Abstract
Lack of insight into mechanisms governing breast cancer metastasis has precluded development of curative therapies. Metastasis-initiating cancer cells (MICs) are uniquely equipped to establish metastases, causing recurrence and therapeutic resistance. Using various metastasis models, we discovered that certain primary tumors elicit a systemic inflammatory response involving IL-1β-expressing innate immune cells that infiltrate distant MIC microenvironments. At the metastatic site, IL-1β maintains MICs in a ZEB1-positive differentiation state, preventing MICs from generating highly proliferative E-cadherin-positive progeny. Thus, when the inherent plasticity of MICs is impeded, overt metastases cannot be established. Ablation of the pro-inflammatory response or IL-1R inhibition relieves the differentiation block and results in metastatic colonization. Among lymph node-positive breast cancer patients, high primary tumor IL-1β expression is associated with better overall survival and distant metastasis-free survival. Our data reveal complex interactions that occur between primary tumors and disseminated MICs that could be exploited to improve patient survival.
Breast cancer patients often exhibit no evidence of disseminated disease at initial diagnosis, yet ~20% of patients ultimately relapse1. Metastatic dissemination often begins at early stages2, 3, yielding many latent micrometastases. By some estimates, less than 0.02% of those disseminated tumor cells will form secondary tumors, indicating that successful metastatic colonization is rare4-6 and ascribed to only specialized minority cancer cells, termed metastasis-initiating cells (MICs)7.
The seemingly simultaneous emergence of clinically detectable metastases has led to the notion that reactivation of secondary lesions from dormancy is triggered systemically8, 9. Pre-clinical modeling has revealed that primary tumors influence metastasis by modulating both systemic and secondary tumor microenvironments before and after dissemination10-15. The role of the immune system during these processes is particularly complex16. Little is known about immune system impact on MIC colonization or the context in which primary tumor-driven pathophysiology will prove pro- or anti-metastatic.
Successful metastatic colonization is also largely dependent upon tumor cell inherent biology. Cellular plasticity is a fundamental component of several leading metastasis models, including co-option of developmental pathways, the epithelial-to-mesenchymal transition (EMT) and cancer stem cell (CSC) models17. Once MICs reach a distant tissue, the necessity of cellular plasticity to developing overt metastatic lesions remains to be determined. Clinical and pre-clinical findings thus provoke the question of whether the success of disseminated MICs is influenced by overall disease pathophysiology.
RESULTS
Identification of Primary Tumors that Inhibit Metastatic Colonization
To determine whether primary tumors influence colonization of disseminated metastasis initiating cells (MICs), we first employed a polyclonal metastatic mammary carcinoma cell line, Met1, derived from a spontaneous lung metastasis in an FVB/N-Tg(MMTV-PyMT) mouse18,.
Met1 cells or PBS vehicle control were injected orthotopically into FVB mice. After two weeks, when primary tumors reached ~100 mm3 (Supplementary Fig. 1a), we synchronized metastasis by injecting the same heterogeneous Met1 population intravenously (Fig.1a), whereby only the MIC subpopulation should be capable of seeding metastases19. Lungs were analyzed after a subsequent 2-week period.
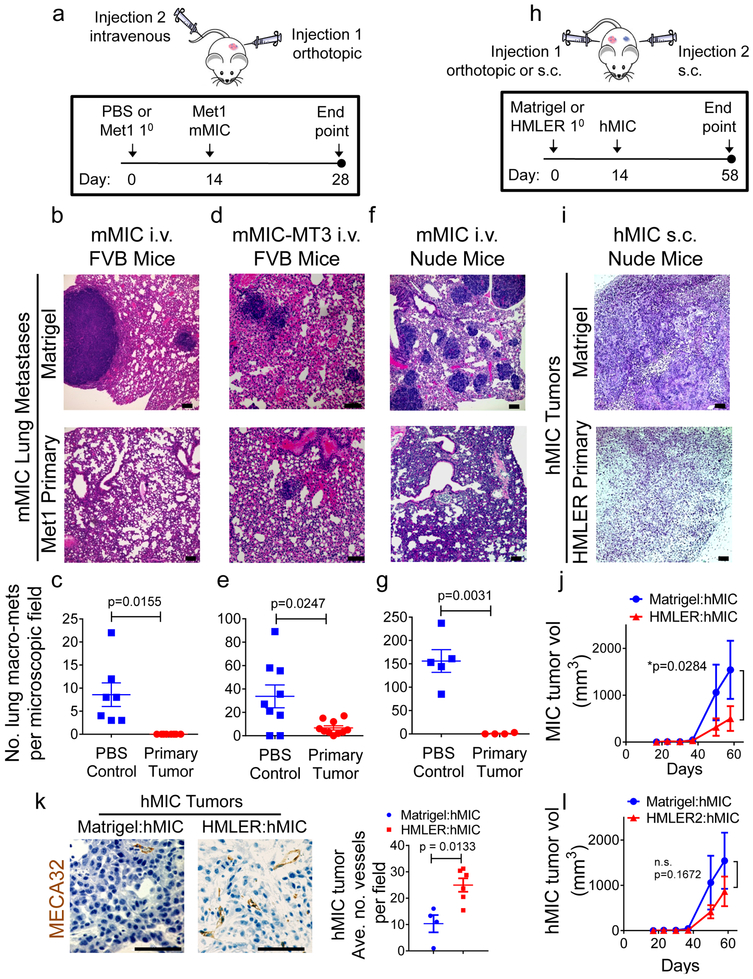
Figure 1. Primary tumors inhibit outgrowth of distant metastasis-initiating cells independently of adaptive immune system.
a,Schematic modeling early stages of murine metastasis-initiating cell (mMIC) lung colonization in presence of Met1 primary tumor in FVB (b-e) or Nude (f-g) mice. PBS vehicle is control for primary tumors. Met1 cells (2.5 × 105/mouse) or PBS control (20 μl) were injected into a single 4th inguinal mammary fat pad at day 0. On day 14, Met1 cells (mMIC) or the Met1-derived clone MT3 (mMIC-MT3) were injected intravenously (tail vein) (7.5 × 105 cells/mouse). Primary tumor growth kinetics were monitored from day 0; pulmonary metastases quantified at experimental end points. b,d,f, Hematoxylin and eosin (H&E) images of lungs from mice bearing Met1 primary tumors or PBS control. c,e,g, Macro-metastases >100 microns quantified from microscopic tissue sections from 4 lung lobes/animal for: mMIC metastases in FVB mice (PBS n=7 animals; Met1 n=8 animals) (c); mMIC-MT3 metastases, FVB mice (PBS n=9 animals, Met1 n=10 animals) (e); mMIC metastases in Nude mice (PBS n=5 animals, Met1 n=4 animals) (g). h, Schematic modeling early stages of human metastasis-initiating cell (hMIC) colonization in presence of HMLER primary tumor (Nude mice). Matrigel vehicle is experimental control for primary tumors. HMLER cells (5.0 × l05/mouse) or Matrigel control (100 μ1) were injected subcutaneously into one flank at day 0. Two weeks later, hMIC (2.5 × l05 cells/mouse) were injected subcutaneously into the contralateral flank. Growth kinetics were monitored for the experimental duration. i, H&E images of hMIC from mice bearing HMLER primary tumors or Matrigel control. j, hMIC tumor growth kinetics in mice bearing Matrigel control (n=9 animals), or HMLER primary tumors (n=10 animals). k, Images: hMIC tumors described in Figure 1i, opposite Matrigel (n=25 images representing 4 tumors) or HMLER primary tumors (n=25 images representing 6 tumors), stained–as indicated; Graph: mean vessel number/microscopic field. l, Tumor growth kinetics of hMIC implanted opposite Matrigel control or an HMLER2 primary tumor (n = 10 animals/cohort), per protocol in 1h. All scale bars=100 μm. Source data for c, e, g, j, k, l provided in Supplementary Table 1. Welch’s 2-sided t test (c, e, g, k); Two-way ANOVA followed by Sidak’s multiple comparison test (j, l).
In three independent experiments, the control cohort developed overt pulmonary metastases while no macrometastases were observed in mice bearing orthotopic Met1 primary tumors (Fig. 1b, c). Importantly, orthotopic Met1 primary tumors did not inhibit Met1 secondary tumors that were injected subcutaneously (Supplementary Fig. 1b), an injection scheme that does not provide selection pressure for mMIC subpopulations19. These results suggested that Met1 primary tumors specifically inhibit growth driven by their MICs and not the bulk, heterogeneous population of tumor cells.
We previously generated a series of single cell-derived clones from the parental Met1 line20. To identify a purified population of Met1 murine MICs (mMICs), we tested the metastatic potential of two of these clones, MT2 and MT3. The MT3 subclone was subsequently defined as a mMIC population due to its enhanced metastatic potential (~90 macrometastases/field; 100% incidence) compared to poorly metastatic MT2 cells (~1 macrometastasis/field; 66.6% incidence) (Supplementary Fig 1c). Hence, Met1 primary tumors or control PBS were orthotopically injected into cohorts of FVB mice followed two weeks later by intravenous injection of the mMIC-MT3 cells (Fig. 1a). At the experimental end point, mMIC-MT3 pulmonary metastases were reduced ~6-fold in the cohort bearing Met1 primary tumors relative to the control cohort (Fig. 1d, e).
To test whether adaptive immunity was necessary for inhibiting mMIC pulmonary metastases, we conducted the same experiments in athymic Nude mice. After two weeks, when primary tumors reached ~200 mm3 (Supplementary Fig. 1d), Met1 cells were injected intravenously (Fig.1a). Again, Met1 primary tumors significantly inhibited pulmonary metastases (Fig. 1f, g), indicating that mMIC inhibition was not T cell-dependent.
The results from immunocompromised mice presented us with the opportunity to test human xenografts. Accordingly, we used the polyclonal human mammary carcinoma cell line, HMLER, which represents heterogeneous cell populations commonly observed in primary breast cancers. We used a well-characterized clonal MIC subpopulation (hMIC) that had been isolated directly from the HMLER cell line; when compared with other HMLER subclones, hMIC is uniquely metastatic21.
Primary HMLER tumors significantly inhibited outgrowth of subcutaneous hMIC secondary tumors (Fig. 1h-j) as well as hMIC pulmonary metastases (Supplementary Fig. 1e). We also tested highly metastatic MDA-MB-231 human breast cancer cells, which are enriched for hMICs22. These 231-MIC secondary tumors were also significantly inhibited by HMLER primary tumors (Supplementary Fig. 1f-i).
We ruled out the possibility that the primary tumors inhibited hMIC outgrowth through release of anti-angiogenic factors23. In fact, hMIC-derived tumors from mice bearing primary tumors contained ~2.5-fold more blood vessels per section than the control cohort (Fig. 1k).
Importantly, we discovered that primary tumors from another HMLER derivative subpopulation, HMLER221, did not inhibit hMIC colonization (Fig. 1l). This finding suggested there are properties specific to inhibitory primary tumors.
These data established several important principles. First, primary tumors that inhibited MIC colonization did not prevent outgrowth of heterogeneous tumor cell populations comprised of MICs and non-MICs, suggesting MIC-specific properties make them susceptible to growth inhibition. Second, systemic growth inhibition of MICs is not tissue-specific, since primary tumors inhibited outgrowth of both subcutaneous and pulmonary MICs. Third, primary tumors systemically inhibited MICs independently of an adaptive immune system without affecting MIC vascularization, pointing to innate immune mechanisms. Fourth, not all primary tumors inhibited distant MIC colonization.
MIC Proliferation and Differentiation are Mechanistically Linked
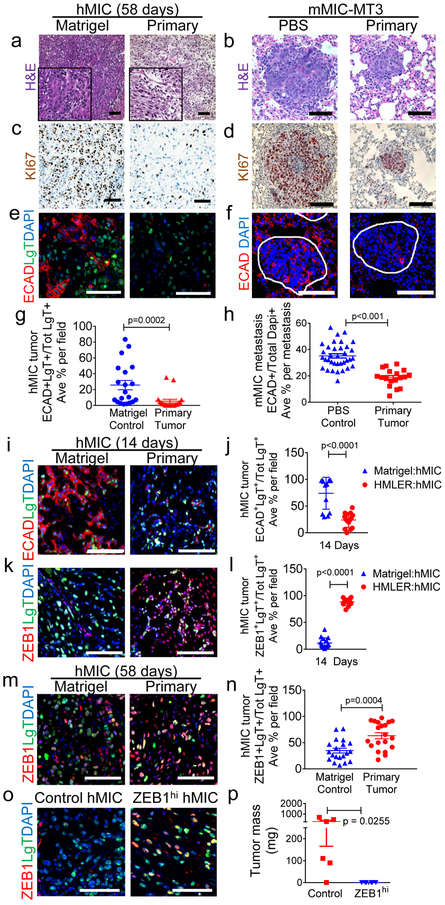
The histopathology of control cohort hMIC tumors was consistent with that of breast ductal adenocarcinomas observed in the clinic; in contrast, cancer cells in hMIC tumors from cohorts bearing primary HMLER tumors appeared mesenchymal-like, resembling breast spindle cell carcinomas (Fig. 2a). Similarly, mMIC-MT3 metastatic foci in primary tumor-bearing mice appeared poorly differentiated (Fig. 2b). hMIC and mMIC proliferation was significantly reduced in cohorts bearing primary tumors relative to their respective control cohorts (Fig. 2c, d).
