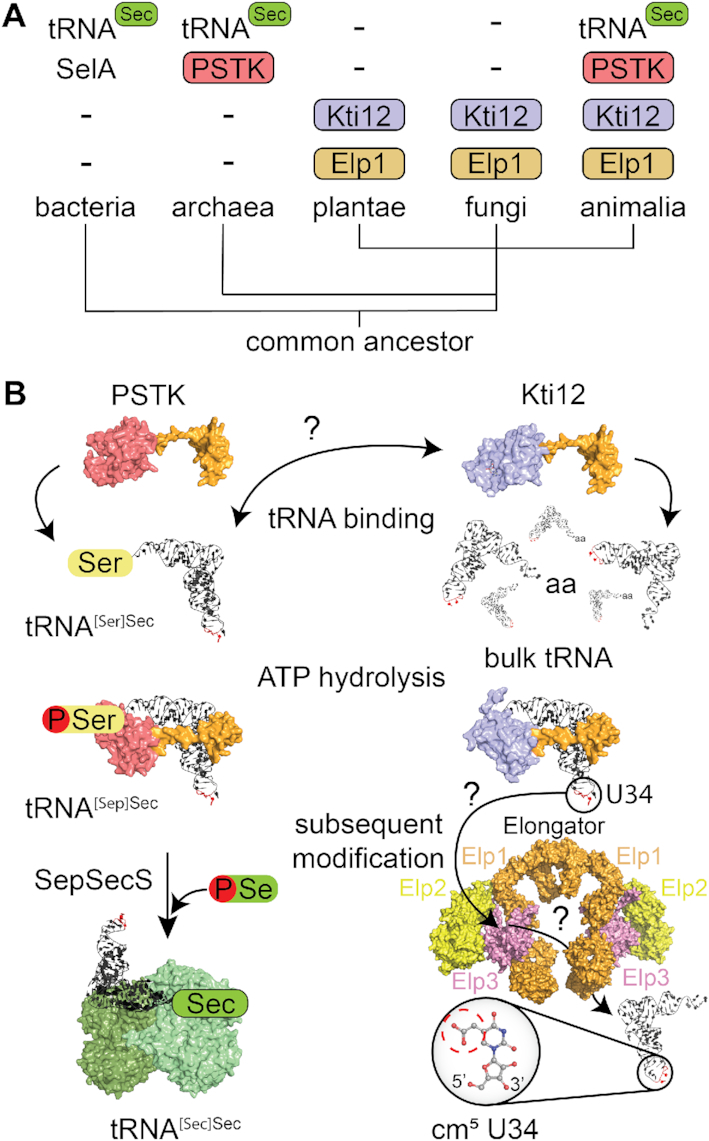
A comparison of Kti12 and PSTK analogs. (A) Schematic overview of the phylogenetic analyses of PSTK and Kti12 distribution. With the exception of bacteria, which use SelA protein for selenocysteine synthesis, PSTK appears together with a tRNASec in archaea and animalia. Plantae and fungi are missing both PSTK as well as tRNASec. Kti12 can be found within all eukaryotic lineages, which is also true for its interaction partner Elp1. (B) PSTK and Kti12 architectures are very similar. Both proteins bind tRNA, however, unlike PSTK, Kti12 is capable of binding to bulk yeast tRNA. Both PSTK and Kti12 utilize tRNA binding for induction of ATP hydrolysis. PSTK transfers gamma phosphate from ATP to tRNA[Ser]Sec generating tRNA[Sep]Sec, which is further used as a substrate by SepSecS, a tetrameric enzyme capable of using selenophosphate as a selenium donor for formation of Sec-tRNASec. Two catalytically active subunits of human SepSecS are shown in light green while catalytically inactive subunits are dark green. In case of Kti12, ATP hydrolysis enables cm5U34 modification in the anticodon of 11 tRNA species by the Elongator complex.
In general, both proteins share multiple similarities, including domain architecture and structure, tRNA binding properties, ATPase activities and downstream signaling to posttranscriptional modification reactions (Figure 7B). PSTK needs to specifically select for tRNA[Ser]Sec at the initial binding step to catalyze the production of tRNA[Sep]Sec and at the same time avoid undesired phosphorylation of tRNA[Ser]Ser. Kti12 binds various uncharged tRNAs, strongly suggesting that Kti12 does not require high specificity and is not likely to carry out specific kinase reactions on these tRNAs. Nevertheless, Kti12 is specifically activated by tRNASecin vitro at an almost identical surface of the NTD, which indicates a similar coupling of tRNA recognition and triggering of ATPase activity.
