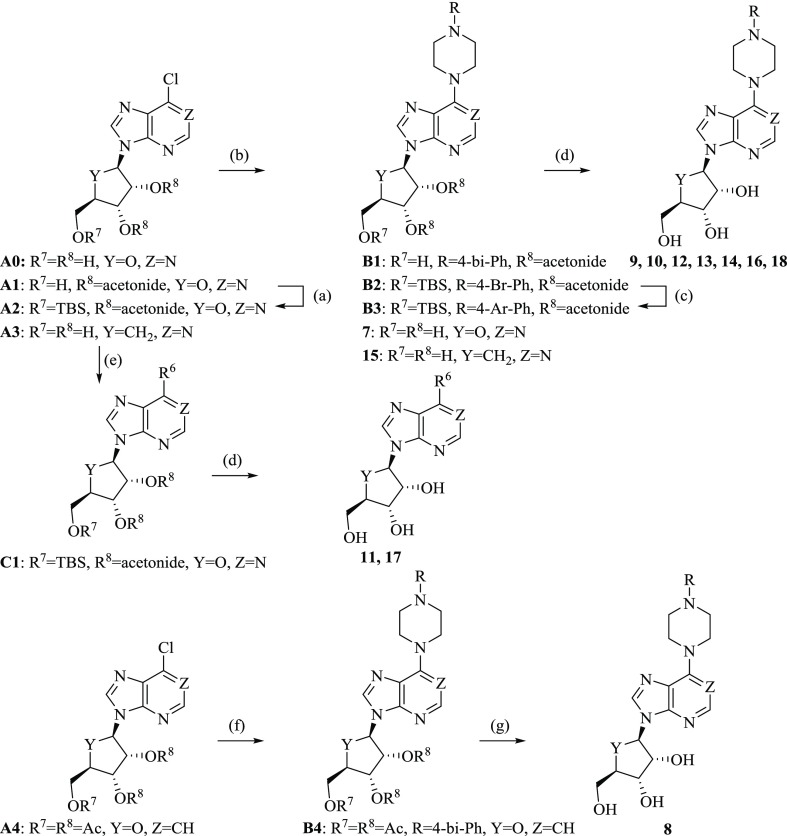
Abstract
Mycobacterium tuberculosis adenosine kinase (MtbAdoK) is an essential enzyme of Mtb and forms part of the purine salvage pathway within mycobacteria. Evidence suggests that the purine salvage pathway might play a crucial role in Mtb survival and persistence during its latent phase of infection. In these studies, we adopted a structural approach to the discovery, structure-guided design, and synthesis of a series of adenosine analogues that displayed inhibition constants ranging from 5 to 120 nM against the enzyme. Two of these compounds exhibited low micromolar activity against Mtb with half maximal effective inhibitory concentrations of 1.7 and 4.0 μM. Our selectivity and preliminary pharmacokinetic studies showed that the compounds possess a higher degree of specificity against MtbAdoK when compared with the human counterpart and are well tolerated in rodents, respectively. Finally, crystallographic studies showed the molecular basis of inhibition, potency, and selectivity and revealed the presence of a potentially therapeutically relevant cavity unique to the MtbAdoK homodimer.
Introduction
Mycobacterium tuberculosis (Mtb), the bacterium that causes pulmonary tuberculosis (TB), represents one of the major leading causes of death worldwide by a single infectious agent. One-third of the world’s population is thought to harbor latent TB, and around 5–10% of these infected individuals are expected to develop the active disease sometime during their lifetime.1 The rapid emergence of multidrug resistant and extensively drug-resistant (XDR) TB demands the development of novel chemotherapeutic agents with novel molecular targets.
The purine salvage pathway is a druggable pathway within mycobacteria. In this pathway, preformed nucleobases from the product of nucleic acid breakdown are converted to their corresponding purine nucleotides by the purine salvage enzymes. Although the de novo and purine salvage pathways have not been extensively studied in Mtb, it is known that Mtb possesses all of the enzymes required for both pathways. It is currently unknown if there is restrictive regulation of the two pathways. However, by switching to the salvage pathway Mtb can bypass several chemically demanding steps.2 This has led to the hypothesis that the salvage pathway might be the most likely source of nucleotides within the hostile and nutrient-deprived microenvironment encountered by Mtb during its latent phase of infection.2,3
M. tuberculosis adenosine kinase (MtbAdoK) performs a critical step in the purine salvage pathway within mycobacteria. The enzyme catalyzes the conversion of adenosine to adenosine monophosphate in a Mg2+ and adenosine 5′-triphosphate (ATP)-dependent manner.4 The crystal structure of MtbAdoK has been previously solved at high resolution with the substrate (adenosine), substrate analogue 2-fluoroadenosine, ATP analogue AMP–PCP and without substrate (apo) at resolutions of 1.90, 1.93, 1.90, and 1.50 Å, respectively.5
The crystallographic data showed that the apo and the AMP–PCP structures adopted the opened conformation of the protein where the active site is solvent exposed. In contrast, the substrate or substrate analogue complexes revealed that the lid domain of MtbAdoK undergoes a 30° movement upon substrate binding.6 This conformational change effectively brings lid domain residues Asp12, Phe116, and Phe102 in close contacts with adenosine, thereby completing the active site (Figure S1a).5,7−9 As previously described, stabilization of the ribose and adenine rings are primarily mediated by π-stacking interactions with residues Phe102 and Phe116, respectively. Hydrogen bonding networks with the adenine moiety were described to occur with residues Gln172, Gln173, Ser8, and Ser36′ while the ribose forms extensive hydrogen bonding interactions with residues Gln172, Asp12, Gly48, Asn52, and catalytic base Asp257 (Figure S1b).5
MtbAdoK is significantly different when compared with eukaryotic adenosine kinases.8 Although the human adenosine kinase (hAdoK) and MtbAdoK are composed of a small-lid like domain and large domain, hAdoK shares less than 20% sequence identity with MtbAdoK. The hAdoK structure has been previously solved at 1.4 Å with substrate bound. The structure was initially solved with two molecules of adenosine embedded within the enzyme, one in the active site and another in cofactor (ATP) site.8 A critical difference between MtbAdoK and its eukaryotic counterparts is that MtbAdoK is a functional homodimer.5,8 In addition, prior to its discovery and characterization by Long et al. in 2003, the adoK gene was thought to be unique to eukaryotic organisms and was annotated as a general carbohydrate kinase (cbhK) in Mtb.4 On the basis of its amino acid sequence, MtbAdoK is more closely related to members of the phosphofructokinase B family of sugars kinases, which includes ribokinase, a homodimer and the prototypical member of this family.6
MtbAdoK is essential for the survival of the bacilli when utilizing cholesterol as a carbon source and in infected mouse models.10,11 Even though the precise role of the purine salvage pathway under these conditions is currently unknown, biochemical and structural characterization suggests that MtbAdoK might represent a new class of bacterial adenosine kinases that differs from its eukaryotic counterparts by its unique quaternary structure and regulatory mechanisms.4,12,13 Furthermore, our preliminary efforts to find novel drugs and drug targets identified MtbAdoK as the target of the nucleoside analogue iodotubercidin. Mtb H37Rv-resistant mutants raised against iodotubercidin identified three nonsynonymous mutations, that is, D251N, Q172P, and G262S; all of which displayed minimum inhibitory concentrations (MICs) > 100 μM when compared with WT (18.0 ± 10.0 μM). The essentiality, differences between hAdoK and MtbAdoK and on-target activity of the nucleoside analogue described above make MtbAdoK an attractive and novel drug target.
To date, most reported inhibitors of MtbAdoK have been designed as substrate surrogates to elicit the production of toxic metabolites.14−18 For example, Parker et al. showed that the H37Ra Mtb strain was able to uptake the adenosine analogue 2-methyl-adenosine and that MtbAdoK was responsible for its activation into the toxic metabolite 2-methyl-AMP.16 Despite having some advantages, AMP or ATP toxic metabolites could inadvertently lead to undesirable and cross-species off-target effects exacerbating the identification and diminishing the value of a potential drug target.
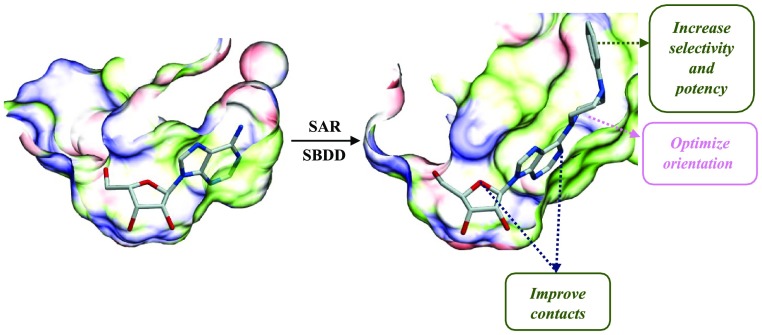
In this work, we adopted a structure-guided approach to the design of very potent and safe adenosine analogues. To gain insights into the chemical space surrounding the active site, we solved the crystal structures of MtbAdoK complexed to several adenosine analogues at resolutions between 1.70 and 2.23 Å. This initial approach laid the foundation for the structure-guided design and synthesis of several very potent 6-substituted adenosine analogues as selective inhibitors of MtbAdoK. Several of the synthesized compounds displayed low micromolar anti-Mtb activity in a whole-cell assay, which supports the concept that inhibition of MtbAdoK can be used as an approach to treat TB. Finally, these compounds exhibited a higher degree of specificity against MtbAdoK when compared with the human counterpart, and a 6-substituted adenosine analogue from our series was well tolerated in Swiss Webster mice at high levels of exposure while demonstrating adequate pharmacokinetic properties.
Results and Discussion
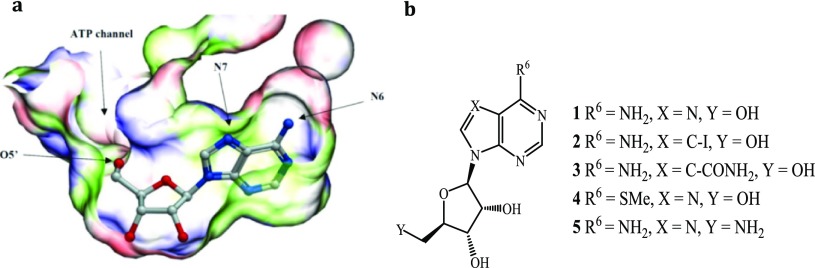
Structural analysis of the MtbAdoK–adenosine (1) complex showed a high potential for the structure-guided drug design (Figure 1a).5 Of particular interest are positions N6 and N7 of the adenine ring and position O5′ of the ribose moiety; all of which have been the focal point of several chemistry-based structural–activity relationship (SAR) studies.12−15 These studies have highlighted that substitutions at the aforementioned positions lead to inhibitory activity against MtbAdoK and hAdoK. The chemistry-based efforts have also suggested that there is a higher degree of selectivity to MtbAdoK when substitutions are located at the N6-position when compared with the N7-analogues. However, the structural and molecular basis for the reported selectivity has remained unexplained. The 5′-position offers a more practical approach to inhibition given that the critical hydroxyl group is required for catalysis.
Figure 1.
Positions of interest for structure-guided drug design and tool compounds utilized in crystallography studies. (a) Adenosine bound to the active site of MtbAdoK (PDB ID 2PKM). (b) Tool compounds used to explore the chemical space surrounding the positions highlighted in (a).
To guide our drug discovery efforts and to investigate the molecular basis of selectivity and inhibition, we solved the crystal structure complexes of several known AdoK inhibitors. Iodotubercidin (2) and sangivamycin (3) were selected to investigate the conformational flexibility and chemical properties surrounding the binding pocket near the N7-position of the adenosine scaffold. Additionally, 6-methylmercaptopurine riboside (4) and 5′-aminoadenosine (5) cocrystal structures were solved to explore the 6- and 5′-positions of the adenosine scaffold, respectively (Figure 1b).
Our structural studies commenced with the cocrystallization of the N7-substituted analogues. Iodotubercidin possesses an iodine group at position 7 and is a semisynthetic derivative of the natural compound tubercidin, whereas sangivamycin is a natural compound derived from Streptomyces rimosus and contains an amide group at position 7.12,19,20 Compound 2 has been noted to be a general kinase inhibitor, displaying an inhibitory activity against MtbAdoK, hAdoK, MAP kinases, and Ser/Thr kinases while compound 3 has been previously observed to be an inhibitor of protein kinase C, MtbAdoK, and hAdoK.19−21
Cocrystallization of MtbAdoK with compounds 2 and 3 was produced via vapor diffusion, and the structures were solved using the molecular replacement (MR) method utilizing the previously reported adenosine-bound structure.5 The crystals of both complexes were determined to be in the P41 crystal space group with two molecules in the asymmetric unit (ASU). The MtbAdoK-2 complex was refined to 2.10 Å resolution and displayed a closed conformation of the enzyme with a backbone rmsd of 1.14 Å amongst all Cα atoms when superimposed to the MtbAdoK–adenosine structure (Figure 2a, Tables S1 and S2). The MtbAdoK-3 complex refined to 2.10 Å resolution displayed a closed conformation of the enzyme with a backbone rmsd of 1.15 Å amongst all Cα atoms when superimposed to the MtbAdoK–adenosine structure (Figure 2b, Tables S3 and S4). As in the case of the hAdoK–adenosine structure, both crystal structures showed positive electron density for the inhibitor bound to the active site in a similar position to adenosine and electron density in the cofactor (ATP) binding site (Figure S2a,b). However, only compound 2 displayed full electron density in the ATP binding site while compound 3 displayed partial electron density for the adenine ring only. Overall, the crystal structures of compounds 2 and 3 complexed to MtbAdoK displayed many similarities with the previously reported MtbAdoK–adenosine complex. The inhibitors were bound in the same conformation and locations as we observed in the adenosine-bound structure. Residues Phe116 and Phe102 were involved in π-stacking interactions with the adenine and ribose rings, respectively. The ribose moiety was found to be stabilized by hydrogen bonding with several residues. Asp12 oxygen OD1 H-bonds with the O2′ and O3′ of ribose, OD2 of catalytic base Asp257 H-bonds with O5′, ND2 of Asn52 H-bonds with O3′, and backbone N of Gly48 H-bonds O2′; all of which are likely to contribute significantly to the binding affinity (Tables S1, S3, and S5).
