Abstract
Background & Aims:
We performed genetic analyses of a multiethnic cohort of patients with idiosyncratic drug-induced liver injury (DILI) to identify variants associated with susceptibility.
Methods:
We performed a genome-wide association study of 2048 individuals with DILI (cases) and 12,429 individuals without (controls). Our analysis included subjects of European (1806 cases and 10,397 controls), African American (133 cases and 1,314 controls), and Hispanic (109 cases and 718 controls) ancestry. We analyzed DNA from 113 Icelandic cases and 239,304 controls to validate our findings.
Results:
We associated idiosyncratic DILI with rs2476601, a nonsynonymous polymorphism that encodes a substitution of tryptophan with arginine in the protein tyrosine phosphatase, non-receptor type 22 gene (PTPN22) (odds ratio [OR], 1.44; 95% CI, 1.28–1.62; P=1.2×10−9 and replicated the finding in the validation set (OR, 1.48; 95% CI, 1.09–1.99; P=.01). The minor allele frequency showed the same effect size (OR > 1) among ethnic groups. The strongest association was with amoxicillin and clavulanate-associated DILI in persons of European ancestry (OR, 1.62; 95% CI, 1.32–1.98; P=4.0×10−6; allele frequency=13.3%), but the polymorphism was associated with DILI of other causes (OR, 1.37; 95% CI, 1.21–1.56; P= 1.5×10−6; allele frequency=11.5%). Among amoxicillin- and clavulanate-associated cases of European ancestry, rs2476601 doubled the risk for DILI among those with the HLA risk alleles A*02:01 and DRB1*15:01.
Conclusions:
In a genome-wide association study, we identified rs2476601 in PTPN22 as a non-HLA variant that associates with risk of liver injury caused by multiple drugs and validated our finding in a separate cohort. This variant has been associated with increased risk of autoimmune diseases, providing support for the concept that alterations in immune regulation contribute to idiosyncratic DILI.
Keywords: amino acid change, GWAS, mutation, inflammation
Introduction
Idiosyncratic drug-induced liver injury (DILI) is a rare adverse drug reaction that is an important cause of acute liver failure in the developed world1, 2. DILI typically occurs in 10 to 20 out of 100,000 treated patients, and while it can lead to death, most cases resolve with discontinuation of the offending drug3, 4. DILI is nonetheless one of the most frequent complications in the development and approval of new drugs, often leading to failure in the later stages of drug development or regulatory actions, including post-marketing withdrawals5.
A number of genome-wide association studies (GWAS) on DILI have been performed, leading to the discovery of significant associations with several HLA alleles that are generally drug-specific. For example, HLA-B*57:01 is associated with DILI in response to flucloxacillin, HLA-A*02:01 and HLA-DRB1*15:01 are associated with amoxicillin-clavulanate (AC), HLA-B*35:02 is associated with minocycline, and HLA-A*33:01 is associated with terbinafine and probably several other drugs as well 6–9. The association of DILI risk with HLA alleles supports a role for adaptive immunity in DILI. Although no confirmed associations outside the HLA region have yet been identified in a GWAS10, a trend toward association with all cause DILI was recently observed with a SNP (rs2476601) in the PTPN22 gene 7. Because this variant has been associated with risk for a variety of autoimmune diseases, confirming this association would provide further support for the immune basis for DILI.
The Drug Induced Liver Injury Network (DILIN) in collaboration with the International Drug Induced Liver Injury Consortium (iDILIC), has assembled a cohort of 2,048 DILI cases and 12,429 population controls across three major ethnic populations (Europeans, African-Americans and Hispanics). After conducting a trans-ethic meta-analysis, we replicated our top associated SNPs on an independent European cohort of cases and performed multiple subset analyses to investigate their relationship with known HLA risk alleles for DILI and their effect sizes within a certain drug or injury type. Here, we confirm a significant association of DILI risk with rs2476601 in the PTPN22 gene. This is the first GWAS significant association outside the HLA region and the first that appears to hold across many different classes of drugs. Our finding supports immune mechanisms having a broad role in DILI.
Materials and Methods
We carried out a case/control association study in three different populations (Europeans, Hispanics and African-Americans) and then performed a meta-analysis.
Cases
In the current study, we analyzed 2048 DILI cases due to multiple causal drugs, including amoxicillin-clavulonate (AC) and flucloxacillin, collected by the iDILIC and DILIN consortia. Causality assessment was performed as previously reported8, 11. 1149 European cases were previously genotyped and analyzed by Urban et al.11 and/or Nicoletti et al.8 and 899 had undergone GWAS genotyping for the first time. Table S1 shows the breakdown of the case cohorts by recruitment center, genotyping chip and ethnicity. Clinical characteristics of the DILI subjects were reported in Table 1.
Table 1:
Clinical characteristics of the samples in the three major DILI case population
| CHARACTERISTICS | Europeans N=1806 |
Hispanics N=109 |
African Americans N=133 |
|---|---|---|---|
| Clinical information | |||
| Mean age, years | 55 | 41 | 47 |
| Female, % | 56.5 | 56.8 | 76.6 |
| Median alanine aminotransferase (range), U/L | 774 (9–15065) | 843 (20–9108 ) | 780 (47–7001) |
| Median alkaline phosphatase (range) , U/L | 290 (11–6239) | 266 (79–2414) | 265 (74–2399) |
| Median latency (range), days | 28 (1–7046) | 58 (3–2789 ) | 51 (3–935) |
| Injury Type | |||
| Cholestatic (%) | 463 (26%) | 12 (11%) | 25 (19%) |
| Hepatocellular (%) | 747 (41%) | 73 (67%) | 80 (60%) |
| Mixed (%) | 465 (26%) | 19(17%) | 22 (16%) |
| Not available (%) | 130 (7%) | 5(5%) | 6 (5%) |
| Total | 1806 | 109 | 133 |
DILIN Cases
A total of 1074 DILIN cases were included, of which 443 DILIN Caucasian cases had been previously reported8, 11, and 631 cases, consisting of 389 European, 133 African-American and 109 Hispanic descendants were newly genotyped12. The DILIN protocol and entrance criteria have been previously published12. All participants provided written informed consent. Causality assessment was performed as previously described, and only cases considered probably, highly likely, or definitely related to the implicated drug were included13. DNA was extracted from lymphocytes and stored at the NIDDK biosample repository at Rutgers University, Piscataway, NJ. Genome-wide genotyping for the 564 DILIN cases was performed with the Multi Ethnic Genome Illumina Array at Duke University and for 32 African Americans and 35 Hispanic DILIN cases was performed with the 1Million Illumina duo Array at Duke University.