It is worth emphasizing that the amino acid phosphorylated by PSTK and the cm5 modified U34 base are located on opposite sides of the tRNA molecule, which make it hard to imagine that the ATPase activity of Kti12 is directly involved in the catalytic modification reaction of Elongator. Nonetheless, our data do show that the function of the Kti12 protein is tightly connected to the tRNA modification activity of the Elongator complex. Strikingly, the PSTK substrate tRNA[Ser]Sec carries U34 modifications (78,88). In addition, opal (UGA) read-trough by a tRNA[Ser]Ser suppressor in yeast (sup9e) has been shown to rely on the activity of the Elongator complex (89). A strong activation of Kti12 by tRNASec could be needed for the Elongator complex to deal with this rather unusual tRNA species, which was reported to have a longer acceptor stem, an extended variable loop and different D- and T-arms in relation to canonical tRNASer (74). The precise role of the tRNA induced ATPase activity of Kti12 in the modification of other tRNAs is unclear. We speculate that Kti12 might trigger the recruitment or release of other regulatory factors of the Elongator complex. For instance, Kti12 might allow the recruitment of the Elongator kinase Hrr25 or the release of the phosphatase Sit4 to promote phosphorylation of Elp1 or vice versa to promote dephosphorylation. Kti12 could in principle employ its tRNA-induced ATP hydrolysis also to directly phosphorylate Elongator upon tRNA binding, but we neither detected any direct phosphorylation of Elp1 in vitro using purified proteins nor Kti12’s ATPase activity seems to be functional in the proximity of Elp1. In addition, it is worth mentioning that Kti12 might have additional or alternative functions independent of Elongator (90).
Last but not least, both Elongator and Kti12 can bind to free tRNA and the CTD of Elp1 is involved in tRNA and Kti12 binding in vivo (7,40,91). To date it is not clear, whether Kti12 could bind a tRNA molecule that is also bound to the Elongator complex and whether PSTK can bind to Elongator in a similar fashion as Kti12. Our data show that the ATPase activity of Kti12, which is essential for the tRNA modification activity of Elongator, does require the 3′CCA tail. This observation could indicate that Kti12 acts as a gatekeeper for Elongator that allows only tRNAs at a certain maturation stage to be modified by the complex. Our data on Elp1-mediated inhibition of Kti12, also indicates that the newly identified surface area of Kti12 that is activated by the tRNA 3′CCA tail might be covered by the binding to Elp1. Though, in this scenario Elongator-bound Kti12 might still interact with tRNA via its flexible C-terminus and it remains to be shown at which stage the ATPase activity is required. In addition, we need to understand if binding of Kti12 to tRNAs or Elongator induces similar rearrangements of the NTD and linker regions as shown for PSTK (71). It is worth mentioning that this mechanism is conserved among organisms that either possess (human) or lack (fungi) the selenocysteine pathways. In summary, our complementary structural and functional characterization of Kti12 indicates strong similarities between the regulatory mechanisms of Elongator mediated tRNA modifications and translational recoding of the seleno-proteome in higher eukaryotes.
DATA AVAILABILITY
The atomic coordinates and structure factors for ChKti12 (PDB ID 6QP0) have been deposited with and validated by the European Protein Data Bank (PDBe).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Annika Kotter for technical assistance with the LC–MS/MS profiles and Jeremy Thorner for the kind gift of the anti-Cdc19 antibodies. We thank Przemyslaw Grudnik and beamline staff of MX-14-2 at BESSY II synchrotron source in Berlin, Germany for help with data collection and analyses. Furthermore, we thank A. Chramiec-Głąbik for preparations of the tRNA samples.
Notes
Present address: Wael Abdel-Fattah, Department of Medical Biochemistry and Biophysics, Umeå University, Sweden.