Figure 2.
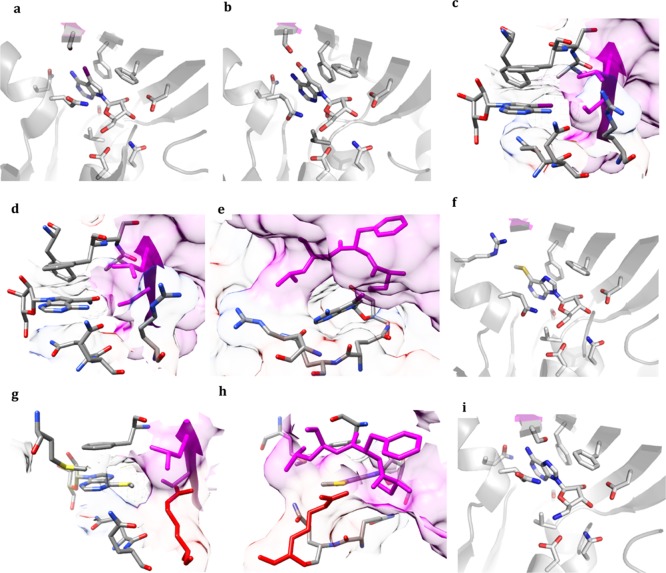
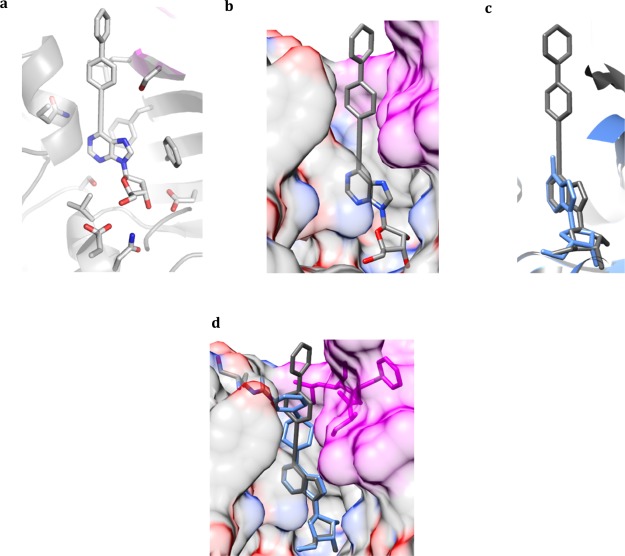
Crystal structure complexes of compounds 2–5. (a) 2 bound to the active site of MtbAdoK. (b) 3 bound to the active site of MtbAdoK. (c,d) N7-substituents of compounds 2–3 are buried in an active site pocket composed of residues from chain A and chain B. (e) Chimney-like cavity observed above position N6 formed by residues from chain A and chain B. (f) 4 bound to the active site of MtbAdoK. (g) Methylmercapto group of 4 is accommodated in a compound-induced pocket formed by the movement of Arg176 (red). (h) Chimney-like cavity observed in the cocrystal structures of 2 and 3 is blocked by the conformational change of Arg176 (red). (i) 5 bound to the active site of MtbAdoK. In all cases, chain A residues are colored by heteroatom and chain B residues are color magenta.
The most notable differences between the cocrystal structures of adenosine, 2, and 3 were observed in the interactions formed by residues Gln172, Gln173, and Ser36′. In the adenosine-bound structure, Gln172 NE2 H-bonds with O5′ (2.62 Å) and O4′ (3.26 Å) while backbone O of Gln172 H-bonds with N6 (3.26 Å). Although interactions with Gln172 were observed in the MtbAdoK-3 complex (NE2–O4′ distance of 3.23 Å and NE2–O5′ distance of 2.83 Å), no interactions formed with Gln172 were observed in the MtbAdoK-2 complex. A similar scenario was seen for Gln173. In the MtbAdoK-1 structure, NE2 of Gln173 H-bonds with N1 (2.99 Å) while OE1 of Gln173 H-bonds with N6 (2.96 Å) of the adenine scaffold. On the other hand, as observed in the MtbAdoK-3 and MtbAdoK-2 binary complexes, Gln173 only formed H-bonds with N6 with distances of 3.25 Å (OE1–N6) and 3.29 Å (OE1–N6), respectively (Tables S1, S3, and S5).
The crystal structures of compounds 2 and 3 showed that the iodine group of 2 and the amide group of 3 was oriented toward Ser36′. One unique aspect of MtbAdoK, being a homodimer, is that residues from one subunit presumably complete the active of the other. It has been shown that Ser36 from chain B completes the active site of chain A and vice versa by hydrogen bonding with positions N6 (OG–N6 distance of 3.43 Å) and N7 (OG–N7 distance of 2.65 Å) of the adenine ring.5 Ser36 forms part of the lid-domain of MtbAdoK, and as reported before, the dimerization interface of MtbAdoK occurs via extensive van der Waals (vdW) contacts formed by the lid-domain of each chain.5 The observed orientation of both N7-substituted adenosine analogues (2–3) suggests that the N7-position is obstructed by chain B. That is, chain B represents an imposing physical barrier at this position, thus limiting drug discovery efforts to the active site of the enzyme (Figure 2c,d). The MtbAdoK-3 structure showed that the −NH2 of the amide group of 3 formed a H-bond with OG of Ser36′ (3.00 Å, Table S3). In contrast, H-bond interactions mediated by Ser36′ with compound 2 were not observed in the MtbAdoK-2 complex. Instead, the interaction is of vdW nature with OG–I distance of 3.07 Å (Table S1). Finally, the N7-substituents of 2 and 3 were observed to be buried in a pocket formed by several conserved active site residues of MtbAdoK including Phe102 (Cys123 in hAdoK) and Phe116 (Phe170 in hAdoK).
Overall, structural analysis of the N7-analogues (2–3) suggested that bulky substitutions at the N7-position are likely to be favorably accommodated in the active site (Figure 2c,d). Our structural findings go well in accordance with previously reported chemistry-based SAR efforts focused on N7-substituted adenosine analogues, where Snášel et al. reported that several N7-anthracene or N7-phenanthrene derivatives displayed high potency against MtbAdoK.15 The same group synthesized several N7-chain-extended bulky groups that displayed low potency against the enzyme more than likely due to occlusion by chain B, validating our structural assessment.15 The bulky iodine group of 2 is observed to be completely buried in a pocket formed by residues Phe116, Phe102, Gln172, Gln173, Ala114, Ser115, Leu38′ and Ser36′ leading to several polar and vdW interactions (Figure 2c). In contrast, the amide group of 3 occupies a smaller portion of the pocket (Figure 2d). We also observed that the amide group of 3 participates in many of the conserved H-bond interactions (Gln172 with O5′ and O4′ and Gln173 with N6) when compared with the MtbAdoK-1 complex (Tables S3 and S5). In contrast, the bulky iodine group of 2 sterically hinders many of the conserved interactions described in the MtbAdoK-1 complex, including all of the H-bonds formed by Gln172 and Ser36′ (Gln172 with O5′, O4′ and N6 and Ser36′ with N6 and N7) and the H-bond between Gln173 and N1 (Tables S1 and S5). Our enzymatic assay also showed that compound 2 is more potent than compound 3 displaying 50% inhibitory concentrations (IC50) of 1.2 and 16.5 μM, respectively, further validating our structural assessment. Finally, the crystal structure complexes of the N7 analogues revealed the presence of a “chimney-like” cavity situated above position N6 of the adenine scaffold and that is formed by residues from both chains of the MtbAdoK homodimer, thereby representing a unique structural feature of MtbAdoK (Figure 2e).
Next, we decided to explore the chemical space surrounding the binding pocket near the N6-position by cocrystallizing MtbAdoK with 6-methylmercaptopurine riboside. Compound 4 is a derivative of mercaptopurine, a drug used to treat acute lymphatic leukemia and possess a methylmercapto group at the N6-position of the adenosine scaffold. The binary complex of MtbAdoK-4 was refined to 2.0 Å resolution with two molecules in the ASU in the P41 crystal space group (Figure 2f, Tables S6 and S7). Like the cocrystals structures of 2 and 3, the MtbAdoK-4 binary complex displayed a closed conformation of the enzyme with a backbone rmsd of 1.46 Å amongst all Cα atoms when compared with the MtbAdoK-1 complex. Positive electron density was also observed for a molecule of 4 bound to the ATP site. As in the case of the MtbAdoK-3 structure, we were only able to model the adenine moiety due to the absence of electron density of the ribose (Figure S3).
The MtbAdoK-4 structure was very similar to the cocrystal structures of compounds 2–3 and MtbAdoK–adenosine complex. The aforementioned binding interactions formed by residues Phe102, Phe116, Asp12, Asp257, Ser8, Asn52, Gly48, and Val49 were maintained. The most notable differences were found with residues Gln173 and Ser36′. Foremost, the critical H-bonds formed by NE2 and OE1 of Gln173 with N1 and N6 of compound 4 were not observed. Also, the H-bonds formed by OG of Ser36′ with N6 and N7 of compound 4 were not observed in the cocrystal structure. The lack of interactions with residues Ser36′ and Gln173 can be attributed to steric shielding by the methylmercapto group of 4. Detailed inspection of the binary complex revealed that the methylmercapto group of 4 was oriented toward a pocket formed by residues Gln173, Gln172, Met121, Phe116, Arg176, Leu38′, and Ser36′ (Figure 2g). Arg176 was observed to be oriented toward the solvent in the MtbAdoK–adenosine and crystals structures of compounds 2 and 3 (Figure 2c,d). However, the MtbAdoK-4 structure revealed that Arg176 had well-defined electron density for the side chain oriented toward the methylmercapto group of 4. The reorientation of Arg176 led to the formation of a new compound-induced pocket between residues Phe116, Met121, Gln172, Gln173, Arg176, Leu38′, and Ser36′, consequently leading to extensive vdW and polar interactions between the residues mentioned above and methylmercapto group (Figure 2g). Remarkably, the compound-induced movement of Arg176 closed the “chimney-like” cavity we saw in the cocrystal structures of compounds 2 and 3, indicating that residues surrounding the N6-position are flexible (Figure 2c–e,g,h). The orientation and proximity (4.4 Å) of the cationic CZ of Arg176 with respect to the sulfur atom of the methylmercapto group suggests the possibility of cation–polar interactions. Finally, it should be noted that the methylmercapto group seemed to occupy a small portion of the pocket, indicating that bulky groups might be good candidates to further improve contacts at the position 6.
Just like compound 2, compound 4 displayed good potency (IC50 of 2.3 μM) in our enzymatic assay. The similar potencies can be attributed to the comparable vdW and polar contacts formed by the substituents within their respective pockets. Although these two inhibitors have substitutions at different positions within the adenine ring, their mode of interaction with the protein is similar. That is, both substitutions prevented H-bonds with positions N1, N6, and N7 of the adenine moiety. Our structure-guided SAR studies revealed that the iodine group of 2 sterically hinders the interactions with the adenine atoms mentioned above and that are formed by residues Ser36′, Gln172, and Gln173. In contrast, the methylmercapto group of 4 not only sterically blocked the aforementioned interactions but also was accommodated in a new compound-induced pocket that is comprised in part by residues from both chains of the MtbAdoK homodimer (Gln173, Gln172, Met121, Phe116, Arg176, Leu38′, and Ser36′). It should be noted that compound 2 has been reported to be a very potent inhibitor of hAdoK (KihAdoK ≈ 30 nM and KiMtbAdoK ≈ 210 nM).4 The different degrees of potencies can be attributed to the way compound 2 interacts with the enzymes. In our MtbAdoK-2 complex, the compound is shown to be partially solvent exposed because of the presence the “chimney-like” cavity. In contrast, the previously solved hAdoK-2 complex showed that the compound is completely buried within the hAdoK monomer because of a unique latch-like region of hAdoK comprised by residues 23–57 of the enzyme’s lid-domain (Figure S4).22 Also, the lack of selectivity of compound 2 can be attributed to the fact that the compound is buried within several conserved residues located in the active site. Overall, the N7 versus N6 substituents, exemplified by compounds 2–3 and 4, seemed to be accommodated in mutually exclusive pockets. The N7 pocket is formed by several conserved active site residues while the N6 pocket is compound-induced and formed by unique MtbAdoK residues Gln172, Gln173, Ser36′, Leu38′, and flexible Arg176.
Finally, to explore the potential for modifying the 5′-position and to evaluate if the ATP channel can be utilized as a viable route for drug design, we solved the crystal structure of 5′-aminoadenosine bound to MtbAdoK (Figure S5). As in the case of the compounds described above, compound 5 has been previously shown to possess inhibitory activity against AdoKs.13 The MtbAdoK-5 cocrystal was obtained in the same condition as the reported adenosine-bound structure and was refined to 1.95 Å resolution in the P3121 crystal space group with one molecule in the ASU. The binary complex displayed a closed conformation of the enzyme with a backbone rmsd of 0.28 Å amongst all Cα backbone atoms when compared with the MtbAdoK–adenosine complex, and the inhibitor was bound in a very similar fashion as adenosine (Figure 2i, Tables S8 and S9, and Figure S6). Unlike, the cocrystal structures of compounds 2–4; the MtbAdoK-5 structure had no electron density for a molecule of 5 bound to the ATP site. The interactions observed between 5 and MtbAdoK were remarkably similar to those previously described in the adenosine-bound complex (Tables S8 and S5).5 Indeed, the main difference was the interaction formed by the 5′-NH2, displaying a H-bond with OD2 of Asp257 (2.61 Å). Here, enzymatic inhibition (IC50 of 8.0 μM) is attributed to the absence of the hydroxyl group that is required for catalysis.