Figure 2. Primary tumors prevent differentiation and proliferation of distant MICs.
a-f, hMIC secondary tumors and mMIC-MT3 pulmonary metastases (per schematics in Fig. 1a, h) stained with: hematoxylin and eosin (H&E) (a, b); Ki67 (brown) and hematoxylin (nuclei; blue) (c, d); Immunofluorescence for E-cadherin (ECAD, red), Large T antigen to identify hMICs (LgT, green), and DAPI (nuclei, blue); lung metastases are circled in white (e, f). g-h, ECAD+ cells as % total number of LgT+ tumor cells (g) or DAPI+ cells (h) per microscopic field (ECAD in hMIC tumors n=20 images, representing 5 tumors/cohort; ECAD in mMIC-MT3 metastasis n=52 images, representing at least 18 metastases/cohort). i, j, Immunofluorescent images of hMIC tumors stained with ECAD (i) and quantified in (j). Matrigel cohort: n=15 images, HMLER cohort: n=13 images. k, l Immunofluorescent images of hMIC tumors stained with ZEB1 (k) and quantified in (l). Matrigel cohort: n=13 images, HMLER cohort: n=16 images. (j,l) presented as % Total number of LgT+ hMIC tumor cells. m, n, hMIC tumors (per Fig. 1a). Immunofluorescent images (m) and quantification (n) of ZEB1 staining (red) in hMIC tumors (positive for LgT antigen, green), as a percentage of total LgT+ cells. DAPI (nuclei, blue). Control n=20 independent images representing 3 tumors; HMLER cohort n=20 independent images representing 3 tumors. o, ZEB1 (red) and LgT+ tumor cells (green) in hMIC tumors from tumors in (p). DAPI (nuclei, blue). p, Final mass of hMIC tumors expressing either doxycycline-inducible control (Control hMIC; n=6 tumors) or ZEB1 cDNA (ZEB1hi hMIC; n=6 tumors). Scale bars=100 μm. Source data for g, h, j, l, n, p provided in Supplementary Table 1. 2-sided Mann-Whitney test (g, l); Welch’s 2-sided t test (h, n); 1-sided Welch’s t test (j,p).
Evidence from multiple groups indicates that MICs reside in a partial-EMT state24. As MICs generate their non-MIC progeny during secondary tumor formation, mesenchymal properties are lost and epithelial phenotypes are reacquired25, 7 Our histopathological observations supported the hypothesis that inhibitory primary tumors maintain the MIC mesenchymal state and prevent differentiation into epithelial progeny.
Consistent with MIC traits, both hMICs and mMICs expressed low or undetectable levels of the epithelial marker E-cadherin (ECAD) in vitro at the time of their injection (Supplementary Fig. 2a, b). In vivo however, MIC-derived metastases and secondary tumors from control cohorts acquired ECAD expression, which was ~5-fold higher in hMICs and ~2-fold higher in mMICs when compared with MICs from primary tumor-bearing mice (Fig. 2e-h). Epithelial phenotypic plasticity was also manifest when we visualized cytokeratin and vimentin (Supplementary Fig. 2c, d).
We next asked whether the block in MIC differentiation and growth inhibition were mechanistically related. We reasoned that these responses should be analyzed from size-matched MIC-derived tumor tissues. We therefore used the hMIC model and injected either HMLER cells or Matrigel control subcutaneously into mice; 14 days later, we injected hMICs into the contralateral flanks of the mice in each cohort and harvested tumors 14 days later (Supplementary Fig. 2e). At this early time point, hMIC tumors were comparable in size (~50 mm3) between cohorts, although hMIC tissues from the mice with primary tumors had ~62% fewer proliferating tumor cells with no significant differences in numbers of Caspase3-positive cells (Supplementary Fig. 2e-i).
We examined these small secondary tumors for ECAD and the human mesenchymal marker, ZEB126, which is highly expressed in hMICs in vitro (Supplementary Fig. 2b). In the control cohort (no primary tumor), the vast majority of hMICs acquired ECAD expression, with only ~10% of the cells expressing ZEB1 (Fig. 2i-l). In striking contrast, hMICs from the mice bearing primary tumors largely maintained ZEB1 expression (~90%) with only a small fraction of tumor cells acquiring ECAD expression (Fig. 2i-l).
The ZEB1hi phenotype persisted through later end points (Fig. 2m-n). We therefore tested the effects of locking MICs in a mesenchymal state by creating and injecting hMICs that stably express either ZEB1 (ZEB1hi hMICs) or an empty vector control (control hMICs) (Supplementary Fig. 2j). After 6-weeks, the ZEB1hi hMIC tumors were 20-fold smaller than control hMIC tumors and indeed maintained high ZEB1 protein expression (Fig. 2o-p). Hence, maintaining high ZEB1 expression in hMICs, either in the presence of a primary tumor or by ZEB1 over expression, severely compromises their tumor-forming ability.
These data demonstrated that proliferation is mechanistically linked to MIC epithelial plasticity, which was critical for robust tumor growth. Specifically, reduced proliferation in the mesenchymal state accounts for the lack of MIC outgrowth when a distantly located primary tumor is present.
Myeloid cells in the metastatic microenvironment prevent MIC differentiation and colonization
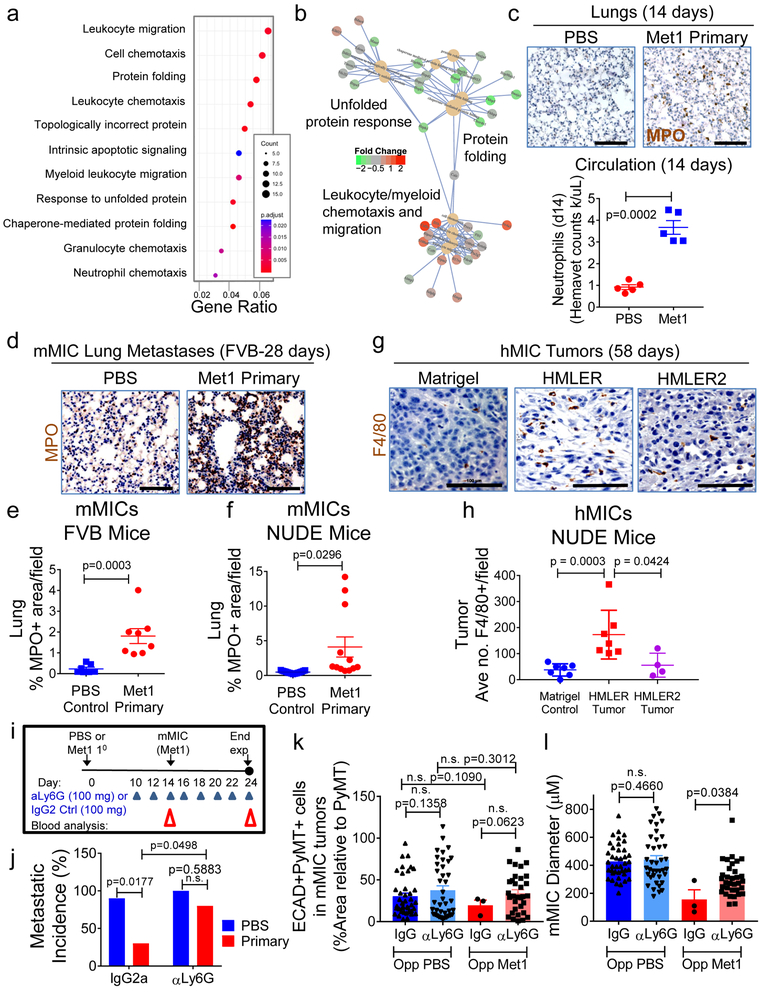
Transcriptomic analysis of lung tissues 14 days after control PBS or Met1 primary tumor initiation – the time point at which mMIC metastases typically encounter the lungs (Supplementary Fig. 3a,b) - revealed a list of significantly differentially expressed genes (DEGs) that clustered by cohort (Supplementary Fig. 3c, d; Supplementary Data Table S2). The MIC-suppressive lung environment was defined by functionally enriched gene ontology (GO) terms and pathways involved in leukocyte (myeloid/neutrophil/granulocyte) migration and chemotaxis and diminished for protein folding responses (Fig. 3a, b).
Figure 3. Myeloid cells are necessary for preventing MIC differentiation and outgrowth.
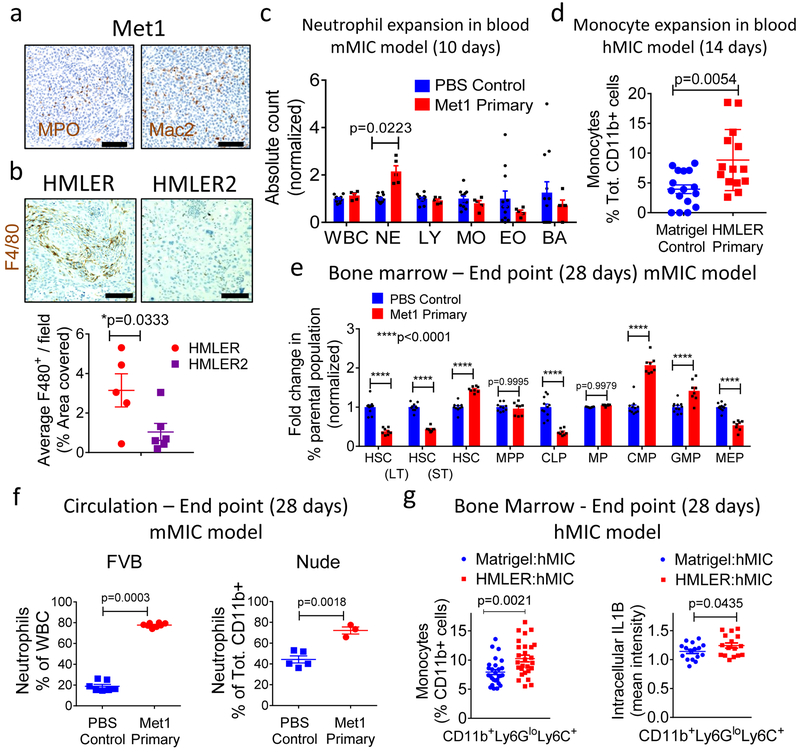
a, Gene ontology (GO) terms found in differentially expressed genes (DEG; DESeq2 adjusted p-value<0.05) between normal or Met 1primary tumour-bearing lungs (n=4 mice/cohort). Y-axis: top 11 enriched (padj<0.01) categories, low (red), high (blue) adjusted p-value; dot size: number of DEGs within GO term; X-axis: ratio of DEGs to total gene number within GO term. b, Connectivity map of top 10 enriched GO terms and DEGs within them. GO term nodes (beige), fold-change (red, up; green, down) between Met1 primary tumor-bearing and control lungs. c, Images: pulmonary neutrophils (myeloperoxidase, MPO; brown) at day 14 from FVB mice bearing Met1 tumor or PBS control. Hematoxylin (nuclei, blue). Graph: circulating neutrophil counts/microliter blood (hemavet). d, Pulmonary neutrophils (MPO; brown) at day 28 from FVB mice bearing PBS control or orthotopic Met1 primary tumors. Hematoxylin (nuclei, blue). e, f, Quantification of pulmonary neutrophils from FVB mice: Control cohort n=21 independent images representing 7 lungs; Met1 primary tumor cohort n=24 independent images representing 8 lungs (e); or Nude mice: Control n=15 independent images representing 5 lungs; Met1 primary cohort n=12 independent images representing 4 lungs (f). g, h Representative immunohistochemistry (g) and corresponding quantification (h) of hMIC tumors grown opposite Matrigel, HMLER, or HMLER2 tumors (per Fig. 1h). Tissues stained with F4/80 (macrophages, brown); hematoxylin (nuclei, blue). Control n=28 independent images representing 7 tumors; HMLER n=24 independent images representing 7 tumors; HMLER2 n=16 independent images representing 4 tumors. (i) Schematic of neutrophil depletion experiments using 100 μg either anti-Ly6G or IgG control. j, Macroscopic pulmonary metastasis incidence (control IgG2a n=4 mice/cohort; anti-Ly6G n=8 mice/cohort). k, ECAD+/PyMT+ staining (% total PyMT+ area) in mMIC lung metastases. l, average mMIC lung metastasis size. mMIC opposite Control PBS: anti-IgG2a n=40 independent images representing 4 mice; anti-Ly6G n=39 independent images representing 4 mice. mMIC opposite Met1 primary tumor: anti-IgG2a n=3 independent images representing 5 mice; anti-Ly6G n=33 independent images representing 4 mice. Scale bars=100 μm. Source data for c, e, f, h, j, k, l in Supplementary Table 1. 1-sided Welch’s t test (c, k); 2sided Welch’s t test (f); 1-sided Mann-Whitney test (h, l); 2-sided Mann-Whitney test (e); 2way- ANOVA with Sidak’s multiple comparison test (j).