Present address: Rene Zabel, serYmun Yeast GmbH, Halle (Saale), Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
OPUS10 grant [UMO-2015/19/B/NZ1/00343 to R.K. and S.G.] from the National Science Centre; TEAM TECH CORE FACILITY/2017–4/6 grant from Foundation for Polish Science (to S.G.), a joint research grant from the Biotechnology and Biological Sciences Research Council (BBSRC) [BB/F0191629/1 to M.J.R.S., BB/F019106/1 to R.S.]; Deutsche Forschungsgemeinschaft (DFG) [SCHA750/18 to to R.S., BR921/9 to K.D.B.]; Priority Program 1784 Chemical Biology of Native Nucleic Acid Modifications [HE3397/13 to to M.H., SCHA750/20 to R.S.]; research consortium PhosMOrg (P/1082: University of Kassel, Germany to R.S.]; STSM fellowship award (to A.H.) in the framework of the European Union Cost Action [EPITRAN CA16120]. Funding for open access charge: Narodowe Centrum Nauki [UMO-2015/19/B/NZ1/00343]; Deutsche Forschungsgemeinschaft [SCHA750/18].
Conflict of interest statement. None declared.
REFERENCES
- 1. Otero G., Fellows J., Li Y., de Bizemont T., Dirac A.M.G., Gustafsson C.M., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell. 1999; 3:109–118. [DOI] [PubMed] [Google Scholar]
- 2. Winkler G.S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J.Q.. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen C., Huang B., Anderson J.T., Bystrom A.S.. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PLoS One. 2011; 6:e20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang B., Johansson M.J.O., Byström A.S.. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005; 11:424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mehlgarten C., Jablonowski D., Wrackmeyer U., Tschitschmann S., Sondermann D., Jager G., Gong Z., Bystrom A.S., Schaffrath R., Breunig K.D. et al.. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants (vol 76, pg 1082, 2010). Mol. Microbiol. 2010; 77:1082–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Glatt S., Letoquart J., Faux C., Taylor N.M., Seraphin B., Muller C.W.. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012; 19:314–320. [DOI] [PubMed] [Google Scholar]
- 7. Di Santo R., Bandau S., Stark M.J.R.. A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol. Microbiol. 2014; 92:1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dauden M.I., Jaciuk M., Muller C.W., Glatt S.. Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett. 2017; 592:502–515. [DOI] [PubMed] [Google Scholar]
- 9. Chen C., Tuck S., Byström A.S.. Defects in tRNA modification associated with neurological and developmental dysfunctions in caenorhabditis elegans elongator mutants. PLoS Genet. 2009; 5:e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johansson M.J.O., Esberg A., Huang B., Bjork G.R., Bystrom A.S.. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell Biol. 2008; 28:3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ranjan N., Rodnina M.V.. Thio-Modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J. Am. Chem. Soc. 2017; 139:5857–5864. [DOI] [PubMed] [Google Scholar]
- 13. Klassen R., Bruch A., Schaffrath R.. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2017; 14:1252–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rezgui V.A., Tyagi K., Ranjan N., Konevega A.L., Mittelstaet J., Rodnina M.V., Peter M., Pedrioli P.G.. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tükenmez H., Xu H., Esberg A., Byström A.S.. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res. 2015; 43:9489–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glatt S., Seraphin B., Müller C.W.. Elongator: transcriptional or translational regulator. Transcription. 2012; 3:273–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bauer F., Matsuyama A., Candiracci J., Dieu M., Scheliga J., Wolf D.A., Yoshida M., Hermand D.. Translational control of cell division by Elongator. Cell Rep. 2012; 1:424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thommen M., Holtkamp W., Rodnina M.V.. Co-translational protein folding: progress and methods. Curr. Opin. Struct. Biol. 2017; 42:83–89. [DOI] [PubMed] [Google Scholar]
- 19. Klassen R., Ciftci A., Funk J., Bruch A., Butter F., Schaffrath R.. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016; 44:10946–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y.T., Hims M.M., Shetty R.S., Mull J., Liu L.J., Leyne M., Slaugenhaupt S.A.. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol. Cell Biol. 2009; 29:736–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kojic M., Gaik M., Kiska B., Salerno-Kochan A., Hunt S., Tedoldi A., Mureev S., Jones A., Whittle B., Genovesi L.A. et al.. Elongator mutation in mice induces neurodegeneration and ataxia-like behavior. Nat. Commun. 2018; 9:3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoo H., Son D., Jang Y.J., Hong K.. Indispensable role for mouse ELP3 in embryonic stem cell maintenance and early development. Biochem. Biophys. Res. Commun. 2016; 478:631–636. [DOI] [PubMed] [Google Scholar]