On the basis of the preliminary structure-guided SAR efforts with compounds 2–5, we screened a focused library of ∼140 nucleoside analogues with substitutions at N6, N7, and/or 5′ positions. The very potent compound identified in this screen (compound 6, IC50 of 196.5 ± 20.3 nM) had a bulky thiophene group at position 6, validating the pharmacological relevance of N6-substituted adenosine analogues (Figure 3a). To investigate the structural basis of the ∼10× increase in potency when compared with compounds 2 and 4 and to advance our structure-guided drug discovery efforts, we crystallized the MtbAdoK-6 binary complex. The cocrystal structure of compound 6 was solved by MR and refined to a resolution of 2.10 Å in the P212121 crystal space group with two molecules in the ASU (Figure 3b, Tables S10 and S11). As observed in the structures described above, the MtbAdoK-6 complex displayed a closed conformation with a backbone rmsd of 2.18 Å amongst all Cα atoms when compared with the MtbAdoK–adenosine complex. Unlike the structures of compounds 2–5, the MtbAdoK-6 complex revealed that the compound binds in a different orientation with respect to adenosine (Figure 3c) and that the bulky substituent prevented full lid domain closure by leaving a gap of 2.94 Å when compared with the fully closed MtbAdoK–adenosine complex. As in the case of the crystal structure complexes of compounds 2–4, the MtbAdoK-6 structure showed electron density in the active site and partial electron density in the ATP site (Figure S7).
Figure 3.
Crystal structure of the MtbAdoK-6 complex. (a) Chemical structure of compound 6. (b) 6 bound to the active site of MtbAdoK. (c) Superimposition of the cocrystal structures of compound 6 (gray) and adenosine (blue). (d) Surface representation of residues forming the pocket where the thiophene group gets accommodated. Chain B is colored magenta, Arg176 is colored red, and chain A is colored by heteroatom. (e) Arg176 adopts a different conformation in the cocrystal structure of 4 (blue) when compared with the cocrystal structure of 6 (gray).
The cocrystal structure of compound 6 showed that the interactions formed by residues Phe116, Phe102, Ser8, Asp12, Asn52, Asp257, Val49, and Gly48 were preserved. The significant differences were found once more with residues Gln172, Gln173, and Ser36′. In fact, none of the H-bond interactions formed between Gln172 (with O4′, O5′ and N6), Gln173 (with N1 and N6), and Ser36′ (with N6 and N7) were observed (Table S10). Instead, these residues were involved in weaker vdW interactions with the adenine ring and ribose rings. The thiophene moiety formed unique contacts. This group was observed to be completely buried within the newly identified and compound-induced pocket that is formed by residues Phe102, Phe116, Gln172, Gln173, Arg176, Ser36′, Leu38′ and Phe37′ (Figure 3d). Closer inspection of the pocket showed that Arg176 reorients once more to accommodate the bulkier thiophene group, confirming the plasticity of this residue (Figure 3e). Furthermore, the orientation of the aromatic thiophene ring with respect to Phe116 suggests the possibility of parallel-displaced π-staking interactions, whereas the orientation of cationic CZ of Arg176 with respect to the sulfur atom of the thiophene group (CZ–S distance of 3.97 Å) suggests the possibility of T-shaped stacking interactions. Overall, the extensive vdW interactions within the pocket coupled to the stacking interactions are the most likely molecular basis for the significant increase in potency of 6 when compared with compounds 2–4. Finally, the orientation of Arg176, when compared with the MtbAdoK-4 structure, suggested that the “chimney-like” cavity we observed in the crystal structure complexes described above might be accessible if larger substitutions are utilized to force Arg176 to give access into the cavity.
To further characterize compound 6, we determined the antimycobacterial and cytotoxic profile of the compound. The antimycobacterial assay with compound 6 showed a half maximal effective concentration (EC50) of 25.6 ± 2.4 μM with a 50% cytotoxic concentration (CC50) > 100 μM against human dermal fibroblasts (HDF) (Figures S8 and S9). Indicating that, in general, 6-substituted adenosine analogues might show selectivity to MtbAdoK over the human counterpart.
Overall, the structure-based SAR efforts showed that the N6, N7, and 5′ positions are indeed conducive to inhibition of the MtbAdoK enzyme through different molecular and structural means. The 5′-position provides a mechanistic approach for inhibition by taking advantage of the critical hydroxyl group required for catalysis and provides a platform to design inhibitors that might take advantage of the ATP channel. On the other hand, the crystal structures of the N7- and N6-substituted adenosine analogues showed that the substitutions sterically hindered conserved H-bonds formed by residues Gln172, Gln173, and Ser36′ with key positions N1, N6, and N7 of the adenine ring; all of which have been noted to be critical for substrate binding and recognition for human, rabbit, and Toxoplasma gondii adenosine kinase.6 The crystal structures of the N7-substituted analogues (2–3) revealed the presence of a “chimney-like” cavity that extends above the N6-position, and that is formed by the MtbAdoK homodimer. However, the MtbAdoK-4 complex showed that the cavity is closed by a conformational movement of Arg176 to interact with the N6-substituent of 4 thereby forming a new and compound-induced pocket. Identification of the very potent compound 6 revealed that the bulky aromatic thiophene group is not only completely buried within the newly identified pocket but also suggested that bulkier substitutions could open back up the “chimney-like” cavity.
Despite offering inhibitory activity against MtbAdoK, the N7-substituents were observed to be buried in a pocket that is formed by several conserved active site residues and that the substituent itself is physically blocked by the dimerization event of MtbAdoK. Consequently, limiting medicinal chemistry efforts to the conserved active site. In contrast, the N6-position offers a better route for structure-guided drug design by providing the possibility of exploiting many contacts formed by the unique oligomerization state of MtbAdoK. In addition, it is possible that the presence of this cavity is the structural basis behind the reported higher degree of specificity conferred by N6-substituted analogues when compared with hAdoK.12 Taken together, we hypothesized that chain extension via bulky substitutions at the N6-position might trigger a conformational change of Arg176, thereby opening the “chimney-like” cavity, thus providing a unique route to increase potency and selectivity.
We next set out to systematically synthesize a series of 6-substituted adenosine analogues. Synthesis of the novel adenosine analogues (7–18) is shown in Scheme 1. Starting from the known 6-chloro-9H-purine A0 or A1 and TBS protected A2, 4-biphenyl and 4-Br-phenylpiperazinyl moieties were introduced by substitution in EtOH to afford B1 and B2 that was converted to variety of 4-arylphenyl(R)piperazine by using different arylboronic acids followed by removal of acetonide with/without the TBS group which gave the triol adenosines, 7, 9, 10, 12, 13, 14, 16, and 18, respectively.23,24 The cyclopentane 15 was directly synthesized from (1R,2S,3R,5R)-3-(6-chloro-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol A3 without alcohol protection.25 The C–C bond at 6-position of purine was formed by Suzuki–Miyaura or Sonogashira cross-coupling to lead to alkyne analogues 11 and 17. To the tri-acetyl protected 7-Cl-imidazopyridine A4, C–N coupling in the presence of second generation RuPhos precatalyst proceeded smoothly and global deprotection of acetyl groups by ammonium hydroxide gave the triol 8 (Scheme 1 and chemistry section of the Supporting Information).26
Scheme 1. Synthesis of Adenosine Analogues.
Reagents and conditions: (a) TBSCl, imidazole, DCM, 15 °C, 17 h, 99%; (b) 1-phenylpiperazine or 1-([1,1′-biphenyl]-4-yl)piperazine or 1-(4-bromophenyl)piperazine, DIEA, EtOH, 70–80 °C, 17 h, 13–90%; (c) ArB(OH)2, K3PO4, XPhos Pd G2, THF/H2O, 70 °C, 17 h, 49–87%; (d) TFA, THF/H2O, 15–25 °C, 2–17 h, 13–67%; (e) 4,4,5,5-tetramethyl-2-(4-(phenylethynyl)phenyl)-1,3,2-dioxaborolane, K3PO4, XPhos Pd G2, THF, 70 °C, 12 h, 95% or 4-ethynyl-1,1′-biphenyl, Cs2CO3, CuI, XPhos Pd G2, CH3CN, 90 °C, 17 h, 27%; (f) 1-([1,1′-biphenyl]-4-yl)piperazine, Cs2CO3, 1-([1,1′-biphenyl]-4-yl)piperazine, RuPhos Pd G2, tert-amyl-OH, 100 °C, 17 h, 50%; and (g) NH4OH, MeOH, 15 °C, 24 h, 21%.
All synthesized adenosine analogues were characterized for inhibitory activity against MtbAdoK and hAdoK. The compounds were also tested against Mtb and HDF cells to profile their antimycobacterial and cytotoxic properties, respectively (Table 1). We began our efforts with compound 7. This compound showed excellent potency and selectivity against MtbAdoK when compared with hAdoK. We hypothesized that the piperazine group might lead to favorable contacts with flexible residue Arg176 while the benzene ring could extend toward the unique cavity, thereby conferring selectivity.
Table 1. SAR Data for Synthesized Adenosine Analogues with Substitutions at Position 6a.
| ID | R or R6 | Y | Z | MtbAdoK Ki (nM) | hAdoK Ki (μM) | Mtb EC50 (μM) | HDF CC50 (μM) |
|---|---|---|---|---|---|---|---|
| 7 | R | O | N | 120.2 ± 0.9 | 5.1 ± 0.5 | ≥65.0 | 30.9 ± 4.3 |
| 8 | R | O | CH | 16.3 ± 0.1 | >50.0 | >50.0 | >50.0 |
| 9 | R | O | N | 18.5 ± 0.06 | >50.0 | >50.0 | >50.0 |
| 10 | R | O | N | 18.9 ± 0.5 | >50.0 | ≥30.0 | >50.0 |
| 11 | R6 | O | N | 19.9 ± 0.03 | >50.0 | ≥50.0 | >50.0 |
| 12 | R | O | N | 21.3 ± 0.2 | >50.0 | >50.0 | >50.0 |
| 13 | R | O | N | 23.0 ± 1.1 | ≥15.1 | >50.0 | >50.0 |
| 14 | R | O | N | 25.5 ± 2.8 | 0.41 ± 0.07 | ≥25.0 | >50.0 |
| 15 | R | CH2 | N | 27.5 ± 3.5 | >50.0 | >50.0 | >50.0 |
| 16 | R | O | N | 32.6 ± 0.3 | 1.6 ± 0.1 | >50.0 | >50.0 |
| 17 | R6 | O | N | 48.0 ± 0.6 | >50.0 | 1.7 ± 0.02 | 3.5 ± 0.4 |
| 18 | R | O | N | 5.3 ± 0.07 | >50.0 | 4.0 ± 0.2 | >50.0 |
The error is reported as ± SD of 2 experiments.
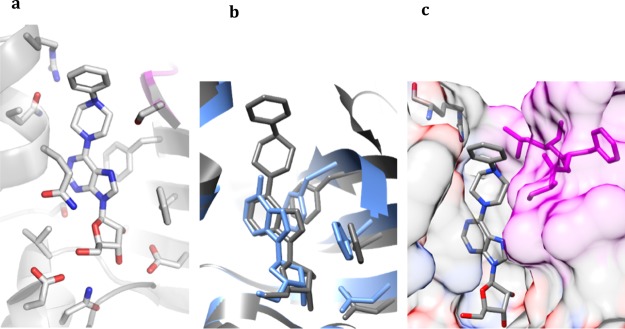
To investigate the structural basis of inhibition and selectivity of 7, we cocrystallized the compound with MtbAdoK. The crystal structure of the MtbAdoK-7 binary complex was solved by MR in the P41 crystal space group with two molecules in the ASU and refined to a resolution of 1.70 Å (Figure 4a–c). Like the structures described above, the MtbAdoK-7 complex displayed a closed conformation of the enzyme with a backbone rmsd of 1.52 Å amongst all Cα atoms when compared with the MtbAdoK–adenosine structure. Unlike the structures described above, the MtbAdoK-7 structure had no electron density for a molecule bound to the ATP site (Figure S10). Overall, the interactions seen between the adenine ring and ribose scaffold were very similar to those observed in the MtbAdoK–adenosine complex (Tables S12 and S13). Interestingly, the most significant differences were found once more with residues Gln172, Gln173, and Ser36′. Here, Gln173 and Ser36′ were not observed to make any of the critical H-bonds with positions N1, N6, and N7 while Gln172 retains the H-bonds with O4′ of ribose (NE2–O4′ distance of 3.03 Å) and O5′ (NE2–O5′ distance of 3.29 Å) but lacks the H-bond with N6. As in the case of the MtbAdoK-6 structure, superimposition of the crystal structure complexes of MtbAdoK–adenosine and MtbAdoK-7 showed that the bulky substitution of 7 causes the compound to bind in a different orientation with respect to adenosine with notable differences in critical lid domain residues Asp12, Phe102, and Phe116 (Figure 4b). Here, the lid domain gap with respect to the MtbAdoK-1 complex was an average of 2.45 Å. We also observed that the phenylpiperazine forces Arg176 to open the “chimney-like” cavity by reorienting the residue back to the solvent. The crystal structure also showed that the piperazine group redirects the distal benzene group within the enzyme’s active site groove and that there was sufficient space to incorporate additional groups at the distal benzene ring to fully occupy the cavity (Figure 4c).
Figure 4.