iDILIC European Cases
A total of 974 European iDILIC cases with a range of causal drugs and recruitment phases were included. Of these, 706 cases had been described previously,8, 11 while 268 cases due to flucloxacillin or AC were newly genotyped. The 268 patients were recruited between May 2009 and May 2013 as a part of an international collaborative study involving international recruitment centers. All participants provided written informed consent, and each study had been approved by the appropriate national or institutional ethical review boards. The clinical inclusion criteria for all cases were those described by Aithal et al.14 The iDILIC cases were evaluated by application of the Council for International Organizations of Medical Science (CIOMS) scale, also called the Roussel Uclaf Causality Assessment Method (RUCAM),15 and by expert review by a panel of three hepatologists. Only cases having at least possible causality (score ≥3) were included in the study. DNA was prepared as described previously6. Genome-wide genotyping of the additional iDILIC cases was performed by the Broad Institute, Boston by Infinium HumanCoreExome BeadChip for 167 cases and by Infinium Human OmniExpress BeadChip for 101 cases.
Clinical characteristics of the DILI cases
We collected additional clinical information to further investigate the relevance of the most significant associations. Time from start of medication to DILI recognition, concomitant medications, and maximum serum levels of alkaline phosphatase (ALP) and (ALT) were available for both DILIN and iDILIC cases. Also available on all cases was whether the injury was hepatocellular, cholestatic or mixed (based on the initial R value15 . Specific diagnosis of autoimmune diseases was recorded for iDILIC cases, while DILIN cases included reports on whether patients had a general history of autoimmune/collagen vascular disease (Yes/No).
Controls
As DILI has a very low incidence, we consider that a large set of general population control samples can overcome the potential bias of inclusion of people who may have experienced unrecorded DILI events. 10,397 European controls described in our previous study8 were used. Moreover, data for 1,314 African American and 718 Hispanic controls were obtained from the MESA study (phs000420.v6.p3) in dbGAP.16 Table S1 also shows the breakdown of the control cohorts by genotyping chip and ethnicity.
Genetic analysis
Quality control (QC) checks on the initial genotype data were performed as summarized in Supplemental Materials and Methods. EIGENSTRAT analysis was used to identify the Caucasian, African-American and Hispanic cohorts. Our final sample sizes were 1,806 cases and 10,397 controls of European ancestry, 133 cases and 1,314 controls of African-American ancestry, and 109 cases and 718 controls of Hispanic ancestry. SNP imputation was performed in batches dividing the samples according to ethnicity and genotyping platforms. For each batch imputation was carried out using Michigan Imputation Server17. as described in the Supplementary Materials and Methods. For HLA genotypes, four digit HLA alleles were inferred using HIBAG18.
Association analyses were performed using logistic regression under the additive model in Plink 19. EIGENSTRAT axes were used as covariates. A meta-analysis of the three ethnic groups was performed using GWAMA fixed effects. Variants that 1) had a concordant effect in at least two of the ethnic groups and that 2) showed final meta p-values below 5×10−8 were considered statistically significant20, 21. The top associated imputed SNPs were genotyped in the available cases using a TaqMan® SNP genotyping assay (ThermoFisher Scientific, Waltham, MA) in accordance with the manufacturer’s recommendations.
Multi-marker association analysis among the combinations of carriage groups of known HLA risk alleles and the PTPN22 variant allele was performed by logistic regression using principal component as covariates and considering the joint negative carriers as the reference group in the drug-specific cohorts (such as AC, Flucloxacillin, Terbinafine and Flupirtine). Moreover, after transforming the quantitative clinical variables (latency, maximum ALP and maximum ALT) to improve normality, we applied a linear regression model to test differences among known HLA risk alleles and PTPN22 variant in AC cohort.
Epistasis analysis was performed by logistic regression, using principal component axes as covariates and considering an interaction term between being a carrier of any HLA risk alleles and being a carrier of the associated variant.
We also performed association analysis between the PTPN22 variant and reported clinical variables. First, we treated latency and other clinical variables as a quantitative trait. After transforming the quantitative variables to improve normality, we applied a linear regression model to test PTPN22 variant effect on clinical trait in a cases-only design. Epistasis, multi-markers and multinomial logistic regression analyses were carried out using STATA15.
Icelandic DILI replication cohort
An independent Icelandic DILI replication cohort was recruited at the National University Hospital of Iceland. The Icelandic DILI cases were evaluated in accordance with iDILIC causality assessment criteria8. Clinical characteristics of the Icelandic DILI subjects are reported in Table S2. The cohort included 113 DILI cases and 239,304 population controls. Geneotyping data from the icelandic sample set was imputed as previously described 22,23, 24. HLA alleles were also imputed by Graphtyper25. Logistic regression under an additive model was used to test for association between variants and DILI. Detailed description of Icelandic analyses is reported in the Supplementary Materials and Methods.