In agreement with RNAseq results, neutrophils were abundant in the lungs and ~4-fold higher in the circulation of the tumor-bearing cohort than in the tumor-free cohort (Fig. 3c). Increased neutrophil infiltration persisted throughout disease progression to the 28-day experimental end point (Fig. 3d, e and Supplementary Fig. 3e) and was also apparent in the lungs of Nude mice (Fig. 3f).
Myeloid cells, including macrophages and neutrophils, can confer either pro- or anti-tumorigenic functions that are governed in tissue- and microenvironment-specific contexts10, 11, 13, 27–29. One particular study reported gene expression signatures of breast cancer metastasis-promoting, immune-suppressive circulating neutrophils (“KEP”)13. Analysis of the reported KEP and normal lung neutrophil signatures revealed that the lungs from mice with Met1 primary tumors had a lower KEP:normal ratio (~1.7) compared to the control cohort (ratio of ~2.8) (Supplementary Fig. 3f). Similarly, leukotrienes expressed by pre-metastatic lung neutrophils to expand the MIC pool in a reported study11 were not differentially expressed in the lungs of the tumor-bearing cohort (see GSE111157). These results suggested that the neutrophils in lungs of Met1 tumor-bearing mice are entrained differently than previously described circulating neutrophils.
Similarly, myeloid cells infiltrated the hMIC tumors from mice bearing inhibitory human primary tumors (HMLER) but not those bearing non-inhibitory primary tumors (HMLER2). In these cases, macrophages were ~4.5-fold more abundant in hMIC secondary tumors from mice with primary HMLER tumors than either Matrigel controls or HMLER2 tumor-bearing mice(Fig. 3g, h).
To determine if neutrophils are necessary for inhibiting mMIC lung colonization, we neutralized Ly6G+ cells. We selected an optimal anti-Ly6G dose that restored circulating Ly6G+ cells to that of the control cohorts, did not affect primary tumor growth, and reduced lung neutrophil infiltration (Supplementary Fig. 3g-l). We then orthotopically injected PBS control or Met1 cells and after 10 days, animals were randomized into two additional cohorts to receive either anti-Ly6G or control IgG2a every 2 days; mMIC cells were then injected intravenously into all cohorts at day 14, and the dosing regimens were continued (Fig. 3i).
As expected, pulmonary macrometastases were reduced ~4-fold in the primary tumor-bearing cohort treated with control IgG2a relative to the control primary tumor-free cohort (Fig. 3j). However, when the primary tumor-bearing cohort was treated with anti-Ly6G and circulating neutrophils were reduced to that of the control cohort without affecting primary tumor mass (Supplementary Fig. 3 m, n), metastatic colonization was no longer inhibited (Fig. 3j). Neutrophil ablation was associated with significantly larger pulmonary metastases that displayed ~2-fold more ECAD compared with the control IgG2a cohort (Fig. 3k, l; Supplementary Fig. 3o-q). We confirmed that lung neutrophils were maintained at baseline levels while lung monocytes were unaffected at the experimental end point (Supplementary Fig. 3r).
IL-1β is Sufficient to Prevent MIC Differentiation
We next interrogated candidate DEGs as drivers of MIC suppression. Among the topmost upregulated DEGs in the lungs of mice with primary tumors, we considered pro-inflammatory cytokines common to both neutrophils and macrophages (Supplementary Fig. 4a)13, 30, 31. One of the most highly upregulated DEGs in lungs of primary tumor-bearing mice was IL-1β (Fig. 4a), which is known to drive ZEB1 expression32, 33. IL-1β was significantly more abundant in the lungs of both FVB and Nude mice bearing Met1 primary tumors (Fig. 4b, c) and mMICs did not secrete appreciable levels of IL-1β in culture (Supplementary Fig. 4b). We confirmed that murine Met1 cells and derivative clones, MT2 and MT3, expressed IL-1R and were responsive to IL-1β in a dose-dependent manner (Supplementary Fig. 4c).
Figure 4. Myeloid cell-derived IL-1β; is sufficient to prevent MIC differentiation.
a, Top 10 differentially up-regulated genes (RNA-seq) in lungs from Met1 primary tumor bearing FVB mice relative to control lungs at day 14 (n=4 mice/cohort). b,qPCR on lungs 14 days following tail vein injection of mMICs into FVB mice bearing Met1 primary tumors or PBS control (per Fig. 1a, b). IL-1β; normalized relative to β-actin; (n=3 lungs/cohort, triplicate). c, Flow cytometric analysis of intracellular IL-1β in CD45+ cells from subset of lungs of Nude mice bearing Met1 primary tumors (n=3 animals) or PBS control (n=5 animals). d,qPCR on hMIC tumors at d28 using indicated human- and mouse-specific primers. n=3 tumors/cohort analyzed in triplicate.e, Flow cytometric analysis of intracellular IL-1β in CD11b+F4/80+ cells harvested from hMIC tumors grown opposite Matrigel control (n=8 tumors) or HMLER primary tumors (n=7 tumors); per Supplementary Fig. 2e. f, Schematic showing single injection xenograft model; applies to (g-i). g, Merged immunofluorescence images of hMIC tumors injected in Matrigel containing PBS (hMIC+PBS) or 10pg/ml IL-1β (hMIC + IL-1β) representing 2 independent experiments. Block arrows: examples of ZEB1+ tumor cells; long arrows: examples of ZEB1-tumour cells; arrowheads: ZEB1+ stromal cells). h, ECAD+LgT+ cells (hMIC+PBS n=10 independent images representing 7 tumors; hMIC+IL-1β n=9 independent images representing 6 tumors) or ZEB1+LgT+ cells (hMIC+PBS n=15 independent images representing 5 tumors; hMIC+IL-1β n=21 independent images representing 7 tumors) as % total number LgT+ tumor cells/microscopic field. i,Representative tumors stained with Mac2 (macrophages, brown) or hematoxylin (nuclei, blue), representing 2 independent experiments. All scale bars = 100 μm. Source data for b, c, d, e, h provided in Supplementary Table 1. 1-sided t test (b); 2-sided Mann- Whitney (c, h); 2-sided Welch’s t test (e); 2-way ANOVA followed by Sidak’s multiple comparison test (d).
We also assessed IL-1 expression in hMIC tumors from HMLER- and Matrigel-bearing mice by species-specific qPCR. Human IL-1B and IL-1A expression were not different between cohorts (Fig. 4d). In fact, hMICs secrete very low levels of IL-1β (<1pg/ml) in culture (Supplementary Fig. 4d). Relative to Matrigel control mice however, murine IL-1 β expression was 2.7-fold elevated in hMIC tumors from mice bearing primary tumors; this was accompanied by a 3.4-fold increase in human IL-1R1 expression (Fig. 4d). Indeed, murine IL-1β can efficiently bind and activate human IL-1R134 and IL-1β induces expression of IL-1R1, thereby amplifying IL-1 signaling35, 36.
Tumor-associated macrophages (CD11b+F4/80+) equivalently infiltrated hMIC tumors from both cohorts at the early time point (Supplementary Fig. 4e). However, macrophages expressed significantly higher levels of intracellular IL-1β protein per cell in the cohort with primary tumors than those from the control cohort (Fig. 4e).
We directly tested effects of IL-1β on MIC plasticity by admixing hMICs with Matrigel containing either IL-1β (hMIC+IL-1β) or PBS control (hMIC+PBS) (Fig. 4f). After 2 weeks, the hMIC+ IL-1β tumors had significantly fewer ECAD+ tumor cells and more ZEB1+ tumor cells than the hMIC+PBS tumors (Fig. 4g-h). Importantly, hMIC+IL-1β tumors displayed significantly enhanced macrophage infiltration (Fig. 4i), demonstrating that a single dose was sufficient to trigger a sustained inflammatory response and maintain the mesenchymal phenotype.
IL-1R1 Signaling is Necessary for Preventing MIC Differentiation
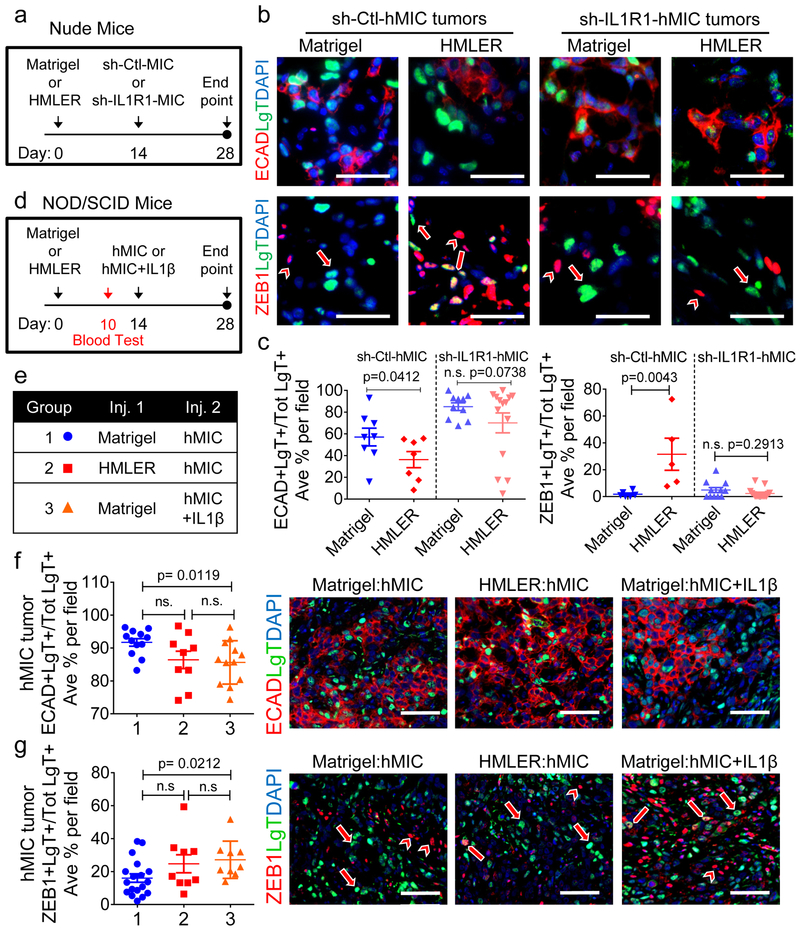
To test whether IL-1R1 signaling is necessary for preventing MIC differentiation, we first generated hMIC-IL-1R1 deficient cells (sh-IL-1R1-hMIC) and scrambled shRNA control cells (sh-Ctl-hMIC). Only 1 of 6 shRNA constructs provided sufficient suppression of IL-1R1 without significantly impacting cell proliferation (Supplementary Fig. 5a, b); hence, we performed all in vivo experiments with shA-IL-1R1-MIC cells only. Cohorts of mice bearing primary HMLER tumors or Matrigel control were injected with sh-IL-1R1-MIC or sh-Ctl-MIC cells and tissues were harvested 2 weeks later (Fig. 5a). hMIC tissue mass was not significantly different between cohorts at this time point (Supplementary Fig. 5c).
Figure 5. IL-1R1 signaling is necessary for preventing MIC differentiation.
a, Experimental schematic: hMICs with or without IL-1R1 suppression (sh-IL-1R1-hMIC and sh-Ctl-hMIC) injected opposite Matrigel control or HMLER primary tumor in Nude mice; applies to (b-c).b, Immunofluorescence images of hMIC tumors. Long arrows: examples of ZEB1-negative tumor cells; block arrows: ZEB1+ tumor cells; arrowheads: ZEB1+ stromal cells. Scale bar=50 μm. sh-Ctl-hMIC opposite Matrigel control, n=7 tumors; opposite HMLER primary tumor, n=8 tumors. Sh-IL-1R1-hMIC opposite Matrigel control and HMLER primary tumor, n=8 tumors/cohort. c, Quantification of ECAD+LgT+ cells (sh-Ctl-hMIC: Matrigel n=8 and HMLER n=7 independent images representing 3 tumors/cohort, sh-IL-1R1-hMIC: Matrigel n=10 and HMLER n=14 independent images representing 5 tumors/cohort) or ZEB1+LgT+ cells (sh-Ctl-hMIC: Matrigel n=6 and HMLER n=5 independent images representing 3 tumors/cohort, sh-IL-1R1-hMIC: Matrigel n=11 and HMLER n=15 independent images representing 5 tumors/cohort) as % of total number of LgT+ human tumor cells /microscopic field. d, e, Experimental schematic modeling early stages of hMIC secondary tumor formation in NOD/SCID mice with HMLER primary tumors or PBS control; secondary injections involve hMICs or hMICs+10pg/ml rIL-1β. (d); Indicated cohorts: Group 1 (n=7 animals); Group 2 (n=7 animals); Group 3 (n=6 animals) (e). f, g,Images: immunofluorescence of hMIC tumors; arrows: ZEB1-negative tumor cells; block arrows: ZEB1+ tumor cells; arrowheads: ZEB1+ stromal cells. Graphs: ECAD+LgT+ cells in hMIC tumors (4 tumors/cohort) opposite Matrigel control (n=12 independent images), HMLER (n = 9 independent images) or hMIC+IL-1β tumors opposite Matrigel (n=12 independent images) (f); or ZEB1+LgT+ cells in hMIC tumors (3 tumors/cohort) opposite Matrigel (n=18 independent images), HMLER (n=9 independent images), or hMIC+IL-1β tumors opposite Matrigel (n=9 independent images) (g). Positive staining represented as % of total number of LgT+ human tumor cells /microscopic field; scale bars=100 μm. Source data for c, f, g provided in Supplementary Table 1. 1-sided Welch’s t test (c -left); 2-sided Mann-Whitney test (c-right); 2-sided Welch’s t test (f, g).