- 23. Close P., Gillard M., Ladang A., Jiang Z., Papuga J., Hawkes N., Nguyen L., Chapelle J.P., Bouillenne F., Svejstrup J. et al.. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of elongator. J. Biol. Chem. 2012; 287:32535–32545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ladang A., Rapino F., Heukamp L.C., Tharun L., Shostak K., Hermand D., Delaunay S., Klevernic I., Jiang Z., Jacques N. et al.. Elp3 drives Wnt-dependent tumor initiation and regeneration in the intestine. J. Exp. Med. 2015; 212:2057–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rapino F., Delaunay S., Rambow F., Zhou Z., Tharun L., De Tullio P., Sin O., Shostak K., Schmitz S., Piepers J. et al.. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature. 2018; 558:605–609. [DOI] [PubMed] [Google Scholar]
- 26. Delaunay S., Rapino F., Tharun L., Zhou Z., Heukamp L., Termathe M., Shostak K., Klevernic I., Florin A., Desmecht H. et al.. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J. Exp. Med. 2016; 213:2503–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bento-Abreu A., Jager G., Swinnen B., Rué L., Hendrickx S., Jones A., Staats K.A., Taes I., Eykens C., Nonneman A. et al.. Elongator subunit 3 (ELP3) modifies ALS through tRNA modification. Hum. Mol. Genet. 2018; 27:1276–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cohen J.S., Srivastava S., Farwell K.D., Lu H.M., Zeng W., Lu H., Chao E.C., Fatemi A.. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. A. 2015; 167:1391–1395. [DOI] [PubMed] [Google Scholar]
- 29. Karlsborn T., Tukenmez H., Chen C., Bystrom A.S.. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcmsU in tRNA. Biochem. Biophys. Res. Commun. 2014; 454:441–445. [DOI] [PubMed] [Google Scholar]
- 30. Alings F., Sarin L.P., Fufezan C., Drexler H.C., Leidel S.A.. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA. 2015; 21:202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Damon J.R., Pincus D., Ploegh H.L.. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol. Biol. Cell. 2015; 26:270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu C., Begley T.J., Dedon P.C.. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014; 588:4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Laxman S., Sutter B.M., Wu X., Kumar S., Guo X., Trudgian D.C., Mirzaei H., Tu B.P.. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell. 2013; 154:416–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Patil A., Dyavaiah M., Joseph F., Rooney J.P., Chan C.T.Y., Dedon P.C., Begley T.J.. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell Cycle. 2012; 11:3656–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Butler A.R., White J.H., Folawiyo Y., Edlin A., Gardiner D., Stark M.J.. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol. Cell Biol. 1994; 14:6306–6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang B., Lu J., Bystrom A.S.. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008; 14:2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bär C., Zabel R., Liu S., Stark M.J.R., Schaffrath R.. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol. Microbiol. 2008; 69:1221–1233. [DOI] [PubMed] [Google Scholar]
- 38. Fichtner L., Schaffrath R.. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol. Microbiol. 2002; 44:865–875. [DOI] [PubMed] [Google Scholar]
- 39. Glatt S., Zabel R., Vonkova I., Kumar A., Netz D.J., Pierik A.J., Rybin V., Lill R., Gavin A.C., Balbach J. et al.. Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure. 2015; 23:149–160. [DOI] [PubMed] [Google Scholar]
- 40. Abdel-Fattah W., Jablonowski D., Di Santo R., Thüring K.L., Scheidt V., Hammermeister A., ten Have S., Helm M., Schaffrath R., Stark M.J.R. et al.. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet. 2015; 11:e1004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jablonowski D., Fichtner L., Stark M.J.R., Schaffrath R.. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol. Biol. Cell. 2004; 15:1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mehlgarten C., Jablonowski D., Breunig K.D., Stark M.J.R., Schaffrath R.. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol. Microbiol. 2009; 73:869–881. [DOI] [PubMed] [Google Scholar]