Results
Overall findings
Our final meta-analysis included 3,622,749 SNPs in 2,048 cases and 12,429 controls (see QQ plots in Figure S1). Clinical characteristics of the well-phenotyped DILI cases across three main ethnicities are reported in Table 1. We identified a significant association with rs2476601 (chr1:114377568>A/G), a polymorphism changing tryptophan to arginine at codon 620 of PTPN22 (OR 1.44 95%CI [1.28–1.62] P=1.2×10−9; Figure 1). The enrichment was observed across all ethnic groups analyzed in our study, although the low number of African-American and Hispanic cases limited the power to identify a significant association for variants with an OR < 2 (Table 2). Similar odds ratios were also evident within subgroups of European ancestry (Table S3). Independent genotyping of available DILIN cases across ethnicities (N=1070) confirmed the GWAS genotypes for rs2476601 with a concordance rate of 100% (Table S4). rs2476601 was also found to increase the risk of DILI in the independent Icelandic replication cohort (AF = 0.13 in 113 cases vs 0.09 in 239,304 controls), having an effect size that was comparable to that of the discovery cohort (OR=1.48, 95%CI [1.09–1.99] P= 0.01).
Figure 1. Manhattan plot displaying the association results of (A) the meta-analysis among the three major populations (Europeans, African Americans and Hispanics) (B) the meta-analysis after conditioning on the four main known HLA DILI risk alleles among the three major populations (Caucasians, African Americans and Hispanics).
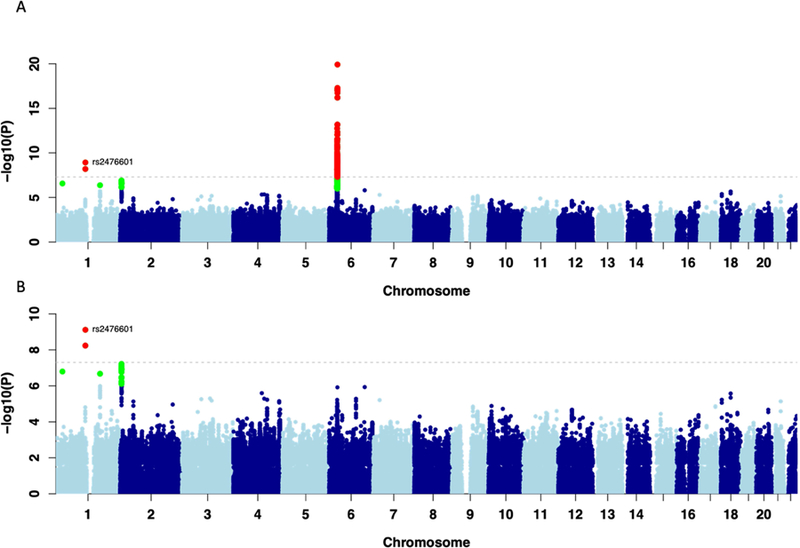
The results are reported for variants which had a consistent effect in Europeans and at least one of the two additional populations. SNPs shown in green have a significance level less than 5×10−6 and red have a significance level less than 5×10−8.
Table 2.
Summary statistics for the univariate trans-ethnic meta-analysis of genome-wide associated variants
| Marker | ALL |
CAU |
AA |
HISP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | |
| rs2476601 | 1.44 | 1.28–1.62 | 1.2×10−9 | 1.42 | 1.27–1.60 | 9.6×10−10 | 1.94 | 0.73–5.18 | 0.19 | 1.91 | 0.94–3.89 | 0.07 |
| rs72631546 | 1.84 | 1.47–2.31 | 1.2×10−7 | 1.79 | 1.41–2.27 | 1.92 ×10−6 | - | - | - | 2.07 | 1.27–3.36 | 0.003 |
| rs3129880 | 1.48 | 1.6–9.32 | 1.2×10−20 | 1.56 | 1.44–1.69 | 5.09 ×10−26 | 0.92 | 0.67–1.27 | 0.62 | 0.97 | 0.68–1.4 | 0.89 |
| HLA-B*57:01 | 2.19 | 1.84–2.61 | 1.40 ×10−18 | 2.24 | 1.94–2.6 | 4.53 ×10−27 | 1.64 | 0.36–7.4 | 0.52 | 0.72 | 0.17–3.02 | 0.65 |
| HLA-DQB1*03:03 | 1.67 | 1.41–1.99 | 7.48 ×10−9 | 1.69 | 1.46–1.96 | 1.27 ×10−12 | - | - | - | 0.97 | 0.29–3.16 | 0.95 |
| HLA-DRB1*15:01 | 1.37 | 1.22–1.53 | 1.03 ×10−7 | 1.40 | 1.27–1.54 | 3.19 ×10−11 | 0.85 | 0.43–1.69 | 0.65 | 1.16 | 0.68–1.99 | 0.58 |
| HLA-C*06:02 | 1.40 | 1.23–1.59 | 1.88 ×10−7 | 1.45 | 1.3–1.62 | 6.47 ×10−11 | 0.92 | 0.58–1.48 | 0.74 | 1.38 | 0.77–2.48 | 0.28 |
| HLA-DQB1*06:02 | 1.31 | 1.17–1.46 | 1.47 ×10−6 | 1.40 | 1.27–1.55 | 3.15 ×10−11 | 0.90 | 0.66–1.21 | 0.48 | 1.11 | 0.64–1.91 | 0.72 |
Odds ratios (OR), confidence intervals (95%CI) and p-values (P) are presented after correcting for population stratification with EIGENSTRAT axes within each major population.
In addition to rs2476601, several variants in the MHC region were found to have genome-wide significant p-values, led by rs3129880 (OR=1.48 95%CI [1.36–1.60] P=1.2×10−20). This variant is a proxy of HLA-DRB1*15:01 (r2 = 0.56) consistent with the large number of AC DILI cases in the cohort. As expected based on inclusion of 195 flucloxacillin and 444 AC Caucasian DILI cases, the most significant HLA risk alleles were HLA-B*57:01, followed by HLA-DRB1*15:01 (OR = 2.19, P = 1.4×10−18, see Table 2). Unlike the rs2476601 association, HLA association signals were specific for the European population in which flucloxacillin and AC cases were the most abundant.