Echoing earlier results, the sh-Ctl-hMIC tumors from mice with primary tumors had significantly fewer ECAD+ cells and more ZEB1+ cells than those from the Matrigel control cohort (Fig. 5b-c). In stark contrast, the sh-IL-1R1-hMIC tumors from both cohorts expressed similar levels of ECAD and ZEB1 and the majority of tumor cells from both cohorts were in ZEB1loECADhi state (Fig. 5b-c).
We also treated mMICs in a 3D tumorsphere assay with IL-1β, anakinra (IL-1R1 antagonist), combination IL-1β+anakinra, or vehicle control. IL-1β treatment activated NF-κB signaling and significantly reduced tumorsphere size whereas anakinra reduced NF-κB activation and increased both tumorsphere size and ECAD protein levels (Supplementary Fig. 5d, e).
In addition to dysfunctional adaptive immunity, NOD/SCID mice have an impaired innate immune system, of which defects in IL-1 signaling are a particular feature37. Hence, we injected cohorts of NOD/SCID mice with either Matrigel or HMLER primary tumors. After 10 days, circulating monocytes were not significantly different between cohorts (Supplementary Fig. 5f). At day 14, we then injected either hMICs or hMICs with rIL-1β (hMICs+IL-1β) as secondary tumors and harvested tissues of equivalent mass after an additional two weeks (Fig. 5d, e and Supplementary Fig. 5g).
The majority of hMIC tumor cells in the control NOD/SCID mice were ECADhiZEB1lo (Fig. 5f, g). However, unlike Nude mice, HMLER primary tumors failed to lock distant hMICs in the ZEB1+ state and these hMIC tumor cells were also ECADhiZEB1lo (Fig. 5f, g). Therefore, ablation of IL-1β-dependent aspects of innate immunity prevented MIC entrapment in the mesenchymal-enriched state, even in the presence of a distant primary tumor. Finally, addition of rIL-1β maintained hMICs in a ZEB1+ECADlo state (Fig. 5f, g), indicating that hMICs were still capable of responding to IL-1β in the NOD/SCID mice.
These observations indicated that the ability of primary tumors to systemically maintain secondary hMIC tumors in the ZEB1+ECADlo state critically depended on eliciting systemic inflammation involving IL-1β-secreting innate immune cells and IL-1R pathway activation in the disseminated MICs.
MIC-inhibitory primary tumors elicit a systemic inflammatory response
Our results thus far suggested that MIC-inhibitory primary tumors elicit a systemic pro-inflammatory response. Indeed, myeloid cells infiltrated the Met1 and HMLER primary tumors but not the HMLER2 primary tumors that did not suppress distant hMIC outgrowth (Fig. 6a, b; Supplementary Fig. 6a). Circulating neutrophils were significantly increased 10 days after disease initiation (i.e., prior to mMIC implantation) in the Met1 primary tumor-bearing FVB cohort and there was a 10% increase in circulating monocytes in HMLER tumor-bearing Nude mice after just 14 days (Fig. 6c, d).
Figure 6. Primary tumors initiate a sustained systemic inflammatory response.
a-b, Met1 primary tumors (Fig. 1a schematic) stained for myeloperoxidase (MPO; brown) to detect neutrophils, Mac2 to detect macrophages (Mac2; brown); hematoxylin (nuclei, blue) (a). HMLER and HMLER2 tumors (Fig. 1h schematic) stained for F4/80 (macrophages, brown) and Hematoxylin (nuclei, blue); quantified in adjacent graph (HMLER n=5 tumors; HMLER2 n=6 tumors) (b). Scale bars=100 μm. c, Absolute blood counts (hemavet) in circulation (FVB mice) 10 days after orthotopic injection of Met1 primary tumors (n=5 animals) or PBS control (n=12 animals). WBC, Whole blood cells; NE, neutrophils; LY, lymphocytes; MO, monocytes; EO, eosinophils; BA, basophils. d, Flow cytometric analysis of circulating monocytes (CD11b+Ly6ChiLy6Glo), Nude mice, 14 days after injection of Matrigel control (n=16 independent images representing 8 tumors) or HMLER primary tumors (n=14 independent images representing 7 tumors), representing 2 independent experiments. e, Fold changes (% parental population) in bone marrow of FVB mice bearing orthotopic Met1 primary tumors (n=8 animals) or PBS control (n=10 annimals) after 28 days. HSC, hematopoietic stem cells; LT-HSC, long-term hematopoietic stem cells; ST-HSC, short term hematopoietic stem cells; MPP, multipotent progenitors; CLP, common lymphoid progenitors; MP, myeloid progenitors, CMP, common myeloid progenitors; GMP, granulocyte-monocyte progenitors; MEP, megakaryocyte/erythroid progenitors. f, Quantification of circulating neutrophils (CD11b+Ly6G+Ly6Clo) at 4-week experimental end points from indicated strains of mice bearing mMIC lung metastases with concurrent orthotopic Met1 primary tumors or PBS control. (FVB mice: Control n=7 animals; Met1 primary n=8 animals); Nude mice: Control n=5 animals; Met1 primary n=3 animals). Representative of 3 (FVB) and 1 (Nude) experiments. g, Monocytes (CD11b+Ly6ChiLy6Glo, n=27 samples/cohort representing 11 tumors, 3 independent experiments) as % total CD11b+ cells in the bone marrow of Nude mice bearing hMIC tumors grown opposite Matrigel control or HMLER primary tumors after 28 days (left). Intracellular IL-1β in bone marrow monocytes (n=16 samples/cohort, representing 8 tumors, 2 independent experiments) (right). Source data for b, c, d, e, f, g provided in Supplementary Table 1. 1-sided Welch’s t test (b, g-right); 2-sided Welch’s t test (d, f-right, g -left); 2-way ANOVA Tukey’s multiple comparison test (c, e); 2-sided Mann-Whitney test (f-left).
In the bone marrow of FVB mice, short-term hematopoietic stem cells (ST-HSCs), common myeloid progenitors (CMP) and granulocyte-monocyte progenitors (GMP) expanded 28 days after Met 1 tumor establishment while production of hematopoietic stem cells (HSC), long-term HSC (HSC LT), common lymphoid progenitors (CLP) and megakaryocyte/erythroid progenitors (MEP) decreased (Fig. 6e). The shift toward CMPs and GMPs reflected a skewing toward the production of neutrophil and macrophage precursors. Circulating neutrophils were still significantly elevated by end-stage in both FVB and Nude mice (Fig. 6f). Similarly, bone marrow monocytes and their expression of intracellular IL-1β were significantly elevated at end-stage of HMLER primary disease (Fig. 6g). These results were indicative of a sustained pro-inflammatory response throughout disease progression.
Inhibiting inflammation at primary tumor site results in distant MIC differentiation and growth
Our results suggested that inhibiting inflammation at the primary tumor site should impact the systemic cascade of events that resulted in MIC suppression at distant sites. Therefore, we analyzed the primary tumors for pro-inflammatory factors that we could interrogate.
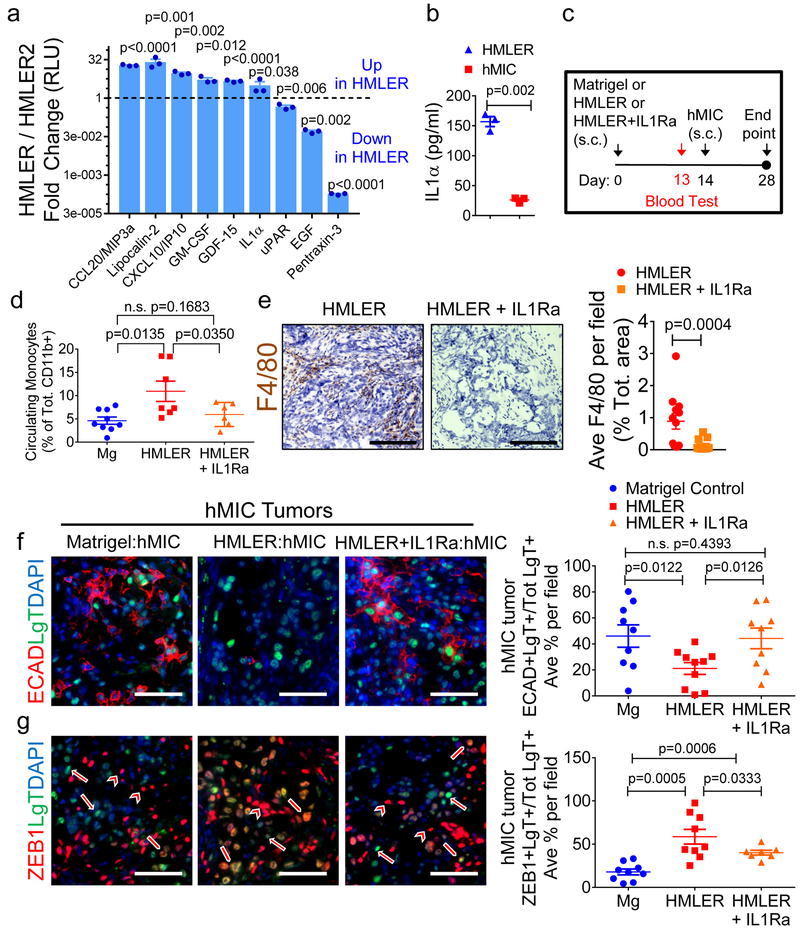
Expression analysis of Met1 primary tumors relative to control tissues (GSE111157) showed enhancement of genes terms related to inflammation and neutrophil recruitment; some of these included IL-1β, LCN2, G-CSF, CCL2, and TNFα (Supplementary Fig. 7a). Cytokine analysis also revealed a number of pro-inflammatory cytokines that were secreted at significantly higher levels (≥3-fold) by the HMLER cells than the non-inhibitory HMLER2 cells, including IL-1α and LCN2 (Fig. 7a). A number of these same cytokines, including IL-1α, LCN2, and G-CSF, were secreted at significantly higher levels (≥2-fold) by the HMLER cells than hMICs (Supplementary Fig. 7b).
Figure 7. Inhibiting inflammation at primary tumor site results in differentiation of disseminated MICs.
a, Inflammatory cytokine array analysis of conditioned media from HMLER and the non-inhibitory primary tumor, HMLER2 cells, 3 days in culture (n=2 biological replicates, 3 technical replicates each). Results are fold change (Log2) HMLER/HMLER2. b, ELISA for IL-1α in conditioned media from HMLER and hMIC cells, 3 days in culture (n=3 biological replicates, 5 technical replicates each). c, Experimental schematic modeling early stages of hMIC colonization (Nude mice) with Matrigel control, HMLER primary tumor, or HMLER primary tumor + IL-1Ra. d, Flow cytometric analysis of CD11b+Ly6C+Ly6Glo circulating monocytes (day 13), prior to contralateral injection of hMIC cells, in mice bearing Matrigel control (n=9 animals), HMLER tumors (n=7 animals), or HMLER+IL-1Ra (100ng/ml) tumors (n=6 animals).e, Images: HMLER and HMLER+IL-1Ra tumors stained with F4/80 (macrophage, brown) and hematoxylin (nuclei, blue). Scale bars=100 μm. Graph: area positively stained for F4/80+ macrophages in HMLER and HMLER+IL-1Ra tumors (n=12 independent images representing 4 tumors/cohort). f, g, Images: Indicated immunohistochemical staining of various hMIC tumors. Arrows: ZEB1-negative tumor cells; block arrows: ZEB1+ tumor cells; arrowheads: ZEB1+ stromal cells; scale bars=100 μm. Graphs: Quantification of ECAD+LgT+ cells in hMIC tumors opposite Matrigel (n=9 independent images), HMLER (n=10 independent images), HMLER+IL-1Ra (n=9 independent images), 3 tumors/cohort (f) or ZEB1+LgT+ cells in hMICs tumors opposite Matrigel (n=9 independent mages), HMLER (n=9 independent images), HMLER+IL-1R (n=7 independent images) 3 tumors/cohort) (g) as % of total number of LgT+ human tumor cells/microscopic field. Source data for a, b, d, e, f, g provided in Supplementary Table 1. Multiple 2-sided t tests (a); 2-sided Welch’s t test (b); 1-sided Welch’s t test (d, f, g); 2-sided Mann-Whitney test (e).