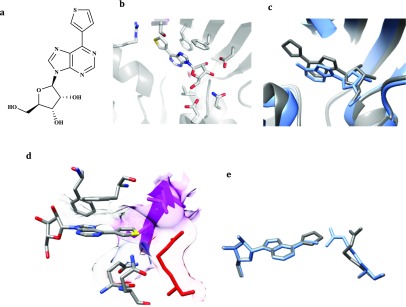
Crystal structure of the MtbAdoK-7 complex. (a) 7 bound to the active site to MtbAdoK. Chain A is colored gray and chain B is colored magenta. (b) The bulky substituent of 7 (gray) forces the compound to bind in a different orientation with respect to adenosine (blue; PDB ID 2PKM). (c) The bulky substituent forces Arg176 to open the cavity while the distal benzene ring is reoriented back into the active site groove that is formed by residues of chain A (heteroatom) and chain B (magenta).
As noted above, another position of interest was the 5′-position of the ribose scaffold. The preliminary structure-based SAR with compound 5 suggested that bisubstrate-like inhibitors might be able to take advantage of the ATP channel. To investigate if the ATP channel is a viable route for drug design, we synthesized several 5′-substituted adenosine analogues (Figure S11 and Table S14). Despite having sufficient space to accommodate large groups, bulky phenyl-substituents at the 5′-position resulted in inhibitors with negligible inhibitory activity. Indeed, only one of the synthesized compounds displayed some measurable inhibitory activity against the enzyme with ∼53% inhibition at 12.5 μM of the compound. The lack of inhibitory activity of these analogues may be due to suboptimal angles adopted by the bulky substitution.
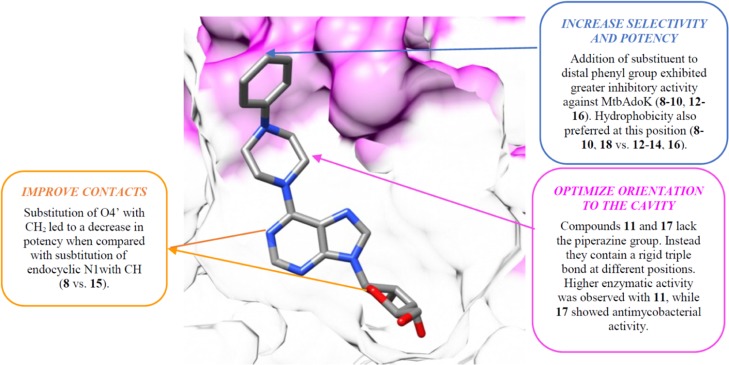
The MtbAdoK-7 complex suggested that further extension at the distal benzene group could be accommodated in the cavity and that rigid substituents at the 6-position might orient the substituent within the cavity. On the basis of these observations, we synthesized compounds 8–17. As shown in Table 1, all compounds displayed high potency against MtbAdoK with Ki values ranging from ∼16 to 48 nM. Overall, the SAR results are in accordance previous observations where it was noted that 6-substituted adenosine analogues displayed a higher degree of specificity to MtbAdoK when compared with hAdoK (Figure 5).12,14 The apparent lack of correlation between enzymatic inhibition and intracellular activity might be due to several reasons such as a lack of permeability, inefficient import, or rapid export. For example, it is known that Mtb possesses over 30 ATP-binding cassettes (ABC) transporters which are involved in the uptake and export of many molecules including nucleosides.27,28 In addition, Mtb possesses several classes of drug efflux pumps including ABC transporters, the small multidrug resistance superfamily, the major facilitator superfamily, and the resistance nodulation cell division superfamily; all of which have been attributed to efflux of many classes of antimycobacterial drugs.29−32 It is currently unknown how Mtb is able to uptake, discriminate, import, and export this series of compounds. Further experiments such as resistant mutant isolation, knockdown or overexpression strains, and systematic interrogation of potential transporters and efflux pumps will be needed to investigate the intracellular fate of the series.
Figure 5.
Summary of SAR results for compounds 8–17. Compound 7 bound to the active site of MtbAdoK. Based on the MtbAdoK-7 complex, compounds 8–17 where synthesized. Chain A is colored white, and chain B is colored magenta.
Compounds 11 and 17 were designed to test different orientations of the 6-substituent within the cavity. These compounds possess a rigid triple bond instead of the flexible piperazine group. Because compound 17 displayed antimycobacterial activity, we decided to determine the cocrystal structure of MtbAdoK with compound 17. The crystal structure of the MtbAdoK-17 complex was solved by MR in the P41 crystal space group with two molecules in the ASU and refined to a resolution of 2.23 Å (Figure 6a–c and Tables S15 and S16). The binary complex displayed a closed conformation of the enzyme with a backbone rmsd of 1.40 Å amongst all Cα atoms when compared with the MtbAdoK–adenosine structure. Just like the MtbAdoK-7 structure, the MtbAdoK-17 binary complex had no electron density for another molecule of 17 bound to the ATP site. Superimposition of the crystal structure complexes of compounds 17 and adenosine also showed that the bulky substituent of 17 causes the compound to bind in a different orientation with respect to adenosine, leaving lid domain gap of 1.81 Å when compared with the MtbAdoK–adenosine complex (Figure 6c).
Figure 6.
Crystal structure of the MtbAdoK-17 complex. (a) 17 bound to the active site of MtbAdoK. (b) The large bulky substitution gets accommodated in the “chimney-like” cavity. (c) Compound 17 (gray) binds in a different orientation with respect to adenosine (blue). (d) Superimposition of crystal structure complexes of 17 (gray) and 7 (blue). Chain B residues forming the distal part of the “chimney-like” cavity colored magenta while chain A residues are colored by heteroatom.
The MtbAdoK-17 binary complex showed that the compound binds to the active site of the enzyme forming similar contacts with the protein as observed with the adenosine-bound complex (Figure 6a, Tables S15 and S5). Structural differences were found again with residues Gln172, Gln173, and Ser36′. Only Gln173 was observed to form a weak H-bond with endocyclic N1 of the adenine ring (NE2–N1 distance of 3.21 Å) while residues Ser36′ and Gln172 were not observed to make any of the critical H-bond interactions described above. The major difference between the MtbAdoK-7 and MtbAdoK-17 structures is the orientation that the 6-substituent adopts as it goes into the “chimney-like” cavity. The cocrystal structure of the MtbAdoK-7 complex suggests that the piperazine-containing adenosine analogues might reorient to the active site groove that is composed of hydrophobic residues from both chains including Ala175.A, Leu38.B, Leu35.B, Phe37.B, and polar residues Ser36.B and Arg176.A. In contrast, the rigidity imparted by the triple bond of compound 17 limits the substitution to adopt a bond angle of 180° with respect to the 6-position. Consequently, one side of the distal benzene ring of 17 was seen to be oriented toward backbone and main chain atoms of Phe37.B while the other side was observed to be partially solvent exposed (Figures 6d and S12).
Finally, on the basis of the chemistry and structure-guided SAR efforts with compounds 7–17, we synthesized compound 18. This compound is an analogue of compounds 8 and 15. The major difference is that 18 contains endocyclic atoms N1 and O4′ which we observed in the structures described above that they were involved in key H-bond interactions with residues Gln172 and Gln173, and the SAR was suggested to be relevant in potency. Indeed, compound 18 was our most potent derivative, exhibiting a Ki of ∼5 nM against MtbAdoK, no cytotoxicity, no activity against hAdoK, and an EC50 of ∼4.0 μM against Mtb. Overall, the observed differences in binding modalities of reorientation within the enzyme versus solvent exposed could explain the overall higher potency of the piperazine-substituted analogues. Furthermore, the predominantly hydrophobic character of the “chimney-like” cavity goes in accordance with the SAR studies where hydrophobic substitutions displayed a higher degree of potency when compared with the polar substituted analogues (Table 1).
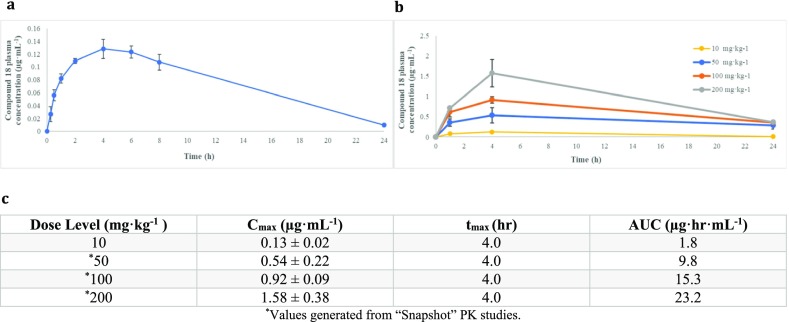
To further characterize our most potent derivative (18), we performed steady-state kinetic experiments and in vivo pharmacokinetics and tolerability studies. The kinetics experiments showed that increasing the concentration of adenosine in the presence of a fixed inhibitor concentration led to an increase of the Km, consistent with a competitive mode of inhibition (Figure S13a,b and Table S17). The Km value for adenosine determined with our enzymatic assay was 1.71 ± 0.02 μM, well in agreement with previously reported values of 0.8–3.4 μM.4,33 To determine in vivo pharmacokinetic parameters, we administered compound 18 to female Swiss Webster mice via oral gavage as a single dose at 10 mg·kg–1 and quantified plasma concentrations at 0, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 h by liquid chromatography–mass spectrometry (LC/MS, Figure 7a).34,35 Compound 18 dosed at 10 mg·kg–1 achieved a Cmax of 0.13 ± 0.02 μg·mL–1 at approximately 4 h with an AUC0–24h of 1.80 μg·h·mL–1. At this dose level, compound 18 was well tolerated.36 In vivo tolerability studies were also performed by administering compound 18 to cohorts of female Swiss Webster mice (n = 3) at 50, 100, and 200 mg·kg–1 via oral gavage once daily for 3 days. We evaluated mice twice daily for signs of acute toxicity and observed no adverse effects or weight loss (Table S18). Blood collected at 0, 1, 4, and 24 h after the first administered dose was used to determine PK for compound 18 at these three higher dose levels (Figure 7b).36 Data points collected from the 10 mg·kg–1 study was included for comparison. Compound 18 achieved a maximum plasma concentration for all dose levels (50, 100, and 200 mg·kg–1) at approximately 4 h with Cmax of 0.54 ± 0.22 μg·mL–1 (1.1 ± 0.4 μM), 0.92 ± 0.09 μg·mL–1 (1.9 ± 0.2 μM), and 1.58 ± 0.38 μg·mL–1 (3.2 ± 0.8 μM), respectively (Figure 7b,c). Calculated exposure for these higher dose levels improved overall exposure to 9.8, 15.3, and 23.2 μg·h·mL–1. On the basis of our preliminary pharmacokinetic data, compound 18 is very well tolerated during acute exposure. Most notably, at the highest concentration tested, 18 falls within the range of EC50 value we determined in our in vitro assay (Table 1).
Figure 7.
Preliminary pharmacokinetics of compound 18. (a) Oral PK study conducted at 10 mg·kg–1 utilizing 9 Swiss Webster Female mice with three animal bleeds per time point. Maximum plasma concentration of 0.13 ± 0.02 μg·mL–1 observed at 4 h following gavage. The AUC0–24h of 1.8 μg·h·mL–1 was calculated using the linear trapezoid rule method. (b) Snapshot PK studies conducted from plasma samples taken from tolerability mice at 50, 100, and 200 mg·kg–1 following first initial dose. 10 mg·kg–1 data included for comparison. (c) Cmax, tmax, and AUC of compound 18 at four dose levels.
Conclusions
These studies represent a systematic structure-guided approach to the rational design of very potent MtbAdoK inhibitors with low micromolar activity against Mtb. Furthermore, our studies offer a structural explanation behind the specificity of previously reported N6-substituted adenosine analogues.12−15 That is, substitutions at the 6-position of the adenine scaffold might be accommodated in a compound-induced pocket that is formed within the unique oligomerization state of MtbAdoK when compared with hAdoK and other eukaryotic adenosine kinases. Unlike the crystal structures of compounds 2–6, the crystal structures of 7 and 17 did not display any electron density for a compound bound to the ATP site suggesting that there might be a size threshold conferring specificity to the active site versus ATP site. Therefore, future strategies to design bisubstrate-like inhibitors should focus on small substituents at the N6-position with bulky groups at the 5′-position.
The crystal structures of the bulky 6-substituted adenosine analogues suggested that the reorientation conferred by the piperazine group might lead to more favorable contacts with the enzyme, thereby leading to a higher degree of potency. The predominantly hydrophobic character of the cavity goes in accordance with the SAR studies demonstrating that hydrophobic substitutions were preferred at the 6-position. Moreover, the lid domain gaps and different orientation of compounds 6, 7, and 17 with respect to the MtbAdoK–adenosine structure suggest that bulky substitutions at position 6 might exert its inhibitory effects by sterically preventing full lid domain closure. Our preliminary in vivo pharmacokinetics and tolerability studies showed that our most potent derivative is very well tolerated and that MIC values are indeed achievable. However, future studies regarding SAR derivatization will be needed to optimize pharmacokinetic parameters. Overall, we showed that the unique “chimney-like” cavity formed by the unique oligomerization state of MtbAdoK could be utilized as a canvas for future medicinal chemistry efforts to further improve the in vitro and in vivo potency and selectivity.
Experimental Section
General Synthetic Chemistry Methods
Solvents, reagents, and intermediates that are commercially available were used as received. Reagents and intermediates that were not commercially available were prepared in the manner as described below. Compounds 1, 3, and 4 were purchased at Sigma, with specific purities of >99.0, >98, and >99%, respectively. Compound 2 was obtained from ChemScene with specific purity of >95%, and compounds 5 and 6 are part of Merck’s nucleoside collection library. 1H NMR spectra were measured on either a Varian VNMR System 400 or Bruker AVANCE 400 spectrometer at 400 MHz and are reported as ppm downfield from Me4Si with number of protons, multiplicities, and coupling constants in hertz indicated parenthetically. Where LC/MS data are presented, analyses were performed using an Agilent 6110A MSD or an Applied Shimadzu 2020MSD. The parent ion is given. Unless otherwise stated, all compounds possess a purity ≥ 95% as determined by LC/MS. Purification was conducted with reversed-phase HPLC (Waters Xbridge Prep OBD C18 (150 × 30 mm, 5 μm), YMC-Actus Pro C18 (150 × 30 mm, 5 μm), and Agela ASB (150 × 25 mm, 5 μm) column) or silica gel column chromatography (Liang Chen Gui Yuan 200–300 mesh) or PTLC (Liang Chen Gui Yuan, acryloid cement; 0.5 × 200 × 200 mm). For the specific chemical synthesis of reaction intermediates, see the Supporting Information.