Subsequent genome-wide conditional analysis incorporating the genotypes of the four well-established DILI HLA risk alleles (HLA-B*57:01, HLA-DRB1*15:01, HLA-A*02:01 and HLA-A*33:01)6–8 as covariates was undertaken to identify novel independent risk factors. The analysis revealed that rs2476601 remained the most significant independent risk variant (OR=1.45 95%CI [1.30–1.64] P = 7.6×10−10, Figure 1B and Table S5). Similarly, the independence between rs2476601 and the main HLA risk alleles was confirmed in the Icelandic cohort in a multivariate regression model (OR=1.54; P = 0.013, Table S6). When controlling for the four major known DILI HLA risk alleles, HLA-C*04:01 was the most significant independent HLA allele associated with DILI risk reaching near statistical significance when corrected for the total number of imputed HLA alleles (OR=1.21; 95%CI [1.09–1.37]; P = 6.3×10−4). HLA-C*04:01 association showed consistent trends across all three ethnicity groups (European P=0.004, OR=1.19; African-American P=0.02, OR=1.42; Hispanic P=0.53, OR=1.13, Table S7). Data for individual drugs in relation to this risk association are shown in Table S8. It is notable that the greatest association was seen with the 58 cases where DILI was attributed to herbal and dietary supplements (OR 2.24, p = 0.0008, individual agents listed in Table S9).
We also found that rs72631546, an intergenic marker on chromosome 2, was the third most significant variant (OR = 1.84, P = 1.2×10−7, Table 2). rs72631546 is in LD (r2 = 0.5) with rs72631567, which was a SNP previously suspected to be associated with DILI risk 8. The rs72631546 association was consistent between Europeans and Hispanics (OR = 1.79 P = 1.9×10−6; OR = 2.07 P = 0.003 respectively) and was independent of the known HLA risk associations (OR = 1.87 P = 6.0×10−8).
Association with PTPN22 rs2476601
In the European cohort, the AC cases showed the most significant association with rs2476601 (OR=1.62 95%CI [1.32–1.98] P = 4.0×10−6) with higher frequency than European controls (AF = 0.13 vs AF = 0.08, respectively). We also found evidence that the association was consistent among the remaining of European DILI cases (N=1362 OR= 1.37 95%CI [1.21–1.56] P= 1.5×10−6, AF= 0.11, Figure S2), and it did not appear to be driven by particular drugs or categories of drugs (Table 3). Significance of P ≤ 0.05 was seen for cases due to several causal drugs including sulfamethoxazole-trimethoprim (P= 0.01) and terbinafine (P= 0.01). On the other hand, cases related to drugs such as flucloxacillin and diclofenac, which were well represented in the cohort as causal agents, showed smaller increases in minor allele frequency (AF) compared with controls, which were not statistically significant (P > 0.05). We also evaluated the relationship of rs2476601 genotype to DILI phenotype. Of our European ancestry cases, 45% had a hepatocellular pattern of injury, and 55% were cholestatic or mixed. We found enrichment compared to controls for rs2476601 in both injury patterns, with similar AF and ORs (hepatocellular AF = 0.12, OR=1.38, P = 0.0001; cholestatic/mixed AF = 0.13, OR = 1.50, P = 6.5×10−8; Table S10). We also found that there was a trend for the frequency of rs2476601 to be higher in the DILI cases most confidently ascribed to the implicated drug (Figure S2). We found that there was no significant association between rs2476601 and time of onset of DILI relative to starting treatment with the implicated drug as well as maximum ALP or maximum ALT values.
Table 3. Association with rs2476601 for drugs with at least 3 case carriers in the European cohort and OR > 1.
Drug results are ordered by p-value
| DRUGS | # Cases | AF | OR | 95%CI | P |
|---|---|---|---|---|---|
| Amoxacillin/Clavulanic Acid | 444 | 0.13 | 1.62 | 1.32–1.98 | 0.000004 |
| Terbinafine | 15 | 0.20 | 3.23 | 1.29–8.1 | 0.01 |
| Sulfamethoxazole/Trimethoprim | 42 | 0.17 | 2.07 | 1.16–3.71 | 0.01 |
| Methotrexate | 9 | 0.22 | 3.34 | 1.09–10.16 | 0.03 |
| Rofecoxib | 6 | 0.25 | 4.08 | 1.05–15.82 | 0.04 |
| Valproic Acid | 16 | 0.18 | 2.43 | 0.99–5.95 | 0.05 |
| Flupirtin | 6 | 0.25 | 4.43 | 0.98–20.05 | 0.05 |
| Fenofibrate | 10 | 0.20 | 2.93 | 0.97–8.87 | 0.06 |
| Erythromycin | 11 | 0.18 | 2.89 | 0.95–8.78 | 0.06 |
| Doxycycline | 6 | 0.25 | 3.21 | 0.85–12.1 | 0.09 |
| Pravastatin | 6 | 0.25 | 3.21 | 0.83–12.47 | 0.09 |
| Nimesulide | 20 | 0.12 | 2.10 | 0.81–5.41 | 0.12 |
| Cefuroxime | 4 | 0.25 | 3.45 | 0.69–17.28 | 0.13 |
| Ethinylestradiol/Levonorgestrel | 7 | 0.21 | 2.53 | 0.71–9.05 | 0.15 |
| Isoniazid | 43 | 0.13 | 1.59 | 0.84–3.02 | 0.16 |
| Celecoxib | 9 | 0.17 | 2.37 | 0.69–8.19 | 0.17 |
| Flucloxacillin | 195 | 0.11 | 1.24 | 0.90–1.71 | 0.18 |
| Nitrofurantoin | 74 | 0.12 | 1.40 | 0.85–2.32 | 0.19 |
| Piroxicam | 5 | 0.20 | 2.85 | 0.60–13.68 | 0.19 |