- 43. Mehlgarten C., Prochaska H., Hammermeister A., Abdel-Fattah W., Wagner M., Krutyhołowa R., Jun S.E., Kim G.-T., Glatt S., Breunig K.D. et al.. Use of a yeast tRNase killer toxin to diagnose Kti12 motifs required for tRNA modification by Elongator. Toxins (Basel). 2017; 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sherrer R.L., Ho J.M.L., Söll D.. Divergence of selenocysteine tRNA recognition by archaeal and eukaryotic O-phosphoseryl-tRNASec kinase. Nucleic Acids Res. 2008; 36:1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carlson Ba., Xu X.M., Kryukov G.V., Rao M., Berry M.J., Gladyshev V.N., Hatfield D.L.. Identification and characterization of phosphoseryl-tRNA(Ser Sec) kinase. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:12848–12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu X.-M., Carlson B.A., Zhang Y., Mix H., Kryukov G.V., Glass R.S., Berry M.J., Gladyshev V.N., Hatfield D.L.. New developments in selenium biochemistry: Selenocysteine biosynthesis in eukaryotes and archaea. Biol. Trace Elem. Res. 2007; 119:234–241. [DOI] [PubMed] [Google Scholar]
- 47. Yuan J., Palioura S., Salazar J.C., Su D., O’Donoghue P., Hohn M.J., Cardoso A.M., Whitman W.B., Söll D.. RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:18923–18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baron C., Westhof E., Böck A., Giegé R.. Solution Structure of Selenocysteine-inserting tRNASec from Escherichia coli. J. Mol. Biol. 1993; 231:274–292. [DOI] [PubMed] [Google Scholar]
- 49. Sturchler C., Westhof E., Carbon P., Krol A.. Unique secondary and tertiary structural features of the eucaryotic selenocysteine tRNA(Sec). Nucleic Acids Res. 1993; 21:1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Beck T., Krasauskas A., Gruene T., Sheldrick G.M.. A magic triangle for experimental phasing of macromolecules. Acta Crystallogr. D Biol. Crystallogr. 2008; 64:1179–1182. [DOI] [PubMed] [Google Scholar]
- 51. Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. IUCr . Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015; 71:3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 55. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 56. Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J.. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009; 25:1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DeLano W.L. The PyMOL molecular graphics system. DeLano Scientific. 2002; Palo Alto: www.pymol.org. [Google Scholar]
- 58. Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N.. ConSurf: Using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013; 53:199–206. [Google Scholar]
- 59. Jurrus E., Engel D., Star K., Monson K., Brandi J., Felberg L.E., Brookes D.H., Wilson L., Chen J.H., Liles K. et al.. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018; 27:112–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H.. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002; 30:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Edelheit O., Hanukoglu A., Hanukoglu I.. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009; 9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E.. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999; 15:963–972. [DOI] [PubMed] [Google Scholar]
- 63. Köhrer C., Rajbhandary U.L.. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods. 2008; 44:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., Eliceiri K.W.. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017; 18:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970; 227:680–685. [DOI] [PubMed] [Google Scholar]
- 66. Frohloff F., Fichtner L., Jablonowski D., Breunig K.D., Schaffrath R.. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001; 20:1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Klassen R., Wemhoff S., Krause J., Meinhardt F.. DNA repair defects sensitize cells to anticodon nuclease yeast killer toxins. Mol. Genet. Genomics. 2011; 285:185–195. [DOI] [PubMed] [Google Scholar]
- 68. Jablonowski D., Zink S., Mehlgarten C., Daum G., Schaffrath R.. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol. Microbiol. 2006; 59:677–688. [DOI] [PubMed] [Google Scholar]
- 69. Kellner S., Neumann J., Rosenkranz D., Lebedeva S., Ketting R.F., Zischler H., Schneider D., Helm M.. Profiling of RNA modifications by multiplexed stable isotope labelling. Chem. Commun. 2014; 50:3516–3518. [DOI] [PubMed] [Google Scholar]