(2R,3R,4S,5R)-2-(7-(4-([1-Phenyl]-4-yl)piperazin-1-yl)-3H-imidazo[4,5-b]pyridin-3-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (7)
To a suspension of 1-phenylpiperazine (210 mg, 1.3 mmol) in anhydrous EtOH (6 mL) were added DIEA (0.3 mL, 1.7 mmol) and 6-chloro-purine riboside A0 (287 mg, 1 mmol) at room temperature, and it was stirred at 70 °C for 17 h after cooling to room temperature. The resulting solid was filtered and washed with dichloromethane (DCM, 6 mL × 2) and dried under reduced pressure to afford (2R,3R,4S,5R)-2-(7-(4-([1-phenyl]-4-yl)piperazin-1-yl)-3H-imidazo[4,5-b]pyridin-3-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (7), (288 mg, 70% yield) as a white solid; LC/MS purity of 97%; Scheme A0 Supporting Information. MS (ESI) m/z: 412.9 [M + H]+. 1H NMR (DMSO-d6, 400 MHz): δ 8.43 (s, 1H), 8.27 (s, 1H), 7.21–7.26 (m, 2H), 6.99–7.01 (m, 2H), 6.81 (m, 1H), 5.93 (d, J = 6.0 Hz, 1H), 5.42 (d, J = 6.0 Hz, 1H), 5.27 (m, 1H), 5.14 (d, J = 4.8 Hz, 1H), 4.58 (m, 1H), 4.38 (br s, 4H), 4.16 (m, 1H), 3.97 (m, 1H), 3.65–3.70 (m, 1H), 3.55–3.60 (m, 1H), 3.20–3.35 (m, 4H).
(2R,3R,4S,5R)-2-(7-(4-([1,1′-Biphenyl]-4-yl)piperazin-1-yl)-3H-imidazo[4,5-b]pyridin-3-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (8)
To a solution of (2R,3R,4R,5R)-2-(7-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-3H-imidazo[4,5-b]pyridin-3-yl)-5-(acetoxymethyl)tetrahydrofuran-3,4-diyl diacetate B4 (130 mg, 0.082 mmol (38.8% purity)) in MeOH (3 mL) was added ammonium hydroxide (1.5 mL) dropwise at 15 °C, and it was stirred at 15 °C for 24 h. The mixture was concentrated and purified by reversed-phase HPLC [Waters Xbridge Prep OBD C18 (150 × 30 mm, 5 μm) column, elution of acetonitrile/water (0.05% ammonia hydroxide)] to afford (2R,3R,4S,5R)-2-(7-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-3H-imidazo[4,5-b]pyridin-3-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol 8 (8.81 mg, 21.3% yield) as a white solid; LC/MS purity of 97%; Scheme B Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.40 (s, 1H), 7.99 (d, J = 5.6 Hz, 1H), 7.62–7.56 (m, 4H), 7.43–7.39 (m, 2H), 7.29–7.27 (m, 1H), 7.10 (d, J = 8.8 Hz, 2H), 6.66 (d, J = 6.0 Hz, 1H), 5.69–5.67 (m, 1H), 5.42 (d, J = 6.0 Hz, 1H), 5.17 (d, J = 4.4 Hz, 1H), 4.66–4.65 (m, 1H), 4.15 (s, 1H), 4.01 (s, 4H), 3.98 (d, J = 2.4 Hz, 1H), 3.66–3.65 (m, 1H), 3.56–3.55 (m, 1H), 3.37 (s, 4H). MS (ESI) m/z: 488.1.
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-(6-(4-(4-(6-methoxypyridin-3-yl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (9)
The mixture of 9-((3aR,4R,6R,6aR)-6-(((tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-6-(4-(4-(6-methoxypyridin-3-yl)phenyl)piperazin-1-yl)-9H-purine B3-1 (80 mg, 0.119 mmol) in THF (1.0 mL) and H2O (0.2 mL) was added TFA (0.5 mL, 6.49 mmol), and the mixture was stirred at 20 °C for 17 h, concentrated, and purified by reversed-phase HPLC [Waters Xbridge Prep OBD C18 (150 × 30 mm, 5 μm) column, elution of acetonitrile/water (0.05% ammonia hydroxide)] to give (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(4-(4-(6-methoxypyridin-3-yl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol 9 (26.4 mg, 42.7% yield) as a yellow solid; LC/MS purity of 99%; Scheme C Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.45–8.42 (m, 2H), 8.28 (s, 1H), 7.96–7.93 (m, 1H), 7.54 (d, J = 8.4 Hz, 2H), 7.09 (d, J = 8.4 Hz, 2H), 6.86 (d, J = 8.4 Hz, 1H), 5.93 (d, J = 5.6 Hz, 1H), 5.48 (d, J = 5.6 Hz, 1H), 5.33 (t, J = 6.0 Hz, 1H), 5.21 (d, J = 4.4 Hz, 1H), 4.59–4.58 (m, 1H), 4.40 (br s, 4H), 4.15 (d, J = 3.2 Hz, 1H), 3.96 (s, 1H), 3.87 (s, 3H), 3.69–3.66 (m, 1H), 3.57–3.56 (m, 1H), 3.35 (s, 4H). MS (ESI) m/z: 520.2 [M + H]+.
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-(6-(4-(4′-(2-hydroxypropan-2-yl)-[1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (10)
Yellow solid. Following the similar procedure shown in Scheme C with different boronic acids (RB(OH)2), compound 10 was synthesized; LC/MS purity of 99%. 1H NMR (DMSO-d6, 400 MHz): δ 8.46 (s, 1H), 8.29 (s, 1H), 7.55–7.48 (m, 6H), 7.09 (d, J = 8.8 Hz, 2H), 5.94 (d, J = 5.6 Hz, 1H), 5.48 (d, J = 5.6 Hz, 1H), 5.33 (t, J = 5.6 Hz, 1H), 5.21 (d, J = 5.2 Hz, 1H), 5.01 (s, 1H), 4.60–4.58 (m, 1H), 4.40 (br s, 4H), 4.16–4.15 (m, 1H), 3.97–3.96 (m, 1H), 3.68–3.67 (m, 1H), 3.58–3.56 (m, 1H), 3.31 (s, 4H), 1.44 (s, 6H). MS (ESI) m/z: 547.2 [M + H]+.
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-(6-(4-(phenylethynyl)phenyl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (11)
To a solution of 9-((3aR,4R,6R,6aR)-6-(((tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-6-(4-(phenylethynyl)phenyl)-9H-purine C1-1 (103 mg, 0.18 mmol) in THF (1.5 mL) and H2O (0.2 mL) was added TFA (2.5 mL), and it was stirred at 20 °C for 17 h. The mixture was diluted with dimethylformamide (DMF, 2 mL), and it was concentrated under reduced pressure. The residue was purified by reversed-phase HPLC [TFA condition: Agela ASB (150 × 25 mm, 5 μm) column, elution of acetonitrile/water (0.1% TFA)] and then lyophilized to give (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(4-(phenylethynyl)phenyl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol 11 (17.48 mg, 23.3% yield) as a yellow solid; LC/MS purity of 99%; Scheme D Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 9.04 (s, 1H), 8.97 (s, 1H), 8.92 (d, J = 8.4 Hz, 2H), 7.80 (d, J = 8.4 Hz, 2H), 7.62–7.61 (m, 2H), 7.47–7.46 (m, 3H), 6.11 (d, J = 5.6 Hz, 1H), 4.67–4.65 (m, 1H), 4.23–4.21 (m, 1H), 4.01 (d, J = 3.6 Hz, 1H), 3.71–3.70 (m, 1H), 3.62–3.59 (m, 1H). MS (ESI) m/z: 429.0 [M + H]+.
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-(6-(4-(4′-(trifluoromethyl)-[1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (12)
Yellow solid. Following the similar procedure shown in Scheme C with different boronic acids (RB(OH)2), compound 12 was synthesized; LC/MS purity of 99%. 1H NMR (DMSO-d6, 400 MHz): δ 8.44 (s, 1H), 8.28 (s, 1H), 7.82 (d, J = 8.4 Hz, 2H), 7.72 (d, J = 8.4 Hz, 2H), 7.64 (d, J = 8.4 Hz, 2H), 7.11 (d, J = 8.4 Hz, 2H), 5.92 (d, J = 6.0 Hz, 1H), 4.58–4.55 (m, 1H), 4.38 (br s, 4H), 4.15 (t, J = 3.2 Hz, 1H), 3.95 (d, J = 3.2 Hz, 1H), 3.65–3.64 (m, 1H), 3.55–3.53 (m, 1H), 3.37 (s, 4H). MS (ESI) m/z: 557.2 [M + H]+.
(2R,3R,4S,5R)-2-(6-(4-(4-(1-(Difluoromethyl)-1H-pyrazol-4-yl)phenyl)piperazin-1-yl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (13)
Yellow solid. Following the similar procedure shown in Scheme C with different boronic acids (RB(OH)2), compound 13 was synthesized; LC/MS purity of 97%. 1H NMR (DMSO-d6, 400 MHz): δ 8.58 (s, 1H), 8.46 (s, 1H), 8.29 (s, 1H), 8.19 (s, 1H), 7.80 (t, J = 59.2 Hz, 1H), 7.57 (d, J = 8.4 Hz, 2H), 7.05 (d, J = 8.8 Hz, 2H), 5.93 (d, J = 5.6 Hz, 1H), 4.59–4.57 (m, 1H), 4.39 (br s, 4H), 4.15 (t, J = 3.2 Hz, 1H), 3.96 (s, 1H), 3.67–3.66 (m, 1H), 3.57–3.54 (m, 1H), 3.32 (s, 4H). MS (ESI) m/z: 529.2 [M + H]+.
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-(6-(4-(3′-(morpholinomethyl)-[1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (14)
Yellow solid. Following the similar procedure shown in Scheme C with different boronic acids (RB(OH)2), compound 14 was synthesized; LC/MS purity of 95%. 1H NMR (CD3OD, 400 MHz): δ 8.34 (d, J = 12.4 Hz, 2H), 7.77–7.75 (m, 2H), 7.64–7.61 (m, 2H), 7.54–7.52 (m, 1H), 7.43–7.42 (m, 1H), 7.16 (d, J = 8.8 Hz, 2H), 6.01 (d, J = 5.6 Hz, 1H), 4.67–4.65 (m, 1H), 4.54 (br s, 4H), 4.42 (s, 2H), 4.33–4.32 (m, 1H), 4.20–4.19 (m, 1H), 4.19–3.98 (m, 2H), 3.89–3.77 (m, 4H), 3.46–3.40 (m, 6H), 3.31–3.20 (2H). MS (ESI) m/z: 588.2 [M + H]+.
(1R,2S,3R,5R)-3-(6-(4-([1,1′-Biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol (15)
The mixture of 1-([1,1′-biphenyl]-4-yl)piperazine (56.3 mg, 0.236 mmol), (1R,2S,3R,5R)-3-(6-chloro-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol A3 (100 mg, 0.197 mmol), and DIEA (0.103 mL, 0.590 mmol) in EtOH (5 mL) was stirred at 70 °C for 17 h. The mixture was cooled and concentrated, purified by reversed-phase HPLC [Waters Xbridge Prep OBD C18 (150 × 30 mm, 5 μm) column, elution of acetonitrile/water (0.05% ammonia hydroxide)] to give (1R,2S,3R,5R)-3-(6-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol 15 (12.26 mg, 12.77% yield) as a white solid; LC/MS purity of 99%; Scheme F Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.29 (s, 1H), 8.27 (s, 1H), 7.62–7.55 (m, 4H), 7.42–7.39 (m, 2H), 7.29–7.27 (m, 1H), 7.09 (d, J = 8.8 Hz, 2H), 4.93 (d, J = 6.8 Hz, 1H), 4.75–4.66 (m, 3H), 4.40–4.15 (m, 5H), 3.84–3.83 (m, 1H), 3.51–3.46 (m, 2H), 3.35 (br s, 4H), 2.25–2.22 (m, 1H), 2.06–2.01 (m, 1H), 1.75–1.70 (m, 1H). MS (ESI) m/z: 487.3 [M + H]+.
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-(6-(4-(4-(6-(trifluoromethyl)pyridin-3-yl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (16)
Yellow solid. Following the similar procedure shown in Scheme C with different boronic acids (RB(OH)2), compound 16 was synthesized; LC/MS purity of 97%. 1H NMR (CD3OD, 400 MHz): δ 8.94 (s, 1H), 8.38 (d, J = 16.4 Hz, 2H), 8.23 (d, J = 7.6 Hz, 1H), 7.83 (d, J = 8.0 Hz, 1H), 7.69 (d, J = 8.8 Hz, 2H), 7.20 (d, J = 8.8 Hz, 2H), 6.01 (d, J = 5.6 Hz, 1H), 4.66–4.65 (m, 1H), 4.56 (br s, 4H), 4.33–4.32 (m, 1H), 4.20–4.19 (m, 1H), 3.90–3.89 (m, 1H), 3.82–3.81 (m, 1H), 3.53–3.50 (m, 5H). MS (ESI) m/z: 558.2 [M + H]+.