| Gabapentin | 5 | 0.20 | 2.79 | 0.58–13.39 | 0.2 |
| Cefazolin | 21 | 0.14 | 1.59 | 0.67–3.8 | 0.3 |
| Mercaptopurine | 10 | 0.15 | 1.72 | 0.50–5.97 | 0.39 |
| Imatinib | 8 | 0.12 | 1.70 | 0.38–7.56 | 0.49 |
| Ticlopidine | 5 | 0.10 | 2.01 | 0.24–16.73 | 0.52 |
| Atorvastatin | 29 | 0.10 | 1.32 | 0.56–3.09 | 0.53 |
| Minocycline | 32 | 0.11 | 1.29 | 0.58–2.86 | 0.53 |
| Interferon Beta-1a | 4 | 0.12 | 1.90 | 0.21–16.79 | 0.57 |
| Amiodarone | 5 | 0.20 | 1.64 | 0.28–9.73 | 0.59 |
| Diclofenac | 66 | 0.10 | 1.17 | 0.67–2.04 | 0.59 |
| Ibuprofen | 15 | 0.10 | 1.36 | 0.41–4.52 | 0.62 |
| Herbal and dietary products | 58 | 0.10 | 1.19 | 0.65–2.17 | 0.58 |
| Disulfiram | 8 | 0.12 | 1.40 | 0.32–6.13 | 0.65 |
| All Other Therapeutic Products | 9 | 0.11 | 1.33 | 0.30–5.93 | 0.71 |
| Nicotinic Acid | 4 | 0.12 | 1.45 | 0.17–12.17 | 0.73 |
| Lisinopril | 5 | 0.10 | 1.45 | 0.17–12.17 | 0.74 |
| Phenytoin | 10 | 0.10 | 1.18 | 0.27–5.11 | 0.82 |
| Rosuvastatin | 4 | 0.12 | 1.27 | 0.14–11.29 | 0.83 |
| Sertraline | 6 | 0.08 | 1.17 | 0.15–9.27 | 0.88 |
| Levofloxacin | 17 | 0.09 | 1.05 | 0.32–3.44 | 0.94 |
Cases= number of cases for each drug
AF = Allele frequcncy; OR = Odd Ratio; ULC= 95% lower Confidence interval of the Odds Ratio; ULC= 95% upper Confidence Interval of the Odds Ratio; P = logistic p-value. In bold drugs previously known to be associated with at least one HLA risk allele.
Assessment of correlation with autoimmune diseases
As rs2476601 has been previously associated with numerous autoimmune diseases26 we investigated whether the presence of autoimmune diseases in our DILI cohort could have contributed to the associations observed. We identified 567 DILI subjects with evidence of autoimmune diseases; 135 of whom had a documented history of autoimmune/collagen vascular disease, and the remaining subjects were suspected to have an autoimmune disease because they had been treated with at least one drug commonly used in these conditions, usually in addition to the agent implicated as causing DILI (list of potential autoimmune treatments is presented in Table S11). When all 567 samples with known or suspected diagnosis of autoimmune disease were excluded from our cohort, the rs2476601 association remained highly significant with the same effect size (n=1245, OR =1.40 95%CI [1.23–1.60] P=6.4*10−7, Table S12).
Assessment of correlation with known HLA risk alleles
We found an enrichment of rs2476601 among European DILI cases due to causal drugs known to have HLA alleles as the main genetic risk association (e.g., flucloxacillin, terbinafine, fenofibrate, minocycline, sertraline, AC) compared to the rest of the cases (OR =1.52 vs OR = 1.38, Table S13). Among these drugs associated with HLA risk alleles, AC was the major causal drug (444 cases) and showed the strongest association with rs2476601 (OR=1.62 95%CI [1.32–1.98] P = 4.0×10−6). AC-drug specific conditional analysis on HLA-A*02:01 and HLA-DRB1*15:01 confirmed that rs2476601 was an independently associated risk factor from the known HLA risk alleles (OR = 1.6 and P = 8.9×10−6, Table S5). Since the three markers were independent among each other, we looked for evidence of co-occurance of rs2476601 and the known HLA risk alleles. We therefore stratified AC cases and controls based on HLA allele carriage (Table S14). There was evidence that carriers of rs2476601 were enriched in AC DILI patients who carried one or both the HLA. In agreement with this finding, multi-marker analysis on the AC cohort confirmed that when rs2476601 co-occurred with either of the two HLA alleles, this consistently enhanced the association with DILI risk by almost two-fold compared to risk associated with the HLA alleles alone or in combination (Table 4). Joint carriage of the three markers was associated with a 13-fold higher DILI risk compared to the negative carriers. We had only 12 AC cases carrying only rs2476601 and neither of the two HLA risk alleles. This limited number did not allow us to capture the association of rs2476601 alone with AC DILI risk, but the 95% confidence interval reported in Table 4 included an OR of 1.5 . Moreover, when we compared the triple positive carrier against HLA-A*02:01 and HLA-DRB1*15:01 positive but rs2476601 negative group we found a significant 1.7 fold increase in the association with DILI risk (OR= 1.8 95%CI[1.24–2.60] ; P= 0.002). This confirmed that rs2476601 is independently associated with AC DILI risk. Finally, we tested the presence of a SNP-HLA interaction effect for AC DILI. The analysis showed that there was an epistatic effect between rs2476601 and the presence of at least one of the HLA risk alleles (OR = 1.9; P = 0.05). In otherwords, the joint effect of one or both HLA risk alleles and rs2476601 was more than additive.
Table 4.