Our earlier results (Figs. 3-5) established that neutrophil neutralization or IL-1R1 suppression within the metastatic niche affects MIC differentiation and colonization; therefore, therapeutic approaches designed to systemically inhibit inflammation would not reveal the necessity or exclusivity of primary tumors in initiating the MIC-inhibitory cascade. Therefore, we sought strategies to inhibit inflammation proximal to the primary tumor. Among the various pro-inflammatory cytokines secreted by primary tumors, IL-1α stood out due to its prominence as a local initiator of systemic inflammatory responses33. Importantly, IL-1R activation triggers induction of some of the pro-inflammatory cytokines that we had observed, including TNFα38, lipocalin 2 (LCN2)39, and CCL240, 41. Indeed, IL-1α was >6-fold more abundant in conditioned medium from HMLERs than hMICs (Fig. 7b).
To determine if primary tumor IL-1R1 signaling initiates the pro-inflammatory cascade, we used a recombinant IL-1 receptor antagonist (IL-1Ra)42. We implanted Matrigel control, HMLER cells, or HMLER cells mixed with IL-1Ra (HMLER+IL-1Ra) into mice and collected blood 13 days later (Fig. 7c). At this time point, circulating monocytes were 2.4-fold elevated in primary tumor-bearing cohort relative to the Matrigel controls; however, in the cohort bearing HMLER+IL-1Ra tumors, circulating monocytes were reduced to that of the control cohort (Fig. 7d). Murine inflammatory plasma cytokines that are commonly triggered by IL-1R1 signaling (e.g., TNFα, G-CSF, and CCL1) were also elevated in mice with HMLER primary tumors but not in the cohort bearing HMLER+IL-1Ra tumors (Supplementary Fig. 7c).
Having confirmed an IL-1-dependent host inflammatory response at day 13, we initiated hMIC secondary tumors in all 3 cohorts the following day (day 14) and continued the experiment for another two weeks (Fig. 7c). Primary tumor masses were equivalent, yet myeloid infiltration into primary HMLER+IL-1Ra tumors was reduced 6.4-fold compared to HMLER controls (Fig. 7e and Supplementary Fig. 7d-g). Macrophages were also less abundant in the secondary hMIC tumors from mice bearing the distant HMLER+IL-1Ra primary tumors (Supplementary Fig. 7h). Confirming earlier findings, hMICs from mice bearing primary tumors were predominantly ZEB1+ECADlo; in contrast, when IL-1R1 signaling was inhibited at the primary tumor site, hMIC secondary tumors acquired the ZEB1loECADhi epithelial phenotype (Fig. 7f-g, and Supplementary Fig. 7i, j).
Low IL-1β expression in primary breast cancer correlates with reduced metastasis free survival
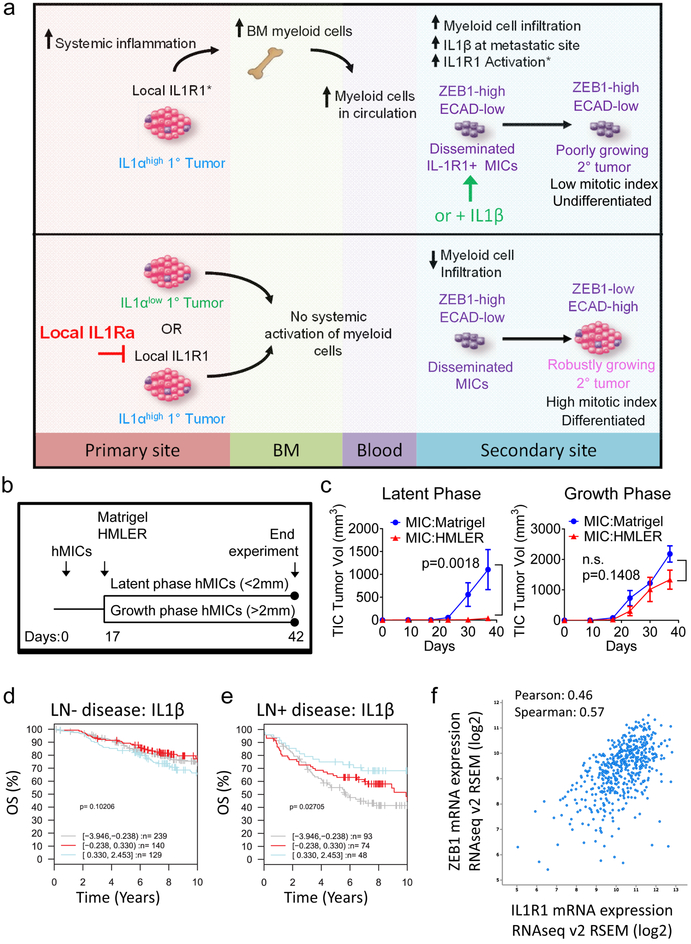
Our studies demonstrated that innate immune cells secreting IL-1β, mobilized by the primary tumor, compromise MIC colonization at secondary sites by preventing their differentiation into epithelial progeny, which is essential for forming actively growing tumors (Fig. 8a). Given that this cascade of events depends on the continued presence of the primary tumor, clinical validation relied on careful selection of appropriate patient populations. Indeed, hMIC-derived metastases were not inhibited if the IL-1β; dependent inflammatory cascade was instigated after MIC dissemination and growth initiation (Fig. 8b-c). hMIC tumors that were <2mm (low mitotic index) at the time of HMLER implantation were significantly suppressed; however, if hMIC tumors had already entered an active growth phase (>2mm) at the time of primary tumor implantation, MIC-derived tumors sustained continued growth (Fig. 8c). These data provided preliminary indication that HMLER tumors do not cause regression of robustly growing hMIC tumors but instead, exert their inhibitory effects at early stages of secondary tumor establishment when MICs are still in the ZEB1+ state.
Figure 8. Low primary tumor IL-1β correlates with reduced overall survival in breast cancer.
a, Model illustrating systemic mechanism by which primary tumors elicit an IL-1β-dependent inflammatory response to suppress MIC colonization. Top panel: Primary tumors that secrete high levels of pro-inflammatory cytokines, e.g., IL-1α, elicit a systemic innate immune response that expands bone marrow and circulating myeloid cells, culminating in increased immune infiltrate into tissues where metastasis-initiating cells (MICs) disseminate. In the metastatic microenvironment, IL-1β acts in a paracrine fashion on IL-1R expressing MICs, causing the MICs to maintain their mesenchymal phenotype of high ZEB1 and/or low E-cadherin (ECAD), thereby preventing MIC differentiation and proliferation. Bottom panel: Preventing inflammation at the primary tumor site or inhibiting IL-1R1 with the antagonist, IL-1Ra, in the metastatic microenvironment causes MICs to differentiate, proliferate and thereby establish robustly growing secondary tumors and metastases. b, Schematic of experiments to test effect of primary HMLER tumors on established hMIC tumors in Nude mice. c, Growth kinetics of hMIC tumors that were either in latent phase (left) or growth phase (right) at day 17 when Matrigel control or HMLER tumor cells were injected contralaterally. (Latent phase: Matrigel n=6 tumors, HMLER n=5 tumors. Growth phase: Matrigel n=4 tumors, HMLER n=5 tumors). One experiment was performed. 2way ANOVA and Sidak’s multiple comparisons test. d, e, Kaplan-Meier analysis using overall survival (OS) as end point with 10-year censoring, based on IL1B gene expression (log2, stratified into three indicated quantiles) in primary tumor tissues from patients with lymph node negative (LN−; n=508 patients) (d)or lymph node positive (LN+; n=215 patients) (e) disease. Logrank p values are shown. Data and analysis obtained from the GOBO database (http://co.bmc.lu.se/gobo/gsa.pl). f,Correlation between IL-1R1 and Zeb1 mRNA expression in 818 tumor tissue samples from patients with invasive breast carcinoma. Data and analysis obtained from the cBioPortal database (http://www.cbioportal.org/index.do).
We therefore compared primary tumor IL-1β expression in breast cancer patients with lymph node-positive (LN+) and LN-negative disease by retrospective gene set analyses using a database of Affymetrix microarray profiles43. Among 508 patients with LN- disease, IL-1β expression did not stratify overall survival (Fig. 8d). However, among 215 patients with LN+ breast cancer, those with high IL-1β had improved overall survival relative to those with low IL-1β expression (Fig. 8e). Interestingly, patients whose primary tumors expressed high IL-1β had improved outcome (distant metastasis-free survival) when we interrogated the entire cohort of 1,379 patients (Supplementary Fig. 8a).
We also analyzed correlations between IL-1R1 and markers of differentiation status that we had observed. In an analysis of 818 tumor tissue samples from patients with invasive breast carcinoma, IL-1R1 expression positively correlated with Zeb1 (Fig. 8f)44-46.
DISCUSSION
The present work demonstrates that MIC plasticity determines metastatic success and agrees with a clinical report that mesenchymal markers are down-regulated in metastases relative to matched primary tumors47. An important implication of our study is that therapies designed to prevent disseminated MIC differentiation compromise their ability to form lethal metastases. Another distinction of our work is that MICs are specifically susceptible to growth inhibition. Interestingly, a recent report indicates that breast carcinomas enriched for mesenchymal markers, similar to our MICs, give rise to immunosuppressive tumors, unlike their more epithelial counterparts48. However, we did not specifically examine hallmarks of immunosuppression, as the MIC-suppressive cascade occurred in a T cell-independent manner.
It is becoming increasingly clear that modulating innate immunity must be included in efforts to improve patient outcome13, 27, 49-51. Tumor-associated lung neutrophils that suppressed MIC colonization in our study appear to be entrained differently than metastasis-promoting neutrophils that have been reported in the circulation13 and pre-metastatic lungs11 in other models. At first glance, the neutrophils in our study may seem anti-metastatic (they prevented MIC colonization) but at the same time, they may also be considered pro-metastatic (they fortified the MIC potential to generate lethal metastases and did not limit primary tumor growth).
Inflammatory processes that initiate primary disease and drive EMT in primary breast cancers, thus causing MICs to disseminate, are not necessarily productive for MIC colonization. For example, IL-1β aids growth of some primary tumors 33, 52 and facilitates invasion and extravasation in early stages of metastasis53, findings that are consistent with the idea that IL-1β promotes the EMT17. However, by specifically examining the role of IL-1β after MIC dissemination, we learned that sustained IL-1β-mediated inflammation or MIC IL-1R signaling prevents colonization and must be shut down for secondary tumor formation. We thus consider IL-1β both dissemination-supportive and colonization-suppressive. These findings are consistent with a recent report that IL-1R inhibition, in combination with paclitaxel, moderately reduced primary breast tumor growth but significantly increased metastasis54. Identifying the appropriate setting to inhibit inflammation is also necessary. For example, a recent study demonstrated that neoadjuvant inhibition of inflammation (achieved by CCL2 blockade) resulted in significantly enhanced mammary carcinoma lung metastasis55.
Such spatial and temporal considerations seem crucial as phase I clinical trials using IL-1R blockade for metastatic disease are being initiated, predominantly supported by pre-clinical studies demonstrating primary tumor inhibition56. Our findings indicate that clinical success of IL-1R inhibition rests upon understanding its role at various stages of disease progression. Hence, IL-1R inhibition therapy may not always confer beneficial effects and further research is required in order to identify appropriate contexts for administering such therapy.
The evolving paradigm of systemic instigation57-61 or inhibition of breast cancer metastasis suggests new directions from which to investigate the interactions between primary tumors and systemic environment during metastatic progression. The fact that inflammatory hallmarks resolved primary tumors that inhibited secondary disease from those that did not, suggests that using primary tumor tissue to better predict metastatic behavior may enable more accurate identification of patients with a high likelihood of relapse. It is therefore reasonable to think that a primary tumor expressing pro-inflammatory cytokines resulting in activation of an IL-1β-dependent innate immune response in the metastatic niche might keep secondary disease at bay and conversely, that primary tumor removal might prompt recurrence. Such concepts are underscored by our clinical finding that among breast cancer patients with LN+ disease, those whose primary tumor expressed high IL-1β had improved outcome relative to those with low IL-1β. Moreover, IL-1R1 associates with expression of mesenchymal factors in our study and a report on patient circulating tumor cells62. The implications of our findings for other cancers26 and patients whose disease mimics the biology we have discovered here remains to be determined.