(2R,3R,4S,5R)-2-(6-([1,1′-Biphenyl]-4-ylethynyl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (17)
Following similar procedure for 11 and Scheme D, compound 17 was synthesized. The reaction of 6-([1,1′-biphenyl]-4-ylethynyl)-9-((3aR,4R,6R,6aR)-6-(((tert-butyldimethylsilyl)oxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-9H-purine C1-2 (36 mg, 0.062 mmol) gave (2R,3R,4S,5R)-2-(6-([1,1′-biphenyl]-4-ylethynyl)-9H-purin-9-yl)-5-(hydroxymethyl) tetrahydrofuran-3,4-diol 17 (9.86 mg, 37.2% yield) as a yellow solid; LC/MS purity of 99%. 1H NMR (DMSO-d6, 400 MHz): δ 8.96 (d, J = 8.4 Hz, 2H), 7.86–7.75 (m, 6H), 7.53–7.50 (m, 2H), 7.42–7.41 (m, 1H), 6.07 (d, J = 5.2 Hz, 1H), 5.61 (d, J = 4.8 Hz, 1H), 5.30 (s, 1H), 5.13 (s, 1H), 4.63 (s, 1H), 4.21 (s, 1H), 4.00 (d, J = 3.6 Hz, 1H), 3.73–3.70 (m, 1H), 3.61–3.58 (m, 1H). MS (ESI) m/z: 429.1 [M + H]+.
(2R,3R,4S,5R)-2-(6-(4-([1,1′-Biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (18)
To a solution of ((3aR,4R,6R,6aR)-6-(6-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methanol B1 (3.1 g, 5.50 mmol) in THF (8 mL) and water (1.5 mL) was added TFA (30 mL) dropwise, and it was stirred at 15 °C for 2 h. After NH4OH was added to adjust pH to 7–8, water (70 mL) was added and the mixture was filtered. The resulting solid was dried under reduced pressure, and the residue was purified by silica gel column chromatography (DCM: MeOH = 100:1 to 30:1) to give (2R,3R,4S,5R)-2-(6-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol (1.327 g, 48.2% yield) as a white solid; LC/MS purity of 98%; Scheme A Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.46 (s, 1H), 8.29 (s, 1H), 7.62–7.55 (m, 4H), 7.43–7.39 (m, 2H), 7.29–7.27 (m, 1H), 7.09 (d, J = 8.4 Hz, 2H), 5.94 (d, J = 6.0 Hz, 1H), 5.48 (d, J = 6.0 Hz, 1H), 5.35–5.32 (m, 1H), 5.20 (d, J = 4.8 Hz, 1H), 4.59 (d, J = 5.2 Hz, 1H), 4.40 (br s, 4H), 4.16 (d, J = 3.6 Hz, 1H), 3.97 (d, J = 2.8 Hz, 1H), 3.70–3.67 (m, 1H), 3.58–3.56 (m, 1H), 3.35 (s, 4H). MS (ESI) m/z: 489.2 [M + H]+.
N-(((2R,3S,4R,5R)-3,4-Dihydroxy-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-2-yl)methyl)-4-methylbenzenesulfonamide (19)
To a solution of N-(((3aR,4R,6R,6aR)-2,2-dimethyl-6-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)-4-methylbenzenesulfonamide D3 (72 mg, 0.090 mmol) in THF (1 mL) and H2O (0.1 mL) was added TFA (1 mL, 12.98 mmol). The mixture was stirred at 15 °C for 17 h and then DMF was added (2 mL), and the mixture was purified by reversed-phase HPLC [TFA condition: Agela ASB (150 × 25 mm, 5 μm) column, elution of acetonitrile/water (0.1% TFA)] and then lyophilized to give N-(((2R,3S,4R,5R)-3,4-dihydroxy-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-2-yl)methyl)-4-methylbenzenesulfonamide (31.74 mg, 38.5% yield) as a white solid; LC/MS purity of 99%; Scheme S1 Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.38 (s, 1H), 8.27–8.21 (m, 2H), 7.65 (d, J = 8.8 Hz, 2H), 7.52 (d, J = 8.4 Hz, 2H), 7.35 (d, J = 8.4 Hz, 2H), 7.11 (d, J = 8.4 Hz, 2H), 5.85 (d, J = 5.6 Hz, 1H), 4.76 (t, J = 5.6 Hz, 2H), 4.36 (br s, 4H), 4.07–3.96 (m, 2H), 3.47 (br s, 4H), 3.07–2.97 (m, 1H), 2.34 (s, 3H). MS (ESI) m/z: 634.2 [M + H]+.
(2R,3S,4R,5R)-2-(((4-Nitrophenyl)amino)methyl)-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (20)
The solution of N-(((3aR,4R,6R,6aR)-2,2-dimethyl-6-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)-4-nitroaniline D4 (50 mg, 0.078 mmol) and TFA (0.5 mL, 6.49 mmol) in THF (1.0 mL) and H2O (0.2 mL) was stirred at 20 °C for 17 h, and then, it was concentrated and purified by reversed-phase HPLC [TFA condition: Agela ASB (150 × 25 mm, 5 μm) column, elution of acetonitrile/water (0.1% TFA)] and then lyophilized to give (2R,3S,4R,5R)-2-(((4-nitrophenyl)amino)methyl)-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol 20 (24.24 mg, 51.5% yield) as a yellow solid; LC/MS purity of 99%; Scheme S2 Supporting Information. 1H NMR (CD3OD, 400 MHz): δ 8.29 (s, 1H), 8.10 (s, 1H), 7.83 (d, J = 9.2 Hz, 2H), 7.50 (d, J = 8.8 Hz, 2H), 7.09 (d, J = 8.8 Hz, 2H), 6.57 (d, J = 9.2 Hz, 2H), 5.96 (d, J = 4.0 Hz, 1H), 4.80–4.74 (m, 1H), 4.51–4.32 (m, 5H), 4.28–4.21 (m, 1H), 3.71–3.62 (m, 1H), 3.56–3.40 (m, 5H). MS (ESI) m/z: 601.2 [M + H]+.
N-(((2R,3S,4R,5R)-3,4-Dihydroxy-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-2-yl)methyl)benzamide (21)
To a solution of N-(((3aR,4R,6R,6aR)-2,2-dimethyl-6-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)benzamide D2 (68 mg, 0.090 mmol) in THF (1.0 mL) and H2O (0.1 mL) was added TFA (1.0 mL, 12.98 mmol). The mixture was stirred at 15 °C for 17 h, and then, it was concentrated under reduced pressure, purified by reversed-phase HPLC (TFA condition: Agela ASB [150 × 25 mm, 5 μm) column, elution of acetonitrile/water (0.1% TFA)] and then lyophilized to give N-(((2R,3S,4R,5R)-3,4-dihydroxy-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-2-yl)methyl)benzamide 21 (25.7 mg, 46.4% yield) as a white solid; LC/MS purity of 95%; Scheme S3 Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.68 (s, 1H), 8.47 (s, 1H), 8.22 (s, 1H), 7.84 (d, J = 7.6 Hz, 2H), 7.55–7.45 (m, 5H), 7.13 (d, J = 8.4 Hz, 2H), 5.91 (d, J = 5.6 Hz, 1H), 4.76 (t, J = 5.6 Hz, 2H), 4.38 (br s, 4H), 4.19–4.10 (m, 2H), 3.65–3.45 (m, 5H). MS (ESI) m/z: 584.3 [M + H]+.
N-(4-((((2R,3S,4R,5R)-3,4-Dihydroxy-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-2-yl)methyl)amino)phenyl)methanesulfonamide (22)
The mixture of N-(4-((((3aR,4R,6R,6aR)-2,2-dimethyl-6-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)amino)phenyl)methanesulfonamide D7 (50 mg, 0.036 mmol) and TFA (0.5 mL, 6.49 mmol) in THF (1.0 mL) and H2O (0.2 mL) was stirred at 20 °C for 17 h. The mixture was concentrated and purified by reversed-phase HPLC [TFA condition: Agela ASB (150 × 25 mm, 5 μm) column, elution of acetonitrile/water (0.1% TFA)] and then lyophilized to give N-(4-((((2R,3S,4R,5R)-3,4-dihydroxy-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-2-yl)methyl)amino)phenyl)methanesulfonamide 21 (15.03 mg, 61.2% yield) as a yellow solid; LC/MS purity of 96%; Scheme S4 Supporting Information. 1H NMR (CD3OD, 400 MHz): δ 8.30 (d, J = 9.2 Hz, 2H), 7.52 (d, J = 7.2 Hz, 2H), 7.28–7.08 (m, 6H), 6.08 (br s, 1H), 4.58–4.26 (m, 6H), 3.89 (br s, 1H), 3.76–3.49 (m, 5H), 2.97 (s, 3H). MS (ESI) m/z: 649.2 [M + H]+.
(2R,3R,4S,5R)-2-(6-(4-([1,1′-Biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-5-((benzyloxy)methyl)tetrahydrofuran-3,4-diol (23)
The solution of 6-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-9-((3aR,4R,6R,6aR)-6-((benzyloxy)methyl)-2,2-dimethyltetrahydrofuro[3,4-d][1,3]dioxol-4-yl)-9H-purine B5 (700 mg, 1.131 mmol) and con. HCl (1 mL, 4.00 mmol) in MeOH (5 mL) was stirred at 25 °C for 1 h. Then, the mixture was neutralized with ammonium hydroxide (5 mL), concentrated, purified by reversed-phase HPLC [neutral condition: Waters Xtimate C18 (150 × 25 mm, 5 μm) column, elution of acetonitrile/water (10 mM NH4HCO3)] to give(2R,3R,4S,5R)-2-(6-(4-([1,1′-biphenyl]-4-yl)piperazin-1-yl)-9H-purin-9-yl)-5-((benzyloxy)methyl)tetrahydrofuran-3,4-diol 23 (600 mg, 88% yield) as a white solid; LC/MS purity of 96%; Scheme S5 Supporting Information. 1H NMR (DMSO-d6, 400 MHz): δ 8.37 (s, 1H), 8.29 (s, 1H), 7.41–7.31 (m, 8H), 7.09 (d, J = 8.4 Hz, 2H), 5.97 (d, J = 6.0 Hz, 1H), 5.55 (d, J = 5.6 Hz, 1H), 5.31 (d, J = 5.2 Hz, 1H), 4.58–4.54 (m, 3H), 4.40 (br s, 4H), 4.22–4.07 (m, 2H), 3.74–3.67 (m, 1H), 3.31 (s, 4H). MS (ESI) m/z: 579.2 [M + H]+.
(2R,3S,4R,5R)-2-(((4-Fluorophenyl)amino)methyl)-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol (24)
The mixture of N-(((3aR,4R,6R,6aR)-2,2-dimethyl-6-(6-(4-(4- (trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuro[3,4-d][1,3]dioxol-4-yl)methyl)-4-fluoroaniline D9 (200 mg, 0.228 mmol) and con. HCl (0.5 mL, 2.0 mmol) in MeOH (5 mL) was stirred at 25 °C for 1 h. Then, the mixture was neutralized with ammonium hydroxide (5 mL), concentrated, purified by reversed-phase HPLC (basic condition: Waters Xbridge Prep OBD C18 (150 × 30 mm, 5 μm) column, elution of acetonitrile/water (0.05% ammonia hydroxide)) to give (2R,3S,4R,5R)-2-(((4-fluorophenyl)amino)methyl)-5-(6-(4-(4-(trifluoromethyl)phenyl)piperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol 23 (43.09 mg, 32.6% yield) as a white solid; LC/MS purity of 99%; Scheme S6 Supporting Information. 1H NMR (CD3OD, 400 MHz): δ 8.31 (s, 1H), 8.14 (s, 1H), 7.50 (d, J = 8.8 Hz, 2H), 7.10 (d, J = 8.8 Hz, 2H), 6.89–6.79 (m, 2H), 6.73–6.62 (m, 2H), 5.97 (d, J = 5.2 Hz, 1H), 4.81 (t, J = 5.6 Hz, 1H), 4.49–4.26 (m, 6H), 3.47–3.41 (m, 6H). MS (ESI) m/z: 574.1 [M + H]+.
Mtb Resistant Mutant Isolation
Isolation of Mtb H37Rv-resistant mutants to iodotubercidin on a solid medium was performed as described by Ioerger et al.37 Three mutants were subjected to whole genome sequencing. Single nucleotide polymorphisms were identified in Mtb adoK and confirmed by sequencing from genomic DNA.38 Resistant mutants were confirmed on the solid medium by confirming the growth on plates containing 5× MIC and in liquid medium using the Microplate Alamar Blue Assay (MABA).39
Cloning, Expression, and Purification of Recombinant MtbAdoK and hAdoK
The WT MtbAdoK gene (Rv2202c) was amplified by the polymerase chain reaction from total genomic DNA of M. tuberculosis H37Rv. The following oligonucleotides were used in the polymerase chain reaction 5′-GGAATTCCATATGGTGACGATCGCGGTAACC-3′ and 5′-CTTAAGCTTCTAGGCCAGCAC-3′, respectively. The amplified DNA fragment was digested with NdeI and HindIII restriction enzymes (New England Biolabs) and subcloned into the corresponding restriction sites of the pET28b vector containing an N-terminal Tobacco Etch Virus cleavable His-tag.5 hAdoK gene was amplified from clone HsCD00042641 (DNASU plasmid repository) using the following synthetic oligonucleotides in the polymerase chain reaction: 5′-GGAATTCCATATGATGACGTCAGTCAGAGAAAATATTC-3′ and 5′-CCCAAGCTTCTAGTGGAAGTCTGGC-3′. The amplified fragment was subsequently cloned into the same pET28b using the same procedures described above. In all cases, gene fidelity was confirmed by DNA sequencing, and sequenced plasmids were used to transform Escherichia coli BL21 (DE3) cells for protein expression. For protein expression, cell cultures were grown in an LB medium at 37.0 °C. Cells were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) when the cell density reached A600 ≈ 0.6–1.0. Cell cultures were incubated for 18 h at 18.0 °C before harvesting.