Summary statistics of the multi marker analysis performed on the carriage of HLA-DRB1*15:01, HLA-A*02:01 and rs2476601 in the European Amoxicillin-Clavulanate-related DILI cohort.
| Cases |
Controls |
||||||
|---|---|---|---|---|---|---|---|
| Carriage group | N | CF | N | CF | OR | 95%CI | P |
| +/+/+ | 49 | 0.11 | 187 | 0.02 | 13.80 | 9.18–20.71 | 1.1*10−36 |
| −/+/+ | 35 | 0.08 | 625 | 0.06 | 3.03 | 1.99–4.63 | 2.3*10−7 |
| +/−/+ | 12 | 0.03 | 206 | 0.02 | 3.28 | 1.74–6.19 | 2.5*10−4 |
| −/−/+ | 12 | 0.03 | 663 | 0.07 | 0.95 | 0.51–1.77 | 0.8 |
| +/+/− | 126 | 0.28 | 895 | 0.09 | 7.68 | 5.63–10.48 | 8.8*10−38 |
| −/+/− | 101 | 0.23 | 3212 | 0.31 | 1.68 | 1.23–2.30 | 1.1*10−3 |
| +/−/− | 41 | 0.09 | 1037 | 0.10 | 2.09 | 1.41–3.11 | 2.7*10−4 |
| −/−/− | 68 | 0.15 | 3531 | 0.34 | - | - | - |
Odds ratios (OR), 95% confidence intervals (95%CI) and p-values (P) are presented after correcting for population stratification and considering the triple negative carriers as the reference group. The three letters in the first column (carriage group) reflects in order the HLA-DRB1*15:01 status, the HLA-A*02:01 status and the rs2476601 status. The risk alleles status is represented by “+ “ = present or “−” = absent N = number of samples in the group; CF = carriage frequency.
Multi-marker analysis on the terbinafine, flucloxacillin and flupirtine cohorts also supported rs2476601 as independently associated with DILI risk , showing consistent ORs in DILI due to flucloxacillin (OR=1.3), terbinafine (OR=3.4) and flupirtine (OR=2.5) in addition to the enhanced the DILI risk associated with the known HLA alleles (Table S15). We also examined whether subjects in the AC cohort who carried any combination of the three risk alleles differed in clinical phenotype (see Materials and Methods) from each other or from individuals not carrying any of the three alleles. No differences were apparent.
Discussion
Here, we report the results of the largest DILI GWAS to date based on 2048 DILI cases and 12,429 controls. Our analyses identified a robust association with a variant in PTPN22, a tyrosine phosphatase that has been linked to numerous autoimmune disorders26. Additionally, the association was not limited to a certain drug or pattern of injury, instead showing associations across the entire cohort. Moreover, we showed that the PTPN22 variant added to the DILI risk associated with known HLA alleles, increasing the association with AC DILI risk almost two-fold and appearing to have a similar effect on other DILI events with known HLA risk alleles. This finding provides new insights into DILI etiology and highlights the potential role of non-HLA variants in immune related genes as risk factors for DILI across a broad spectrum of causal drugs.
rs2476601 is associated with increased risk of type 1 diabetes mellitus, rheumatoid arthritis, systemic lupus erythematosus, vitiligo and Graves’ disease, among others, but is also associated with decreased risk of Crohn’s disease and Behçet disease26. This is the first confirmed genome-wide association with DILI risk that lies outside the MHC locus and is the first variant that appears to generally predispose to DILI as opposed to DILI due to specific drugs. The replication of the association in separate Icelandic cases and controls where the PTPN22 variant is relatively common confirmed the association.
Our previous study also suggested that rs2476601 might be associated with risk for DILI, but the variant did not meet the criteria for genome-wide significance7. The present study confirmed that association, and with a larger sample size the strength of the association exceeded the required statistical threshold for a variant of this moderate effect size. It should be noted that the effect size seen for this variant with DILI was similar to those seen for other disease conditions where rs2476601 is associated with increased risk27. The frequency of rs2476601 varies greatly from population to population, being as high as 15% in Finns and as low as <0.01% in East Asians28. Here, we found that the frequency of the variant in DILI cases was higher in each population studied relative to the frequency in a matched control population, with the odds ratio remaining similar among different ethnic and racial populations. Despite this consistency of the odds ratio, the relatively small effect size of this variant means that larger sample sizes are needed to confirm the associations as real in all populations and with different agents responsible for causing DILI. The largest subset of the current study was Northern Europeans (n=1,107 cases and 5,090 controls), where a very strong signal for this variant was seen (P=3.6×10−6, OR=1.41, Table S3). The other analyzed ethnicities, which had much smaller sample sizes, did not produce p-values below 0.05 despite their similar odds ratios for the effect of this variant.
Because patients with autoimmune disease may take more medications than others, there was a possibility that the association we observed with rs2476601 could actually be due to an increased prevalence of autoimmune diseases in DILI patients. To address this possibility, we identified cases with a recorded or suspected (based on concomitant medications) diagnosis of autoimmune diseases. Although rs2476601 in these patients had a slightly higher effect size, the association remained comparably strong even among patients not known or suspected to have underlying autoimmune conditions. Because history of autoimmune diseases was not systematically collected in all subjects, and because patients with autoimmune diseases may not be taking medication treatment for these conditions at the time of the DILI event, we cannot rule out the possibility that some of our DILI patients had undiagnosed and untreated autoimmune conditions.
The association with rs2476601 was consistent across various phenotypes in our cohort, including injury patterns (cholestatic or mixed vs. hepatocellular), causal drugs, and strength of causality assessment. However, we did not find any association with other features, including DILI latency. We found that AC cases and other cases with the highest causality scores tended to have the highest frequencies for rs2476601. As DILI is a diagnosis of exclusion, it is unavoidable to have some uncertainty about the true cause of liver injury in certain patients, and so for real risk associations, we expect to see the strongest associations in those cases with the highest causality score. In our cohort, most AC cases were classified as having a high likelihood of DILI since the AC-DILI characteristic phenotypes have been well defined29. The higher allele frequency in AC cases likely reflects a higher proportion of patients who truly have DILI due to this drug. However, there may be an AC-specific genetic effect of rs2476601, as AC cases had a higher allele frequency than did other DILI cases even when restricting to the same causality probability categories.