Supplementary Material
Supplementary Figure 1. Primary tumors inhibit outgrowth of distant metastasis-initiating cells
a, Met1 orthotopic primary tumor growth kinetics, FVB mice, represented in Fig. 1b (n=10 animals). b, Schematic of sequential subcutaneous experimental model (left) and growth kinetics (right) of Met1 secondary tumors (2.5 × 105/mouse; implanted at day 14) in mice with PBS control or Met1 primary tumor (2.5 × 105/mouse; n=5 animals/cohort). c, Numbers of pulmonary macrometastases following tail vein injection of MT2 or MT3 cells (7.5×105cells; n=3 animals). Macrometastases >100 microns were quantified on microscope tissue sections from 4 lung lobes per mouse. d, Met1 orthotopic tumor growth kinetics, Nude mice, represented in Fig. 1f (n=5 animals). e, Schematic of experimental model (left), and number of pulmonary macrometastases following tail vein injection of 2.5×105 hMICs into Nude mice with either Matrigel (n = 10 animals) or HMLER primary tumors (n=9 animals; original injection of 5.0×105 cells/mouse) (right). Macrometastases (>100 microns) or micrometastases (>5 cells or <5 cells) were quantified from microscopic whole lung tissue sections. f, Schematic of experimental model (applies to g and h). g, Growth kinetics of HMLER primary tumors, Nude mice, described in Figure 1h (n=10 animals). h, MIC-231 tumor growth kinetics, Nude mice, opposite Matrigel control (n=12 animals) or HMLER primary tumors (n=5 animals). Representative of 2 experiments. i, Images: representative immunofluorescent images of 231-MIC tumors grown opposite Matrigel control or an HMLER primary tumor (represented in Supplementary Fig. 1h) stained with Ki67 (red), hMIT to identify human mitochondria (green), DAPI (nuclei, blue); Scale bars=100 μm. Graph: Quantification of Ki67+hMit+ cells as a percentage of the total number of hMit+ tumor cells/microscopic field (n=9 independent images representing 3 tumors/cohort). Source data for a, b, c, d, e, g, h, i in Supplementary Table 1. 2-way ANOVA, followed by Sidak’s multiple comparison test (b, g, h); 1-sided Welch’s t test (e); 2-sided Welch’s t test (c, i).
Supplementary Figure 2. MIC Differentiation is Perturbed by the Presence of a Primary Tumor
a In vitro immunocytochemical flourescence showing E-cadherin (ECAD, red) and DAPI (nuclei, blue) in Met1 parental cell line (mMIC) and Met1-derived clones, MT2 and MT3 (mMIC-MT3). b Images: Immunofluorescence showing ZEB1 and ECAD expression in cultured hMICs prior to xenotransplantation. Western blot: mesenchymal marker Vimentin (VIM) and epithelial marker ECAD protein in polyclonal HMLER cells and derivative hMIC and HMLER2 cells. GADPH shown as internal control. Positive controls: Ctrl E (epithelial-MCF7Ras); Ctrl M (mesenchymal CD44hi HMLER cells). c, Merged immunofluorescent images of mMIC-MT3 tumors (described in Fig. 1d) stained for basal cytokeratin 14 (CK14, red), luminal CK8 (green) or PyMT antigen (expressed by tumor cells only-green). Arrows - CK14+ tumor cells. d, Images: hMIC tumors (from Fig 1i) stained with CK14 (red), VIM (green) and DAPI (blue); Graph: quantification of indicated stains on hMIC tumors grown opposite Matrigel (n=4 tumors) or primary tumor (n=5 tumors). e, Schematic: modeling early stages of hMIC colonization. Graph: hMIC tumor growth kinetics opposite Matrigel control or HMLER primary tumor (n=4 tumors/group); differences not statistically significant. f, g, Immunofluorescent images (f) and quantification (g) of hMIC tumors stained for ki67 (red), LgT antigen (tumor cells, green), and DAPI (nuclei, blue) as a percentage of total LgT+ cells. Control, n=10 independent images representing 4 tumors; HMLER cohort, n=9 independent images representing 4 tumors. h, i, Immunofluorescent images (h) and quantification (i) of staining hMIC tumors for cleaved caspase3 (CASP3, red), human-specific mitochondria (hMIT, green), and DAPI (nuclei, blue) grown in mice with Matrigel control (n=6 independent images representing 4 tumors) or HMLER primary tumors (n=5 independent images representing 4 tumors). j, Expression of ZEB1 (ZEB1-GFP construct) or HRAS (HRAS-tomato construct) analyzed by FACS (1.0×105 cells) in Control hMIC or ZEB1hi hMIC (from Fig. 2n-p). All scale bars=100 μm. Source data for d, e, g, i in Supplementary Table 1 and d on Supplementary Figure 9. 2-way ANOVA (e); 2-sided Welch’s t test (d, i); 2-sided Mann-Whitney test (g).
Supplementary Figure 3. Innate Inflammatory Cells are Necessary for MIC Colonization
a, Experimental schematic for RNA-seq tissue analysis (Fig. 3a-c and Supplementary Fig. 3b-e). b, Met1 primary tumor mass in FVB mice (n=5 animals). c, d, RNA-seq analysis on lungs from mice with PBS control (n=4 animals) or a Met1 primary tumor (n=4 animals). Heatmap (c): top 50 differentially expressed genes (adjusted p-value, DESeq2). Blue=low, green=mean, and yellow=high relative expression levels. PBS control lungs (yellow), Met1 primary tumor-bearing lungs (purple). Volcano plot (d): DESeq2 comparison Single gene with Padj<0.05 and absolute log2(FoldChange)>1 (green). e, Experimental schematic and flow cytometric quantification of immune cell populations in lungs of indicated FVB mice at 28-day end point (see Fig. 1a). f, Ratio of genes expressed by pro-metastatic immunosuppressive neutrophils from K14cre;Cdh1F/F;Trp53F/F (KEP) mice to control neutrophils from wild type littermates (KEP:Normal)13 extrapolated onto our signatures from control (blue) primary tumor-bearing lungs (red). Higher ratios indicate higher pro-metastatic KEP signature. Box plot: median, 25th and 75th percentiles, whiskers extend to minimum and maximum values. g, Experimental design to identify optimal anti-Ly6G dose for neutrophil depletion. h, Primary tumor mass in Control anti-IgG2a (n=3 mice/cohort) and anti-Ly6G (n=4 mice/cohort). i, Flow cytometric gating strategy for neutrophils (CD45+CD11b+Gr1+Ly6Clo) and monocytes (CD45+CD11b+Gr1+Ly6Chi). j-l, Flow cytometric analysis of blood at 14d (j), 20d (k), and lungs 20d (l) in experiment described in (g). Control anti-IgG2a n=3 mice/cohort; anti-Ly6G n=4 mice/cohort. m, Schematic modeling neutrophil depletion experiment (n-r). n, Primary tumor mass from Fig. 3i (control IgG2a n=4 mice/cohort; anti-Ly6G n=8 mice/cohort). o, Immunofluorescent images of Met1 lung metastasis. Scale bars=200 μm. p, Metastases at higher magnification (indicated in circles in (o) analyzed by ImageJ to measure shortest-length diameter). q, Single channel ECAD or PyMT staining measured by ImageJ analysis (Fig. 3k, l). r, End point flow cytometric analysis of neutrophils and monocytes in lungs of mice treated with control IgG2a (n=4 mice/cohort) or anti-Ly6G (n=8 mice/cohort). Source data for b, e, h, j, k, l, n, r in Supplementary Table 1. DESeq2’s Wald test two-sided (c), 2-sided Welch’s t-test (f); 2-sided Welch’s t test (n); 2-way ANOVA followed by Sidak’s multiple comparison test (f, h, j, k, l, o).
Supplementary Figure 4. Innate Inflammatory Response is sufficient for MIC colonization
a, Some of the predominant cytokines expressed by monocytes, neutrophils or by both. b, ELISA to detect cumulative IL-1β in the conditioned media of Met1 cell line and its derived clones after 3 days in vitro (n=5 biological replicates/group). c, Met1 parental cells and the MT2 and MT3 subclones stained as indicated (upper panel). Scale bars= 100 μm. Western blot detecting phospho-p65 (p-p65; activated form of NF-kB p65) or total NF-kB p65 (p65) in Met1, MT2 and MT3 treated in vitro with indicated doses of IL-1β at indicated time points (lower panel). d, ELISA to detect cumulative IL-1β in the conditioned media of HMLER and hMIC cell lines over 3 consecutive days of in vitro culture (n=4 biological replicates/group). e, Experimental schematic and flow cytometric quantification of immune cell populations in digested hMIC tumors from indicated cohorts in Nude mice at the 28-day experimental end point (see Fig. 2i) (n=3 hMIC tumors in Matrigel control and 4 hMIC tumors in HMLER cohort). Source data for b, d, e in Supplementary Table 1. 2-sided Welch’s t test (b, e); Two-way ANOVA, Sidak’s multiple comparisons test (d).
Supplementary Figure 5. IL-1R1 Pathway is sufficient to block MICs colonization
a, qRT-PCR to determine IL-1R1 mRNA levels in hMIC expressing shRNA against IL-1R1 (shA-IL-1R1-MIC and shB-IL-1R1-MIC) compared to hMIC expressing a scrambled shRNA (sh-Ctl-MIC). B-actin (ACTB) served as the internal control (n=3 PCR technical triplicates). b, Proliferation analysis (CyQuant assay) comparing growth of MIC-control cells to those expressing shRNA against IL-1R1 (shA-IL-1R1-MIC and shB-IL-1R1-MIC; n=3 in triplicate. One experiment, representative of 5 independent experiments). c, hMIC tumor mass (mg) at the 4-week experimental end point (n=8 animals/cohort, except sh-Ctl-MIC n = 7 animals). d, Met1 tumorspheres generated in 3 independent experiments, treated with IL-1β(10 ng/mL), Anakinra (500 ng/ml) or IL-1β + Anakinra combination (n=3 replicates, 28 technical replicates) every 48h for 7d. e, Western blot detecting phospho-p65, total p65, or ECAD in Met1 cells treated with 10ng/ml of IL-1β and increasing doses of ANAKINRA (IL-1Ra) for 48h. B-actin (ACTB) was used as a loading control. f, Flow cytometric analysis of monocytes (CD11b+Ly6C+Ly6Glo) presented as % CD11b+ cells in blood at 10 days, prior to the injection of contralateral tumors (Matrigel n=11 animals (combined from Groups 1 and 3); HMLER n = 7 animals). g, Final mass (mg) of hMIC tumors from indicated cohorts (differences not statistically significant, Control n=7 animals, HMLER n = 6 animals). One experiment performed. Source data from a, b, c, d, f, g in Supplementary Table 1. 2-way ANOVA and Sidak’s multiple comparisons test (a, b); 1-way ANOVA (c, g); 2-sided Student’s t tests (d); 2-sided Mann-Whitney test (f).
Supplementary Figure 6. Primary tumors induce an inflammatory response.
a, Representative images of F4/80 staining and ImageJ analysis used to quantify area covered by positive F4/80 staining in Figure 6b. Scale bar= 100 μm. Representative of n=5 HMLER tumors and n=6 HMLER2 tumors.
Supplementary Figure 7. Inhibition of IL-1R1 at Primary Tumor Site Enables MIC Colonization
a, KEGG IL-17 signaling pathway (mmu04657). The log2 fold change values comparing primary tumor to control lungs. Red, higher expression in primary tumors; green, higher expression in normal lungs (n=3 lungs from 3 animals for Met1 primary tumors, and n=4 control lungs from 4 animals). b, Inflammatory cytokine array analysis of conditioned media from HMLER and hMIC cells after 3 days in culture (n=2 biological replicates, 3 technical replicates). Results are fold change (Log2) HMLER/MIC. c, Mouse inflammatory cytokine array results showing differences between plasma from mice bearing Matrigel control, HMLER, or HMLER+IL-1Ra tumors after 2 weeks in vivo (n=2, plasma pooled from 3 individual animals per sample). Graph: cytokines statistically different between cohorts. d, hMIC tumor mass (4 weeks) grown opposite Matrigel control (n=8 tumors), HMLER primary tumor (n=7 tumors), or HMLER+IL-1Ra antagonist (IL-1Ra) (n=8 tumors). One experiment was performed. e, Graph: HMLER primary tumor mass embedded in control Matrigel (n=5 animals) or Matrigel with 100ng/ml IL-1Ra (n=6 animals). Incidence (%) marked on graphs. f, Hematoxylin and eosin (H&E) stains from HMLER and HMLER+IL1Ra tumors (Figure 7c-g), low and high magnification. g, F4/80 staining and ImageJ analysis to quantify area covered by F4/80+ staining in Figure 7e. Boxes indicate area shown in Figure 7e. h, Immunohistochemistry showing macrophage infiltration (F4/80 stain) in hMIC tumors grown opposite Matrigel control, HMLER, or HMLER+IL-1Ra primary tumors. i, j, Photomicrographs of hMIC tumors grown opposite Matrigel control, HMLER primary tumor, or HMLER + IL-1Ra primary tumors. ECAD (red), LgT (green – to detect human tumor cells), DAPI (blue). (h) ZEB1 (red), LgT (green), DAPI (blue). Boxes indicate area shown in Figure 7f-g. Scale bars= 100 μm. Source data for b, c, d, e in Supplementary Table 1. DESeq2’s Wald test two-sided (a); 2-sided Mann-Whitney test (d); 2-sided Welch’s t test (e).