Harvested cells were lysed using a French press, and the lysate was centrifuged at 17 000 rpm for 1 h. Recombinant MtbAdoK and hAdoK were purified by using a HisTrap HP Nickel column (GE Healthcare). Purification buffers A and B contained, 50.0 mM HEPES, pH 7.5, 500.0 mM NaCl, 500.0 mM imidazole (buffer B only), and 5.0% glycerol. For crystallization studies, MtbAdoK was dialyzed in 20.0 mM HEPES, pH 7.5, 50.0 mM NaCl, 2.0 mM DTT, and 5.0% glycerol. For enzymatic assays, the proteins were dialyzed in 50.0 mM HEPES, 50.0 mM NaCl, 100.0 mM KCL, 4.0 mM DTT, and 20.0% glycerol. Finally, the proteins were aliquoted and stored in −80.0 °C for subsequent crystallization and enzymatic assays.
Enzymatic Assay, IC50 Determination, Steady-State Kinetics, and Ki Determination
Compounds were tested against MtbAdoK using the pyruvate kinase–lactate dehydrogenase coupled assay system in a Cary100 UV–Vis spectrophotometer by monitoring the decrease in absorbance of NADH at 340 nm.40 The reaction was started by the addition of 60 nM of enzyme into a final volume of 200 μL master mix containing 50.0 mM HEPES pH 7.5, 50.0 mM KCl, 6.0 mM MgCl2 (4.0 mM MgCl2 for hAdoK), 3.0 mM ATP (2.0 mM ATP for hAdoK), 200.0 μM NADH, 1.0 mM phosphoenolpyruvate, 1.0 mM DTT, 12.0 U/mL pyruvate kinase, 12.0 U/mL lactate dehydrogenase, and 15.0 μM adenosine. IC50 values for each compound were determined by varying the concentration of the inhibitor at fixed concentrations of the enzyme and by fitting the dose–response data into the four-parameter logistic curve (eq 1) model of GraphPad prism 7.02, as follows
| 1 |
where I is the logarithm of inhibitor concentration, H is the Hill slope and Y, Ymax, and Ymin are the specific activity, maximum specific activity, and minimum specific activity, respectively. Kinetic assays for MtbAdoK were performed essentially as described above with the following exceptions: the master mix contained 60.0 nM MtbAdoK, and the reaction was started by the addition of varying concentrations of adenosine in the presence of constant concentrations of compound (0.0, 20.0, and 40.0 nM). Kinetic data were obtained by fitting the initial velocity data into GraphPad Prism 7.02 nonlinear regression function of Michaelis–Menten model (eq 2), as follows
| 2 |
where Vo is the initial velocity, Ymax is the maximum specific activity, S is the substrate concentration, and Km is the Michaelis–Menten constant.
Specific activity values for kinetics and IC50 measurements were determined using (eq 3)
| 3 |
where a is the change in absorbance over time, εNADH is the millimolar extinction coefficient of NADH, b is the pathlength, d is the dilution factor of the enzyme in the assay, and c is the concentration of enzyme stock used for the assay.
Finally, the inhibition constant (Ki) was determined by using the Cheng–Prusoff relationship for competitive inhibition (eq 4).41,42
| 4 |
In all cases, the compounds were serially diluted in 100.0% DMSO and added to the respective enzymatic reaction to a final concentration of 2.5% DMSO.
Crystallization, Data Collection, and Crystal Structure Determination
Purified MtbAdoK was concentrated to 18 mg/mL before crystallization trials. All cocrystals were obtained by the vapor diffusion method. MtbAdoK-2, 3, 4, and 7 cocrystals were achieved by mixing 2.0 μL of protein solution, preincubated with 4.0 mM of the compound for 1.0 h at 25.0 °C, with 1.0 μL of 100.0 mM HEPES pH 7.5, 2.0 M ammonium sulfate and 2.0% PEG 400. MtbAdoK-5 cocrystals were obtained by mixing 2.0 μL of protein solution, preincubated with 4.0 mM of 5 for 1.0 h at 25.0 °C, with 1.0 μL of 100.0 mM HEPES pH 7.5 and 1.2 M sodium citrate tribasic dihydrate. MtbAdoK-6 cocrystals were obtained by mixing 2.0 μL of protein solution, preincubated with 4.0 mM of the compound for 1.0 h at 25.0 °C, with 1.0 μL of 5.0 M sodium formate. Finally, MtbAdoK-17 cocrystals were obtained by mixing 2.0 μL of protein solution, preincubated with 2.0 mM 17 for 1 h at 25 °C, with 1.0 μL of 100 mM Bis–Tris pH 6.5, 2.0 M ammonium sulfate and 2.0% PEG 400. In all cases, crystals were grown at 20.0 °C. Prior to data collection, crystals were cryoprotected with Paratone (Hampton research) and flash frozen in liquid nitrogen. X-ray diffraction data were collected at Argonne’s National Lab Advanced Photon Source beamlines 19ID and 23ID. Diffraction data were indexed, scaled, and integrated using HKL2000.43 Initial phases were obtained by MR in MOLREP using the high-resolution structure of the MtbAdoK-1 complex with PDB accession code 2PKM.44 Refinement was performed in PHENIX followed by iterative runs of inspection and manual modification using coot.45,46 Ligand model and restraint files were created in ELBOW from the PHENIX suite and fitted into the electron density using COOT. Images and figures were rendered using Molsoft ICM, Chimera and PyMOL.47−49
Antimycobacterial Assay
Antitubercular testing and EC50 determination was performed using the MABA assay in a 96-well format as previously described.50,51 Starter culture of Mtb mc2 7000 was grown in 7H9 media supplemented with OADC (Middlebrook), 0.5% dextrose, 0.085% NaCl, 0.05% Tyloxapol (Sigma), 0.25 μg/mL malachite green (Sigma), and 25 μg/mL pantothenate. Once cells reached an optical density of OD600 ≈ 1.5, cells were diluted to an OD600 of 0.01 in the same media composition without OADC. Compounds were serially diluted in 100.0% DMSO and added to the cells to a final concentration of DMSO of 2.5%. Plates were incubated for 6 days before staining with resazurin (Sigma). After staining, plates were incubated for 2 additional days for developing. Finally, developed plates were read using a POLARstar Omega spectrophotometer (BMG Labtech) and by monitoring the fluorescence emission of resazurin (excitation = 570.0 nm, emission = 585.0 nm). In all cases, rifampicin was used as a negative control. The anti-Tb activity was determined as a percent control, and EC50 values for each compound were determined by utilizing eq 1.
Human Dermal Fibroblast Cytotoxicity Assay
HDFs were purchased from ATCC (Manassas, VA). HDF cells were cultured in DMEM (Lonza) media supplemented with 10.0% fetal bovine serum (Lonza) and penicillin/streptomycin (Lonza). For the cytotoxicity assay, compound stocks were serially diluted in phosphate-buffered saline plus 10.0% DMSO. On the day of assay, HDF cells were trypsinized, counted, and resuspended at a concentration of 64 000 cells/mL in media. Cells were plated, overlaid with the compound serial dilutions, and incubated at 37.0 °C. After 48.0 h, resazurin dye was added, and the assay plates cultured for another 24.0 h. On the next day, the absorbance of the resazurin was measured on a microplate reader to assess cell death. Cytotoxicity was determined as a percent control, and CC50 values for each compound were determined by utilizing eq 1.
Pharmacokinetic and Tolerability Studies
***All animal experiments described in this manuscript followed protocols approved by the Texas A&M University Institutional Animal Care and Use Committee under Animal Use Protocol “Preliminary in vivo PK/Tox on newly developed drug compounds” #2017-0368.***
Eight-week old female ND4 Swiss Webster mice (∼28 g each) (Envigo, Indianapolis, IN) were used for PK and combination tolerability-snapshot PK studies. Compound 18 was formulated in 5% DMSO, 95% canola oil and administered as a single 100 μL oral gavage at 10 mg·kg–1 for PK and as a 200 μL oral gavage at 50, 100, or 200 mg·kg–1 QD × 3 days for tolerability-snapshot PK. Mice were anesthetized using isoflurane and 50 μL collected at 0, 0.25, 0.5, 1, 2, and 4 h survival bleed times for traditional PK or 0, 1, 4, and 24 h for tolerability-snapshot PK studies after the first administered dose. Around 200 μL was collected at 6, 8, and 24 h terminal bleed times for the traditional PK. Survival bleeds were drawn from the facial vein and terminal bleeds via cardiac puncture following CO2 asphyxiation.35 Three mice were used per time point.
Blood samples were centrifuged (3000g, 15 min) for plasma separation. Aliquots of plasma (10 μL) were extracted with two 500 μL equivalents of methanol containing 0.1% formic acid to ensure maximum recovery of compound 18 from plasma. The supernatant was collected from precipitated protein pellet following centrifugation (3000g, 5 min) and evaporated to dryness using a temperature-controlled Eppendorf Vacufuge at 30 °C for 4 h. The dry samples were reconstituted with 100 μL of methanol (1:10 original analyte dilution factor) spiked with 250 ng·mL–1 verapamil (Sigma-Aldrich) as an internal standard and subjected to LC/MS analysis on a Bruker Daltronics micrOTOF-Q II mass spectrometer coupled with an Agilent 1200 Infinity series HPLC with temperature controlled autosampler (24 °C) and photodiode array detector. Standard solutions of compound 18 ranging from 1000 to 7.8125 ng·mL–1 spiked into mouse control plasma was also analyzed to generate a calibration curve corrected for matrix effects. A linear regression generated from calibration samples was used to quantify samples.34 The LOD of the LC/MS method was approximately 2 ng·mL–1. A 4.6 × 100 mm Kinetex 2.6 μm EVO C18 100 Å column at a flow rate of 0.5 mL·min–1 was used in the analysis. The mobile phase consisted of water containing 0.1% formic acid as solvent A and acetonitrile with 0.1% formic acid as solvent B. The gradient conditions were maintained as follows: 90% A, 10% B to 100% B in 8 min; 100% held for 4 min as a washing step; 100% B back to 90% A in 2 min; 90% A, 10% B held for 3 min to reset and equilibrate the column. Each analytical run was automatically calibrated using a secondary line injection of 10 μL of sodium acetate external standard at 13 min. The injection volume of the analyte was 10 μL, and MS was operated in positive mode with electrospray ionization at source. Mass spectra were monitored in a range of 50–1000m/z. Data were processed using DataAnalysis 4.1 and QuantAnalysis (Bruker Daltronics).
Acknowledgments
The authors would like to acknowledge the Argonne National Lab staff at beamlines 19-ID and 23-ID, the TB Drug Accelerator Program (TBDA), the Bill and Melinda Gates Foundation and the Welch Foundation for their financial and/or intellectual support.
Glossary
Abbreviations
- Mtb
Mycobacterium tuberculosis
- MtbAdoK
Mycobacterium tuberculosis adenosine kinase
- hAdoK
human adenosine kinase
- vdW
van der Waals
- HDF
human dermal fibroblasts
- MR
molecular replacement
- TBSCl
tert-butyldimethylsilyl chloride
- EtOH
ethanol
- DIEA
N,N-diisopropylethylamine
- ArB(OH)2
aryl boronic acids
- XPhos Pd G2
chloro(2-dicyclohexylphosphino-2′,4′,6′-triisopropyl-1,1′-biphenyl)[2-(2′-amino-1,1′-biphenyl)]palladium(II)
- MeOH
methanol
- RuPhos
2-dicyclohexylphosphino-2′,6′-diisopropoxybiphenyl
- AUC
area under the curve
- PK
pharmacokinetic
- Cmax
maximum concentration
- tmax
time at maximum concentration.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.9b00020.
Accession Codes
Authors will release the atomic coordinates and experimental data upon article publication: MtbAdoK-2 complex-PDB ID 6C67, MtbAdoK-3 complex-PDB ID 6C9N, MtbAdoK-4 complex-PDB ID 6C9P, MtbAdoK-5 complex-PDB ID 6C9Q, MtbAdoK-6 complex-PDB ID 6C9R, MtbAdoK-7 complex-PDB ID 6C9V, MtbAdoK-17 complex-PDB ID 6C9S.
Author Present Address
# Lilly China R&D Center, Eli Lilly and Company, Shanghai China.
Author Present Address
¶ GSK, Swedeland Road, King of Prussia, PA.
This work was supported by grants from the Bill and Melinda Gates Foundation (OPP1024055), Welch Foundation (A-0015), NIAID-NIH (TB Structural Genomics grant P01A1095208) to J.C.S. and the Bill and Melinda Gates Foundation (OPP1024038) to T.P.
The authors declare no competing financial interest.
Supplementary Material
References
- WHO . Global Tuberculosis Report, 2016.