- 70. Sherrer R.L., O’Donoghue P., Söll D.. Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 2008; 36:1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sherrer S.R.L., Araiso Y., Aldag C., Ishitani R., Ho J.M., Söll D., Nureki O.. C-terminal domain of archaeal O-phosphoseryl- tRNA kinase displays large-scale motion to bind the 7-bp D-stem of archaeal tRNA. Nucleic Acids Res. 2011; 39:1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Moser J., Lange C., Krausze J., Rebelein J., Schubert W.-D., Ribbe M.W., Heinz D.W., Jahn D.. Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:2094–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fichtner L., Frohloff F., Bürkner K., Larsen M., Breunig K.D., Schaffrath R.. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol. Microbiol. 2002; 43:783–791. [DOI] [PubMed] [Google Scholar]
- 74. Chiba S., Itoh Y., Sekine S., Yokoyama S.. Structural Basis for the Major Role of O-Phosphoseryl-tRNA Kinase in the UGA-Specific Encoding of Selenocysteine. Mol. Cell. 2010; 39:410–420. [DOI] [PubMed] [Google Scholar]
- 75. Glatt S., Zabel R., Kolaj-Robin O., Onuma O.F., Baudin F., Graziadei A., Taverniti V., Lin T.Y., Baymann F., Seraphin B. et al.. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016; 23:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Araiso Y., Sherrer R.L., Ishitani R., Ho J.M.L., Söll D., Nureki O.. Structure of a tRNA-dependent kinase essential for selenocysteine decoding. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:16215–16220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fisher A.J., Smith C.A., Thoden J.B., Smith R., Sutoh K., Holden H.M., Rayment I.. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995; 34:8960–8972. [DOI] [PubMed] [Google Scholar]
- 78. Carlson B.A., Lee B.J., Tsuji P.A., Copeland P.R., Schweizer U., Gladyshev V.N., Hatfield D.L.. Selenocysteine tRNA([Ser]Sec), the central component of selenoprotein Biosynthesis: Isolation, identification, modification, and sequencing. Methods Mol. Biol. 2018; 1661:43–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Klassen R., Grunewald P., Thüring K.L., Eichler C., Helm M., Schaffrath R.. Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PLoS One. 2015; 10:e0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lu J., Huang B., Esberg A., Johansson M.J.O., Bystrom A.S.. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005; 11:1648–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jablonowski D., Schaffrath R.. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem. Soc. Trans. 2007; 35:1533–1537. [DOI] [PubMed] [Google Scholar]
- 82. Fichtner L., Frohloff F., Jablonowski D., Stark M.J.R., Schaffrath R.. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol. Microbiol. 2002; 45:817–826. [DOI] [PubMed] [Google Scholar]
- 83. Dauden M.I., Kosinski J., Kolaj-Robin O., Desfosses A., Ori A., Faux C., Hoffmann N.A., Onuma O.F., Breunig K.D., Beck M. et al.. Architecture of the yeast Elongator complex. EMBO Rep. 2017; 18:264–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Setiaputra D.T., Cheng D.T., Lu S., Hansen J.M., Dalwadi U., Lam C.H., To J.L., Dong M., Yip C.K.. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep. 2017; 18:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. van Tran N., Muller L., Ross R.L., Lestini R., Létoquart J., Ulryck N., Limbach P.A., de Crécy-Lagard V., Cianférani S., Graille M.. Evolutionary insights into Trm112-methyltransferase holoenzymes involved in translation between archaea and eukaryotes. Nucleic Acids Res. 2018; 46:8483–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Palasca O., Santos A., Stolte C., Gorodkin J., Jensen L.J.. TISSUES 2.0: an integrative web resource on mammalian tissue expression. Database. 2018; 2018:bay003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A. et al.. Proteomics. Tissue-based map of the human proteome. Science. 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 88. Diamond A.M., Choi I.S., Crain P.F., Hashizume T., Pomerantz S.C., Cruz R., Steer C.J., Hill K.E., Burk R.F., McCloskey J.A. et al.. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec). J. Biol. Chem. 1993; 268:14215–14223. [PubMed] [Google Scholar]
- 89. Zabel R., Bär C., Mehlgarten C., Schaffrath R.. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol. Microbiol. 2008; 69:175–187. [DOI] [PubMed] [Google Scholar]
- 90. Schaffrath R., Leidel S.A.. Wobble uridine modifications-a reason to live, a reason to die. RNA Biol. 2017; 14:1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Frohloff F., Jablonowski D., Fichtner L., Schaffrath R.. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (gamma-toxin target (TOT)) complex. J. Biol. Chem. 2003; 278:956–961. [DOI] [PubMed] [Google Scholar]
- 92. Laskowski R.A., Swindells M.B.. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011; 51:2778–2786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors for ChKti12 (PDB ID 6QP0) have been deposited with and validated by the European Protein Data Bank (PDBe).