Supplementary Figure 8. Analysis of IL1 pathway components as predictors of disease outcome in clinical breast cancer specimens.
a, Kaplan-Meier analysis using distant metastasis-free survival (DMFS) as end point with 10-year censoring based on stratification of IL1B gene expression (log2; low, grey line, n=740; high, red line, n=639) in tumor tissue from 1,379 breast cancer patients. Logrank p value is shown. Analysis performed using the GOBO database (http://co.bmc.lu.se/gobo/gsa.pl).
Supplementary Figure 9. Western blot scanned films. Boxes highlight lanes used in figures.
Supplementary Table 1. Source Data. Raw data for Figs 1c, 1e, 1g, 1j, 1k, 1l, 2g, 2h, 2j, 2l, 2n 2p, 3c, 3e, 3f, 3h, 3j, 3k, 3l, 4b, 4c, 4d, 4e, 4h, 5c, 5f, 5g, 6b, 6c, 6d, 6e, 6f, 7a, 7b, 7d, 7e, 7f, 7g, Supplementary Figs 1b, 1c, 1d, 1e, 1g, 1h, 1i, 2d, 2e, 2g, 2i, 3b, 3e, 3h, 3j, 3k, 3l, 3n, 3r, 4b, 4d, 4e, 5a, 5b, 5c, 5d, 5f, 5g, 7b, 7c, 7d, 7e, 8b.
Supplementary Table 2. List of differentially expressed genes (DEGs) in lungs from mice bearing primary Met1 tumors versus control lungs. Significantly upregulated genes after RNA sequencing analysis on lungs from mice 14 days after control PBS or Met1 primary tumor initiation.
ACKNOWLEDGMENTS
This work was supported by funding from the International Mentoring Foundation for the Advancement of Higher Education (IMFAHE) (AGR); Center for Stem Cell Bioinformatics at the Harvard Stem Cell Institute (JNH); Samuel Waxman Cancer Research Foundation, Breast Cancer Research Foundation, and Ludwig Center for Molecular Oncology (RAW); Advanced Medical Research Foundation and Nelune Foundation (CLC); National Institutes of Health (NCI) RO1 CA166284, Presidential Early Career Award for Scientists and Engineers, American Cancer Society Research Scholar award, and Department of Defense BCMRP Era of Hope Scholar Award W81XWH-14-1-0191 (SSM). We thank Dr. Johanna Joyce for the Met1 cell line (with permission from Dr. Alexander Borowsky); Drs. Jaclyn Sceneay, Brian Bierie, Yuanbo Qin, and Kristin Tracy for technical expertise; Jessica Olive for editorial assistance; Elizabeth Frank and the Dana-Farber/Harvard Cancer Center Breast Cancer Advocate group for critical discussion and input; Zach Herbert of the Molecular Biology Core Facilities, Dana-Farber Cancer Institute; Drs. George Bell and Bernard Rosner for statistical advice; The Specialized Histopathology Core at Dana-Farber/Harvard Cancer Center (supported in part by an NCI Cancer Center Support Grant # NIH 5 P30 CA06516); Histology Core Facility at the KI Swanson Biotechnology Center, MIT; Flow Cytometry Core Facility at the Whitehead Institute for Biomedical Research. Finally we thank GG, Alice, and Jonathan Bernath, Zaina Corsino-Gupta, and Poly, Bower, and Kerley Nguyen for their support during the preparation of this manuscript.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no conflict of interest
REFERENCES
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2016. CA: a cancer journal for clinicians 66, 7–30 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Klein CA Parallel progression of primary tumours and metastases. Nat Rev Cancer 9, 302–312 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Pantel K, Brakenhoff RH & Brandt B Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat Rev Cancer 8, 329–340 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Luzzi KJ et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. The American journal of pathology 153, 865–873 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron MD et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer research 60, 2541–2546 (2000). [PubMed] [Google Scholar]
- 6.Chambers AF, Groom AC & MacDonald IC Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2, 563–572 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Celia-Terrassa T & Kang Y Distinctive properties of metastasis-initiating cells. Genes & development 30, 892–908 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghajar CM Metastasis prevention by targeting the dormant niche. Nat Rev Cancer 15, 238–247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redig AJ & McAllister SS Breast cancer as a systemic disease: a view of metastasis. Journal of internal medicine 274, 113–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finisguerra V et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature 522, 349–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wculek SK & Malanchi I Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 528, 413–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granot Z et al. Tumor entrained neutrophils inhibit seeding in the premetastatic lung. Cancer cell 20, 300–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffelt SB et al. IL-17-producing gammadelta T cells and neutrophils conspire to promote breast cancer metastasis. Nature 522, 345–348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murgai M, Giles A & Kaplan R Physiological, Tumor, and Metastatic Niches: Opportunities and Challenges for Targeting the Tumor Microenvironment. Critical reviews in oncogenesis 20, 301–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAllister SS & Weinberg RA The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 16, 717–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Visser KE & Coussens LM The inflammatory tumor microenvironment and its impact on cancer development. Contributions to microbiology 13, 118–137 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Chaffer CL, San Juan BP, Lim E & Weinberg RA EMT, cell plasticity and metastasis. Cancer metastasis reviews 35, 645–654 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Borowsky AD et al. Syngeneic mouse mammary carcinoma cell lines: two closely related cell lines with divergent metastatic behavior. Clin Exp Metastasis 22, 47–59 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Werbeck JL et al. Tumor microenvironment regulates metastasis and metastasis genes of mouse MMTV-PymT mammary cancer cells in vivo. Veterinary pathology 51, 868–881 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olive JF et al. Accounting for tumor heterogeneity when using CRISPR-Cas9 for cancer progression and drug sensitivity studies. PloS one 13, e0198790 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaffer CL et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A 108, 7950–7955 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bierie B et al. Integrin-beta4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc Natl Acad Sci U S A 114, E2337–E2346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naumov GN, Folkman J & Straume O Tumor dormancy due to failure of angiogenesis: role of the microenvironment. Clin Exp Metastasis 26, 51–60 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Ye X & Weinberg RA Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends in cell biology 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattabiraman DR et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumorinitiating ability. Science 351, aad3680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellner U et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11, 1487–1495 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Palucka AK & Coussens LM The Basis of Oncoimmunology. Cell 164, 1233–1247 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagerling C & Werb Z Neutrophils: Critical components in experimental animal models of cancer. Semin Immunol 28, 197–204 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeNardo DG et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov 1, 54–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani A, Cassatella MA, Costantini C & Jaillon S Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol 11, 519–531 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Gordon S Alternative activation of macrophages. Nat Rev Immunol 3, 23–35 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Muir AB et al. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Exp Cell Res 319, 850–859 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apte RN et al. The involvement of IL-1 in tumorigenesis, tumor invasiveness, metastasis and tumor-host interactions. Cancer metastasis reviews 25, 387–408 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Hou Z, Falcone DJ, Subbaramaiah K & Dannenberg AJ Macrophages induce COX-2 expression in breast cancer cells: role of IL-1beta autoamplification. Carcinogenesis 32, 695–702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellehumeur C, Blanchet J, Fontaine JY, Bourcier N & Akoum A Interleukin 1 regulates its own receptors in human endometrial cells via distinct mechanisms. Hum Reprod 24, 2193–2204 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Windheim M & Hansen B Interleukin-1-induced activation of the small GTPase Rac1 depends on receptor internalization and regulates gene expression. Cell Signal 26, 49–55 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Shultz LD et al. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol 154, 180–191 (1995). [PubMed] [Google Scholar]
- 38.Garlanda C, Dinarello CA & Mantovani A The interleukin-1 family: back to the future. Immunity 39, 1003–1018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y et al. Lipocalin 2 Upregulation Protects Hepatocytes from IL1-beta-Induced Stress. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 36, 753–762 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Deshmane SL, Kremlev S, Amini S & Sawaya BE Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 29, 313–326 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei X et al. The PSMP-CCR2 interactions trigger monocyte/macrophage-dependent colitis. Sci Rep 7, 5107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dinarello CA Immunological and inflammatory functions of the interleukin-1 family. Annual review of immunology 27, 519–550 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Ringner M, Fredlund E, Hakkinen J, Borg A & Staaf J GOBO: gene expression-based outcome for breast cancer online. PLoS One 6, e17911 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ciriello G et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–519 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cejalvo JM et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res 77, 2213–2221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dongre A et al. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res 77, 3982–3989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerriero JL et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruffell B & Coussens LM Macrophages and therapeutic resistance in cancer. Cancer cell 27, 462–472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu H et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol 16, 1105–1117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid MC et al. Combined blockade of integrin-alpha4beta1 plus cytokines SDF-1alpha or IL-1βeta potently inhibits tumor inflammation and growth. Cancer research 71, 6965–6975 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spiegel A et al. Neutrophils Suppress Intraluminal NK Cell-Mediated Tumor Cell Clearance and Enhance Extravasation of Disseminated Carcinoma Cells. Cancer discovery 6, 630–649 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voloshin T et al. Blocking IL1beta Pathway Following Paclitaxel Chemotherapy Slightly Inhibits Primary Tumor Growth but Promotes Spontaneous Metastasis. Molecular cancer therapeutics 14, 1385–1394 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Kersten K et al. Mammary tumor-derived CCL2 enhances pro-metastatic systemic inflammation through upregulation of IL1beta in tumor-associated macrophages. Oncoimmunology 6, e1334744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinarello CA Why not treat human cancer with interleukin-1 blockade? Cancer metastasis reviews 29, 317–329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castano Z et al. Stromal EGF and igf-I together modulate plasticity of disseminated triple-negative breast tumors. Cancer discovery 3, 922–935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elkabets M et al. Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. The Journal of clinical investigation 121, 784–799 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuznetsov HS et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer discovery 2, 1150–1165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McAllister SS et al. Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell 133, 994–1005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.De Cock JM et al. Inflammation Triggers Zeb1-Dependent Escape from Tumor Latency. Cancer Res 76, 6778–6784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu M et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Primary tumors inhibit outgrowth of distant metastasis-initiating cells
a, Met1 orthotopic primary tumor growth kinetics, FVB mice, represented in Fig. 1b (n=10 animals). b, Schematic of sequential subcutaneous experimental model (left) and growth kinetics (right) of Met1 secondary tumors (2.5 × 105/mouse; implanted at day 14) in mice with PBS control or Met1 primary tumor (2.5 × 105/mouse; n=5 animals/cohort). c, Numbers of pulmonary macrometastases following tail vein injection of MT2 or MT3 cells (7.5×105cells; n=3 animals). Macrometastases >100 microns were quantified on microscope tissue sections from 4 lung lobes per mouse. d, Met1 orthotopic tumor growth kinetics, Nude mice, represented in Fig. 1f (n=5 animals). e, Schematic of experimental model (left), and number of pulmonary macrometastases following tail vein injection of 2.5×105 hMICs into Nude mice with either Matrigel (n = 10 animals) or HMLER primary tumors (n=9 animals; original injection of 5.0×105 cells/mouse) (right). Macrometastases (>100 microns) or micrometastases (>5 cells or <5 cells) were quantified from microscopic whole lung tissue sections. f, Schematic of experimental model (applies to g and h). g, Growth kinetics of HMLER primary tumors, Nude mice, described in Figure 1h (n=10 animals). h, MIC-231 tumor growth kinetics, Nude mice, opposite Matrigel control (n=12 animals) or HMLER primary tumors (n=5 animals). Representative of 2 experiments. i, Images: representative immunofluorescent images of 231-MIC tumors grown opposite Matrigel control or an HMLER primary tumor (represented in Supplementary Fig. 1h) stained with Ki67 (red), hMIT to identify human mitochondria (green), DAPI (nuclei, blue); Scale bars=100 μm. Graph: Quantification of Ki67+hMit+ cells as a percentage of the total number of hMit+ tumor cells/microscopic field (n=9 independent images representing 3 tumors/cohort). Source data for a, b, c, d, e, g, h, i in Supplementary Table 1. 2-way ANOVA, followed by Sidak’s multiple comparison test (b, g, h); 1-sided Welch’s t test (e); 2-sided Welch’s t test (c, i).