- Ducati R. G.; Breda A.; Basso L. A.; Santos D. S. Purine Salvage Pathway in Mycobacterium tuberculosis. Curr. Med. Chem 2011, 18, 1258–1275. 10.2174/092986711795029627. [DOI] [PubMed] [Google Scholar]
- Villela A. D.; Sanchez-Quitian Z. A.; Ducati R. G.; Santos D. S.; Basso L. A. Pyrimidine Salvage Pathway in Mycobacterium tuberculosis. Curr. Med. Chem 2011, 18, 1286–1298. 10.2174/092986711795029555. [DOI] [PubMed] [Google Scholar]
- Long M. C.; Escuyer V.; Parker W. B. Identification and Characterization of a Unique Adenosine Kinase from Mycobacterium tuberculosis. J. Bacteriol. 2003, 185, 6548–6555. 10.1128/jb.185.22.6548-6555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy M. C. M.; Palaninathan S. K.; Shetty N. D.; Owen J. L.; Watson M. D.; Sacchettini J. C. High Resolution Crystal Structures ofMycobacterium tuberculosisAdenosine Kinase. J. Biol. Chem. 2007, 282, 27334–27342. 10.1074/jbc.m703290200. [DOI] [PubMed] [Google Scholar]
- Park J.; Gupta R. S. Adenosine kinase and ribokinase - the RK family of proteins. Cell. Mol. Life Sci. 2008, 65, 2875–2896. 10.1007/s00018-008-8123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M. A.; Scott D. M.; Mathews I. I.; Ealick S. E.; Roos D. S.; Ullman B.; Brennan R. G. Crystal Structures of Toxoplasma gondii Adenosine Kinase Reveal a Novel Catalytic Mechanism and Prodrug Binding. J. Mol. Biol. 2000, 298, 875–893. 10.1006/jmbi.2000.3753. [DOI] [PubMed] [Google Scholar]
- Mathews I. I.; Erion M. D.; Ealick S. E. Structure of Human Adenosine Kinase at 1.5 Å Resolution†,‡. Biochemistry 1998, 37, 15607–15620. 10.1021/bi9815445. [DOI] [PubMed] [Google Scholar]
- Romanello L.; Bachega J. F. R.; Cassago A.; Brandão-Neto J.; DeMarco R.; Garratt R. C.; Pereira H. D. M. Adenosine kinase fromSchistosoma mansoni: structural basis for the differential incorporation of nucleoside analogues. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 126–136. 10.1107/s0907444912044800. [DOI] [PubMed] [Google Scholar]
- Griffin J. E.; Gawronski J. D.; DeJesus M. A.; Ioerger T. R.; Akerley B. J.; Sassetti C. M. High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 2011, 7, e1002251 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J.; Reddy M. C.; Ioerger T. R.; Rothchild A. C.; Dartois V.; Schuster B. M.; Trauner A.; Wallis D.; Galaviz S.; Huttenhower C.; Sacchettini J. C.; Behar S. M.; Rubin E. J. Tryptophan Biosynthesis Protects Mycobacteria from CD4 T-Cell-Mediated Killing. Cell 2013, 155, 1296–1308. 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M. C.; Parker W. B. Structure-activity relationship for nucleoside analogs as inhibitors or substrates of adenosine kinase from Mycobacterium tuberculosis. Biochem. Pharmacol. 2006, 71, 1671–1682. 10.1016/j.bcp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Long M. C.; Shaddix S. C.; Moukha-Chafiq O.; Maddry J. A.; Nagy L.; Parker W. B. Structure-activity relationship for adenosine kinase from Mycobacterium tuberculosis. Biochem. Pharmacol. 2008, 75, 1588–1600. 10.1016/j.bcp.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnuit V.; Slavětínská L. P.; Nauš P.; Džubák P.; Hajdúch M.; Stolaříková J.; Snášel J.; Pichová I.; Hocek M. 2-Substituted 6-(Het)aryl-7-deazapurine Ribonucleosides: Synthesis, Inhibition of Adenosine Kinases, and Antimycobacterial Activity. ChemMedChem 2015, 10, 1079–1093. 10.1002/cmdc.201500081. [DOI] [PubMed] [Google Scholar]
- Snášel J.; Nauš P.; Dostál J.; Hnízda A.; Fanfrlík J.; Brynda J.; Bourderioux A.; Dušek M.; Dvořáková H.; Stolaříková J.; Zábranská H.; Pohl R.; Konečný P.; Džubák P.; Votruba I.; Hajdúch M.; Řezáčová P.; Veverka V.; Hocek M.; Pichová I. Structural Basis for Inhibition of Mycobacterial and Human Adenosine Kinase by 7-Substituted 7-(Het)aryl-7-deazaadenine Ribonucleosides. J. Med. Chem. 2014, 57, 8268–8279. 10.1021/jm500497v. [DOI] [PubMed] [Google Scholar]
- Parker W. B.; Barrow E. W.; Allan P. W.; Shaddix S. C.; Long M. C.; Barrow W. W.; Bansal N.; Maddry J. A. Metabolism of 2-methyladenosine in Mycobacterium tuberculosis. Tuberculosis 2004, 84, 327–336. 10.1016/j.tube.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Chen C.-K.; Barrow W. W.; Parker W. B.; Barrow E. W.; Maddry J. A.; Suling W. J.; Allan P. W.; Bansal N. The Metabolism of 2-methyladenosine in Mycobacterium smegmatis. Microbiology 2002, 148, 289–295. 10.1099/00221287-148-1-289. [DOI] [PubMed] [Google Scholar]
- Barrow E. W.; Westbrook L.; Bansal N.; Suling W. J.; Maddry J. A.; Parker W. B.; Barrow W. W. Antimycobacterial Activity of 2-methyl-adenosine. J. Antimicrob. Chemother. 2003, 52, 801–808. 10.1093/jac/dkg444. [DOI] [PubMed] [Google Scholar]
- Massillon D.; Stalmans W.; van de Werve G.; Bollen M. Identification of the Glycogenic Compound 5-iodotubercidin as a General Protein Kinase Inhibitor. Biochem. J. 1994, 299, 123–128. 10.1042/bj2990123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkar B. G.; DaRe J. M.; Kopcho J. J.; Browne C. E.; Schanzer J. M.; Wiesner J. B.; Erion M. D. Adenosine Kinase Inhibitors. 1. Synthesis, Enzyme Inhibition, and Antiseizure Activity of 5-Iodotubercidin Analogues. J. Med. Chem. 2000, 43, 2883–2893. 10.1021/jm000024g. [DOI] [PubMed] [Google Scholar]
- Loomis C. R.; Bell R. M. Sangivamycin, a Nucleoside Analogue, is a Potent Inhibitor of Protein Kinase C. J. Biol. Chem. 1988, 263, 1682–1692. [PubMed] [Google Scholar]
- Muchmore S. W.; Smith R. A.; Stewart A. O.; Cowart M. D.; Gomtsyan A.; Matulenko M. A.; Yu H.; Severin J. M.; Bhagwat S. S.; Lee C.-H.; Kowaluk E. A.; Jarvis M. F.; Jakob C. L. Crystal Structures of Human Adenosine Kinase Inhibitor Complexes Reveal Two Distinct Binding Modes. J. Med. Chem. 2006, 49, 6726–6731. 10.1021/jm060189a. [DOI] [PubMed] [Google Scholar]
- Zemlicka J.; Endo T. O6-(4-Nitrophenyl)inosine and -Guanosine as Chromogenic Substrates for Adenosine Deaminase. Nucleosides Nucleotides 1996, 15, 619–629. 10.1080/07328319608002410. [DOI] [Google Scholar]
- Camp D.; Li Y.; McCluskey A.; Moni R. W.; Quinn R. J. Diimidazo[1,2-c:4′,5′-e]pyrimidines: N6-N1 conformationally restricted adenosines. Bioorg. Med. Chem. Lett. 1998, 8, 695–698. 10.1016/s0960-894x(98)00101-2. [DOI] [PubMed] [Google Scholar]
- Katagiri N.; Matsuhashi Y.; Kokufuda H.; Takebayashi M.; Kaneko C. A Highly Efficient Synthesis of the Antiviral Agent (+)-cyclaradine Involving the Regioselective Cleavage of Epoxide by Neighboring Participation. Tetrahedron Lett. 1997, 38, 1961–1964. 10.1016/s0040-4039(97)00234-7. [DOI] [Google Scholar]
- Itoh T.; Sugawara T.; Mizuno Y. A Novel Synthesis of 1-Deazaadenosine. Heterocycles 1982, 17, 305–309. 10.3987/s-1982-01-0305. [DOI] [Google Scholar]
- Wilkens S. Structure and Mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. 10.12703/p7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braibant M.; Gilot P.; Content J. The ATP Binding Cassette (ABC) Transport Systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2000, 24, 449–467. 10.1111/j.1574-6976.2000.tb00550.x. [DOI] [PubMed] [Google Scholar]
- Sandhu P.; Akhter Y. The Drug Binding Sites and Transport Mechanism of the RND Pumps from Mycobacterium tuberculosis: Insights from Molecular Dynamics Simulations. Arch. Biochem. Biophys. 2016, 592, 38–49. 10.1016/j.abb.2016.01.007. [DOI] [PubMed] [Google Scholar]
- De Rossi E.; Aínsa J. A.; Riccardi G. Role of Mycobacterial Efflux Transporters in Drug Resistance: an Unresolved Question. FEMS Microbiol. Rev. 2006, 30, 36–52. 10.1111/j.1574-6976.2005.00002.x. [DOI] [PubMed] [Google Scholar]
- Balganesh M.; Dinesh N.; Sharma S.; Kuruppath S.; Nair A. V.; Sharma U. Efflux Pumps of Mycobacterium tuberculosis Play a Significant Role in Antituberculosis Activity of Potential Drug Candidates. Antimicrob. Agents Chemother. 2012, 56, 2643–2651. 10.1128/aac.06003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pule C. M.; Sampson S. L.; Warren R. M.; Black P. A.; van Helden P. D.; Victor T. C.; Louw G. E. Efflux Pump Inhibitors: Targeting Mycobacterial Efflux Systems to Enhance TB Therapy. J. Antimicrob. Chemother. 2016, 71, 17–26. 10.1093/jac/dkv316. [DOI] [PubMed] [Google Scholar]
- Park J.; Singh B.; Gupta R. S. Mycobacterial Adenosine Kinase is not a Typical Adenosine Kinase. FEBS Lett. 2009, 583, 2231–2236. 10.1016/j.febslet.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Ahmed T. R.; Shashank R. Application of LC-MS in Supporting PK/PD Studies During Drug Discovery and Development. Pharm. Res. 2012, 5, 2514–2526. [Google Scholar]
- Diehl K.-H.; Hull R.; Morton D.; Pfister R.; Rabemampianina Y.; Smith D.; Vidal J.-M.; Vorstenbosch C. V. D. A Good Practice Guide to the Administration of Substances and Removal of Blood, Including Routes and Volumes. J. Appl. Toxicol. 2001, 21, 15–23. 10.1002/jat.727. [DOI] [PubMed] [Google Scholar]
- Liu B.; Chang J.; Gordon W. P.; Isbell J.; Zhou Y.; Tuntland T. Snapshot PK: a Rapid Rodent in vivo Preclinical Screening Approach. Drug Discovery Today 2008, 13, 360–367. 10.1016/j.drudis.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Ioerger T. R.; O’Malley T.; Liao R.; Guinn K. M.; Hickey M. J.; Mohaideen N.; Murphy K. C.; Boshoff H. I. M.; Mizrahi V.; Rubin E. J.; Sassetti C. M.; Barry C. E. III; Sherman D. R.; Parish T.; Sacchettini J. C. Identification of New Drug Targets and Resistance Mechanisms in Mycobacterium tuberculosis. PLoS One 2013, 8, e75245 10.1371/journal.pone.0075245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger T. R.; Feng Y.; Ganesula K.; Chen X.; Dobos K. M.; Fortune S.; Jacobs W. R.; Mizrahi V.; Parish T.; Rubin E.; Sassetti C.; Sacchettini J. C. Variation among Genome Sequences of H37Rv Strains of Mycobacterium tuberculosis from Multiple Laboratories. J. Bacteriol. 2010, 192, 3645–3653. 10.1128/jb.00166-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.; Lee H. S.; Franzblau S.. Microplate Alamar Blue Assay (MABA) and Low Oxygen Recovery Assay (LORA) for Mycobacterium tuberculosis. In Mycobacteria Protocols; Parish T., Roberts D. M., Eds.; Springer New York: New York, NY, 2015; pp 281–292. [DOI] [PubMed] [Google Scholar]
- Blondin C.; Serina L.; Wiesmuller L.; Gilles A. M.; Barzu O. Improved Spectrophotometric Assay of Nucleoside Monophosphate Kinase Activity Using the Pyruvate Kinase/Lactate Dehydrogenase Coupling System. Anal. Biochem. 1994, 220, 219–221. 10.1006/abio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Cer R. Z.; Mudunuri U.; Stephens R.; Lebeda F. J. IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 2009, 37, W441–W445. 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung-Chi C.; Prusoff W. H. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z.; Minor W.. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In Methods in Enzymology; Carter C. W. Jr., Sweet R. M., Eds.; Academic Press: New York, 1997; Vol. 276: Macromolecular Crystallography, Part A, pp 307–326. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994, 50 (), 760. 10.1107/s0907444994003112 [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Grosse-Kunstleve R. W.; Hung L.-W.; Ioerger T. R.; McCoy A. J.; Moriarty N. W.; Read R. J.; Sacchettini J. C.; Sauter N. K.; Terwilliger T. C. PHENIX: Building New Software for Automated Crystallographic Structure Determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002, 58, 1948–1954. 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development ofCoot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- PyMOL The PyMOL Molecular Graphics System, version 1.8; Schrödinger, LLC.
- Abagyan R.; Totrov M.; Kuznetsov D. ICM?A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. J. Comput. Chem. 1994, 15, 488–506. 10.1002/jcc.540150503. [DOI] [Google Scholar]
- Franzblau S. G.; Witzig R. S.; McLaughlin J. C.; Torres P.; Madico G.; Hernandez A.; Degnan M. T.; Cook M. B.; Quenzer V. K.; Ferguson R. M.; Gilman R. H. Rapid, Low-Technology MIC Determination with Clinical Mycobacterium tuberculosis Isolates by Using the Microplate Alamar Blue Assay. J. Clin. Microbiol 1998, 36, 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger I. V.; Freundlich J. S.; Gawandi V. B.; Roberts J. P.; Gawandi V. B.; Sun Q.; Owen J. L.; Fraile M. T.; Huss S. I.; Lavandera J.-L.; Ioerger T. R.; Sacchettini J. C. Structure-Guided Discovery of Phenyl-diketo Acids as Potent Inhibitors of M. tuberculosis Malate Synthase. Chem. Biol. 2012, 19, 1556–1567. 10.1016/j.chembiol.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.