Our analysis showed that rs2476601 appears to be associated with DILI risk regardless of which HLA alleles are associated with DILI risk. This is also the case with autoimmune diseases where rs2476601 is associated across diseases that are themselves associated with different HLA alleles26. This effect is consistent with the fact that PTPN22 controls events downstream from HLA presentation of neoantigen as summarized in Burn et al 30. PTPN22 encodes lymphoid protein tyrosine phosphatase (Lyp) which is expressed exclusively in immune cells. Although the mechanisms whereby the rs2476601 variant reduces immune tolerance are not clear, Lyp is involved in T-cell receptor signaling, acting at several intermediate points in the signaling cascade. Lyp also appears to influence regulatory T-cell function. Considerable data now support the concept that DILI can result from a T-cell-mediated immune attack on the liver, presumably directed at HLA-presented neoantigens. This response may be initiated relatively frequently during treatment with drugs capable of causing DILI. However, clinically important liver injury does not occur in most of these patients because immune tolerance is activated, 31, 32.
We therefore suggest that rs2476601 predisposes to DILI by reducing immune tolerance. Our finding supports this hypothesis, since we found a significant genetic interaction effect among HLA risk alleles and rs2476601, detecting that the AC DILI risk associated with the joint carriage of HLA risk alleles and the variant is more that the sum of the risks associated with each single risk factor. Most of the currently reported associations for other diseases and the PTPN22 variant also show HLA associations, particularly HLA class II associations which is consistent with the strong association seen with AC-DILI but only a slightly increased frequency of the variant with flucloxacillin-DILI where the associated allele is class I. Moreover, the increased frequency of the rs2476601 variant in DILI cases where there was no apparent HLA association suggests the possibility of other HLA risk alleles yet to be discovered and/or a potential role for PTPN22 in non-T cell-mediated forms of DILI where other immune cells might be involved.
Although the association with rs2476601 was robust, its effect size was modest. The odds ratio averaged about 1.3 in the various ethnic groups identified here. However, in the 10% of the subjects who also carried the two known HLA risk alleles (HLA-A*02:01 and HLA-DRB1*15:01), the risk of DILI due to AC was increased over 13-fold. Given the rarity of serious liver injury due to AC, despite its widespread use, genotyping for risk management is probably not realistic. There may be instances when genotyping for this variant together with the identified HLA risk alleles could improve confidence in the causality assessment but this testing is not currently commercially available to our knowledge. It should also be noted that with drugs causing more frequent and severe liver injury, genotyping to identify 10% of a patient population at 13-fold increased risk of DILI might be reasonable.
In addition to the rs2476601 association, we also found evidence for a novel independent HLA risk factor in HLA-C*04:01. This association is interesting as the allele may be a risk factor across ethnicities and ADR clinical phenotypes. In fact, HLA-C*04:01 has previously been shown to be associated with nevirapine hypersensitivity in a Malawian population,33 and in our cohort, the association was concordant across all three populations (Table S8) and across multiple drugs and herbal preparations (Table S7).
In conclusion, we have identified the variant rs2476601 in the PTPN22 gene as the first robust genetic risk association for DILI lying outside the MHC region. In addition, this association is the first to be associated across a broad range of implicated drugs and across different ethnic backgrounds. rs2476601 is therefore the first identified general risk association for DILI. The prior well-established association of this polymorphism with the risk of autoimmune diseases broadens the role of the immune system in DILI pathogenesis and may help inform future treatment and prevention efforts.
Supplementary Material
Acknowledgments
We are extremely grateful to Daniele Cusi (Hypergenes), Patrik K. Magnusson (Swedish Twin Registry) and Javier Martin (Spanish DNA bank) for provision of control data. The iDILIC team are very grateful to Arthur Holden (iSAEC) for his continuing support. Contributors to sample collection via DILIN, iDILIC, the Spanish DILI registry, EUDRAGENE, and DILIGEN are listed in the Appendix.
Funding
The DILIN (https://dilin.dcri.duke.edu/) is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH) as a Cooperative Agreement (U01s) under grants: U01-DK065176 (Duke), U01-DK065201 (UNC), U01-DK065184 (Michigan), U01-DK065211 (Indiana), U01DK065193 (UConn), U01-DK065238 (UCSF/CPMC), U01-DK083023 (UTSW), U01-DK083027 (TJH/UPenn), U01-DK082992 (Mayo), U01-DK083020 (USC), U01-DK100928 (Icahn). Additional funding is provided by CTSA grants UL1 RR025761 (Indiana), UL1 RR025747 (UNC), and UL1 UL1 RR024986 (UMich). The iDILIC study was supported by the International Serious Adverse Events Consortium which received funding from Abbott, Amgen, Daiichi-Sankyo, GlaxoSmithKline, Merck, Novartis, Pfizer, Roche, Sanofi-Aventis, Takeda, and the Wellcome Trust. DILIGEN and iDILIC sample collection was funded by the National Institute for Health Research (NIHR) Nottingham Digestive Diseases Biomedical Research Unit at the Nottingham University Hospitals NHS Trust and University of Nottingham. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. GPA is the gastrointestinal and liver disorder theme lead for the NIHR Nottingham BRC (Reference no: BRC-1215–20003). The EUDRAGENE collaboration received support from the EC 5th Framework program (QLRI-CT-2002–02757). The Spanish DILI Registry is partly funded by the Spanish Medicine Agency, Fondo Europeo de Desarrollo Regional - FEDER (FIS PI16_01748, PI15 _01440). CIBERehd is funded by Instituto de Salud Carlos III. The Swedish case collection (SWEDEGENE) has received support from the Swedish Medical Products Agency, the Swedish Society of Medicine (2008–21619), Swedish Research Council (Medicine 521–2011-2440), and Swedish Heart and Lung Foundation (20120557). MM was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London.