Supplementary Figure 2. MIC Differentiation is Perturbed by the Presence of a Primary Tumor
a In vitro immunocytochemical flourescence showing E-cadherin (ECAD, red) and DAPI (nuclei, blue) in Met1 parental cell line (mMIC) and Met1-derived clones, MT2 and MT3 (mMIC-MT3). b Images: Immunofluorescence showing ZEB1 and ECAD expression in cultured hMICs prior to xenotransplantation. Western blot: mesenchymal marker Vimentin (VIM) and epithelial marker ECAD protein in polyclonal HMLER cells and derivative hMIC and HMLER2 cells. GADPH shown as internal control. Positive controls: Ctrl E (epithelial-MCF7Ras); Ctrl M (mesenchymal CD44hi HMLER cells). c, Merged immunofluorescent images of mMIC-MT3 tumors (described in Fig. 1d) stained for basal cytokeratin 14 (CK14, red), luminal CK8 (green) or PyMT antigen (expressed by tumor cells only-green). Arrows - CK14+ tumor cells. d, Images: hMIC tumors (from Fig 1i) stained with CK14 (red), VIM (green) and DAPI (blue); Graph: quantification of indicated stains on hMIC tumors grown opposite Matrigel (n=4 tumors) or primary tumor (n=5 tumors). e, Schematic: modeling early stages of hMIC colonization. Graph: hMIC tumor growth kinetics opposite Matrigel control or HMLER primary tumor (n=4 tumors/group); differences not statistically significant. f, g, Immunofluorescent images (f) and quantification (g) of hMIC tumors stained for ki67 (red), LgT antigen (tumor cells, green), and DAPI (nuclei, blue) as a percentage of total LgT+ cells. Control, n=10 independent images representing 4 tumors; HMLER cohort, n=9 independent images representing 4 tumors. h, i, Immunofluorescent images (h) and quantification (i) of staining hMIC tumors for cleaved caspase3 (CASP3, red), human-specific mitochondria (hMIT, green), and DAPI (nuclei, blue) grown in mice with Matrigel control (n=6 independent images representing 4 tumors) or HMLER primary tumors (n=5 independent images representing 4 tumors). j, Expression of ZEB1 (ZEB1-GFP construct) or HRAS (HRAS-tomato construct) analyzed by FACS (1.0×105 cells) in Control hMIC or ZEB1hi hMIC (from Fig. 2n-p). All scale bars=100 μm. Source data for d, e, g, i in Supplementary Table 1 and d on Supplementary Figure 9. 2-way ANOVA (e); 2-sided Welch’s t test (d, i); 2-sided Mann-Whitney test (g).
Supplementary Figure 3. Innate Inflammatory Cells are Necessary for MIC Colonization
a, Experimental schematic for RNA-seq tissue analysis (Fig. 3a-c and Supplementary Fig. 3b-e). b, Met1 primary tumor mass in FVB mice (n=5 animals). c, d, RNA-seq analysis on lungs from mice with PBS control (n=4 animals) or a Met1 primary tumor (n=4 animals). Heatmap (c): top 50 differentially expressed genes (adjusted p-value, DESeq2). Blue=low, green=mean, and yellow=high relative expression levels. PBS control lungs (yellow), Met1 primary tumor-bearing lungs (purple). Volcano plot (d): DESeq2 comparison Single gene with Padj<0.05 and absolute log2(FoldChange)>1 (green). e, Experimental schematic and flow cytometric quantification of immune cell populations in lungs of indicated FVB mice at 28-day end point (see Fig. 1a). f, Ratio of genes expressed by pro-metastatic immunosuppressive neutrophils from K14cre;Cdh1F/F;Trp53F/F (KEP) mice to control neutrophils from wild type littermates (KEP:Normal)13 extrapolated onto our signatures from control (blue) primary tumor-bearing lungs (red). Higher ratios indicate higher pro-metastatic KEP signature. Box plot: median, 25th and 75th percentiles, whiskers extend to minimum and maximum values. g, Experimental design to identify optimal anti-Ly6G dose for neutrophil depletion. h, Primary tumor mass in Control anti-IgG2a (n=3 mice/cohort) and anti-Ly6G (n=4 mice/cohort). i, Flow cytometric gating strategy for neutrophils (CD45+CD11b+Gr1+Ly6Clo) and monocytes (CD45+CD11b+Gr1+Ly6Chi). j-l, Flow cytometric analysis of blood at 14d (j), 20d (k), and lungs 20d (l) in experiment described in (g). Control anti-IgG2a n=3 mice/cohort; anti-Ly6G n=4 mice/cohort. m, Schematic modeling neutrophil depletion experiment (n-r). n, Primary tumor mass from Fig. 3i (control IgG2a n=4 mice/cohort; anti-Ly6G n=8 mice/cohort). o, Immunofluorescent images of Met1 lung metastasis. Scale bars=200 μm. p, Metastases at higher magnification (indicated in circles in (o) analyzed by ImageJ to measure shortest-length diameter). q, Single channel ECAD or PyMT staining measured by ImageJ analysis (Fig. 3k, l). r, End point flow cytometric analysis of neutrophils and monocytes in lungs of mice treated with control IgG2a (n=4 mice/cohort) or anti-Ly6G (n=8 mice/cohort). Source data for b, e, h, j, k, l, n, r in Supplementary Table 1. DESeq2’s Wald test two-sided (c), 2-sided Welch’s t-test (f); 2-sided Welch’s t test (n); 2-way ANOVA followed by Sidak’s multiple comparison test (f, h, j, k, l, o).
Supplementary Figure 4. Innate Inflammatory Response is sufficient for MIC colonization
a, Some of the predominant cytokines expressed by monocytes, neutrophils or by both. b, ELISA to detect cumulative IL-1β in the conditioned media of Met1 cell line and its derived clones after 3 days in vitro (n=5 biological replicates/group). c, Met1 parental cells and the MT2 and MT3 subclones stained as indicated (upper panel). Scale bars= 100 μm. Western blot detecting phospho-p65 (p-p65; activated form of NF-kB p65) or total NF-kB p65 (p65) in Met1, MT2 and MT3 treated in vitro with indicated doses of IL-1β at indicated time points (lower panel). d, ELISA to detect cumulative IL-1β in the conditioned media of HMLER and hMIC cell lines over 3 consecutive days of in vitro culture (n=4 biological replicates/group). e, Experimental schematic and flow cytometric quantification of immune cell populations in digested hMIC tumors from indicated cohorts in Nude mice at the 28-day experimental end point (see Fig. 2i) (n=3 hMIC tumors in Matrigel control and 4 hMIC tumors in HMLER cohort). Source data for b, d, e in Supplementary Table 1. 2-sided Welch’s t test (b, e); Two-way ANOVA, Sidak’s multiple comparisons test (d).
Supplementary Figure 5. IL-1R1 Pathway is sufficient to block MICs colonization
a, qRT-PCR to determine IL-1R1 mRNA levels in hMIC expressing shRNA against IL-1R1 (shA-IL-1R1-MIC and shB-IL-1R1-MIC) compared to hMIC expressing a scrambled shRNA (sh-Ctl-MIC). B-actin (ACTB) served as the internal control (n=3 PCR technical triplicates). b, Proliferation analysis (CyQuant assay) comparing growth of MIC-control cells to those expressing shRNA against IL-1R1 (shA-IL-1R1-MIC and shB-IL-1R1-MIC; n=3 in triplicate. One experiment, representative of 5 independent experiments). c, hMIC tumor mass (mg) at the 4-week experimental end point (n=8 animals/cohort, except sh-Ctl-MIC n = 7 animals). d, Met1 tumorspheres generated in 3 independent experiments, treated with IL-1β(10 ng/mL), Anakinra (500 ng/ml) or IL-1β + Anakinra combination (n=3 replicates, 28 technical replicates) every 48h for 7d. e, Western blot detecting phospho-p65, total p65, or ECAD in Met1 cells treated with 10ng/ml of IL-1β and increasing doses of ANAKINRA (IL-1Ra) for 48h. B-actin (ACTB) was used as a loading control. f, Flow cytometric analysis of monocytes (CD11b+Ly6C+Ly6Glo) presented as % CD11b+ cells in blood at 10 days, prior to the injection of contralateral tumors (Matrigel n=11 animals (combined from Groups 1 and 3); HMLER n = 7 animals). g, Final mass (mg) of hMIC tumors from indicated cohorts (differences not statistically significant, Control n=7 animals, HMLER n = 6 animals). One experiment performed. Source data from a, b, c, d, f, g in Supplementary Table 1. 2-way ANOVA and Sidak’s multiple comparisons test (a, b); 1-way ANOVA (c, g); 2-sided Student’s t tests (d); 2-sided Mann-Whitney test (f).
Supplementary Figure 6. Primary tumors induce an inflammatory response.
a, Representative images of F4/80 staining and ImageJ analysis used to quantify area covered by positive F4/80 staining in Figure 6b. Scale bar= 100 μm. Representative of n=5 HMLER tumors and n=6 HMLER2 tumors.
Supplementary Figure 7. Inhibition of IL-1R1 at Primary Tumor Site Enables MIC Colonization
a, KEGG IL-17 signaling pathway (mmu04657). The log2 fold change values comparing primary tumor to control lungs. Red, higher expression in primary tumors; green, higher expression in normal lungs (n=3 lungs from 3 animals for Met1 primary tumors, and n=4 control lungs from 4 animals). b, Inflammatory cytokine array analysis of conditioned media from HMLER and hMIC cells after 3 days in culture (n=2 biological replicates, 3 technical replicates). Results are fold change (Log2) HMLER/MIC. c, Mouse inflammatory cytokine array results showing differences between plasma from mice bearing Matrigel control, HMLER, or HMLER+IL-1Ra tumors after 2 weeks in vivo (n=2, plasma pooled from 3 individual animals per sample). Graph: cytokines statistically different between cohorts. d, hMIC tumor mass (4 weeks) grown opposite Matrigel control (n=8 tumors), HMLER primary tumor (n=7 tumors), or HMLER+IL-1Ra antagonist (IL-1Ra) (n=8 tumors). One experiment was performed. e, Graph: HMLER primary tumor mass embedded in control Matrigel (n=5 animals) or Matrigel with 100ng/ml IL-1Ra (n=6 animals). Incidence (%) marked on graphs. f, Hematoxylin and eosin (H&E) stains from HMLER and HMLER+IL1Ra tumors (Figure 7c-g), low and high magnification. g, F4/80 staining and ImageJ analysis to quantify area covered by F4/80+ staining in Figure 7e. Boxes indicate area shown in Figure 7e. h, Immunohistochemistry showing macrophage infiltration (F4/80 stain) in hMIC tumors grown opposite Matrigel control, HMLER, or HMLER+IL-1Ra primary tumors. i, j, Photomicrographs of hMIC tumors grown opposite Matrigel control, HMLER primary tumor, or HMLER + IL-1Ra primary tumors. ECAD (red), LgT (green – to detect human tumor cells), DAPI (blue). (h) ZEB1 (red), LgT (green), DAPI (blue). Boxes indicate area shown in Figure 7f-g. Scale bars= 100 μm. Source data for b, c, d, e in Supplementary Table 1. DESeq2’s Wald test two-sided (a); 2-sided Mann-Whitney test (d); 2-sided Welch’s t test (e).
Supplementary Figure 8. Analysis of IL1 pathway components as predictors of disease outcome in clinical breast cancer specimens.
a, Kaplan-Meier analysis using distant metastasis-free survival (DMFS) as end point with 10-year censoring based on stratification of IL1B gene expression (log2; low, grey line, n=740; high, red line, n=639) in tumor tissue from 1,379 breast cancer patients. Logrank p value is shown. Analysis performed using the GOBO database (http://co.bmc.lu.se/gobo/gsa.pl).
Supplementary Figure 9. Western blot scanned films. Boxes highlight lanes used in figures.
Supplementary Table 1. Source Data. Raw data for Figs 1c, 1e, 1g, 1j, 1k, 1l, 2g, 2h, 2j, 2l, 2n 2p, 3c, 3e, 3f, 3h, 3j, 3k, 3l, 4b, 4c, 4d, 4e, 4h, 5c, 5f, 5g, 6b, 6c, 6d, 6e, 6f, 7a, 7b, 7d, 7e, 7f, 7g, Supplementary Figs 1b, 1c, 1d, 1e, 1g, 1h, 1i, 2d, 2e, 2g, 2i, 3b, 3e, 3h, 3j, 3k, 3l, 3n, 3r, 4b, 4d, 4e, 5a, 5b, 5c, 5d, 5f, 5g, 7b, 7c, 7d, 7e, 8b.
Supplementary Table 2. List of differentially expressed genes (DEGs) in lungs from mice bearing primary Met1 tumors versus control lungs. Significantly upregulated genes after RNA sequencing analysis on lungs from mice 14 days after control PBS or Met1 primary tumor initiation.