Abbreviations:
- DILI
drug-induced liver injury
- GWAS
genome wide association study
- OR
Odd Ratio
- RUCAM
Roussel Uclaf Causality Assessment Method
- MHC
Major Histocompatibility Complex
- AF
allele frequency
- HLA
Human leukocyte antigen
- SNP
Single Nucleotide Polymorphism
- AC
Amoxicillin-clavulanate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: The authors disclose the following: Dr. Nelson is an employee of GlaxoSmithKline. Dr. Nicoletti is an employee of Sema4. Drs. Chalasani, Fontana, and Watkins report consulting agreements and research grants with several pharmaceutical companies but none represent potential conflicts for this paper. Dr. Rafnar and Dr. Stefansson are employees of deCODE genetics/Amgen. The DILIN causality committee considers potential conflicts while assigning cases for adjudication to individual investigators. The remaining authors disclose no conflicts.
References
- 1.Gulmez SE, Larrey D, Pageaux GP, et al. Transplantation for acute liver failure in patients exposed to NSAIDs or paracetamol (acetaminophen): the multinational case-population SALT study. Drug Saf 2013;36:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuben A, Koch DG, Lee WM. Drug-induced acute liver failure: results of a U.S. multicenter, prospective study. Hepatology 2010;52:2065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjornsson ES, Bergmann OM, Bjornsson HK, et al. Incidence, presentation, and outcomes in patients with drug-induced liver injury in the general population of Iceland. Gastroenterology 2013;144:1419–25, 1425 e1–3; quiz e19–20. [DOI] [PubMed] [Google Scholar]
- 4.Sgro C, Clinard F, Ouazir K, et al. Incidence of drug-induced hepatic injuries: a French population-based study. Hepatology 2002;36:451–5. [DOI] [PubMed] [Google Scholar]
- 5.Stevens JL, Baker TK. The future of drug safety testing: expanding the view and narrowing the focus. Drug Discov Today 2009;14:162–7. [DOI] [PubMed] [Google Scholar]
- 6.Daly AK, Donaldson PT, Bhatnagar P, et al. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet 2009;41:816–9. [DOI] [PubMed] [Google Scholar]
- 7.Lucena MI, Molokhia M, Shen Y, et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology 2011;141:338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicoletti P, Aithal GP, Bjornsson ES, et al. Association of Liver Injury From Specific Drugs, or Groups of Drugs, With Polymorphisms in HLA and Other Genes in a Genome-Wide Association Study. Gastroenterology 2017;152:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urban TJ, Nicoletti P, Chalasani N, et al. Minocycline hepatotoxicity: Clinical characterization and identification of HLA-B *35:02 as a risk factor. J Hepatol 2017;67:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban TJ, Goldstein DB, Watkins PB. Genetic basis of susceptibility to drug-induced liver injury: what have we learned and where do we go from here? Pharmacogenomics 2012;13:735–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urban TJ, Shen Y, Stolz A, et al. Limited contribution of common genetic variants to risk for liver injury due to a variety of drugs. Pharmacogenet Genomics 2012;22:784–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana RJ, Watkins PB, Bonkovsky HL, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf 2009;32:55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi PH. Drug-Induced Liver Injury Network Causality Assessment: Criteria and Experience in the United States. Int J Mol Sci 2016;17:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aithal GP, Watkins PB, Andrade RJ, et al. Case Definition and Phenotype Standardization in Drug-Induced Liver Injury. Clinical Pharmacology and Therapeutics 2011;89:806–815. [DOI] [PubMed] [Google Scholar]
- 15.Danan G, Benichou C. Causality assessment of adverse reactions to drugs--I. A novel method based on the conclusions of international consensus meetings: application to drug-induced liver injuries. J Clin Epidemiol 1993;46:1323–30. [DOI] [PubMed] [Google Scholar]
- 16.Tryka KA, Hao L, Sturcke A, et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res 2014;42:D975–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng X, Shen J, Cox C, et al. HIBAG--HLA genotype imputation with attribute bagging. Pharmacogenomics Journal 2014;14:192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–69. [DOI] [PubMed] [Google Scholar]
- 22.Gudbjartsson DF, Helgason H, Gudjonsson SA, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet 2015;47:435–44. [DOI] [PubMed] [Google Scholar]
- 23.Kong A, Masson G, Frigge ML, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet 2008;40:1068–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature 2009;462:868–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eggertsson HP, Jonsson H, Kristmundsdottir S, et al. Graphtyper enables population-scale genotyping using pangenome graphs. Nat Genet 2017;49:1654–1660. [DOI] [PubMed] [Google Scholar]
- 26.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol 2014;10:602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho JH, Feldman M. Heterogeneity of autoimmune diseases: pathophysiologic insights from genetics and implications for new therapies. Nat Med 2015;21:730–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.deLemos AS, Ghabril M, Rockey DC, et al. Amoxicillin-Clavulanate-Induced Liver Injury. Dig Dis Sci 2016;61:2406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burn GL, Svensson L, Sanchez-Blanco C, et al. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett 2011;585:3689–98. [DOI] [PubMed] [Google Scholar]
- 31.Mosedale M, Watkins PB. Drug-induced liver injury: Advances in mechanistic understanding that will inform risk management. Clin Pharmacol Ther 2017;101:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho T, Uetrecht J. How Reactive Metabolites Induce an Immune Response That Sometimes Leads to an Idiosyncratic Drug Reaction. Chem Res Toxicol 2017;30:295–314. [DOI] [PubMed] [Google Scholar]
- 33.Carr DF, Chaponda M, Jorgensen AL, et al. Association of human leukocyte antigen alleles and nevirapine hypersensitivity in a Malawian HIV-infected population. Clin Infect Dis 2013;56:1330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


