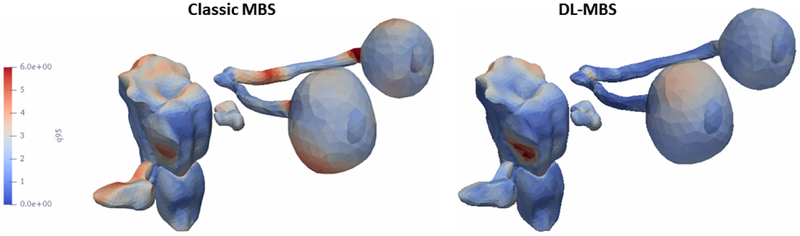Abstract
Organ-at-risk (OAR) segmentation is a key step for radiotherapy treatment planning. Model-based segmentation (MBS) has been successfully used for the fully automatic segmentation of anatomical structures and it has proven to be robust to noise due to its incorporated shape prior knowledge. In this work, we investigate the advantages of combining neural networks with the prior anatomical shape knowledge of the model-based segmentation of organs-at-risk for brain radiotherapy (RT) on Magnetic Resonance Imaging (MRI). We train our boundary detectors using two different approaches: classic strong gradients as described in [4] and as a locally adaptive regression task, where for each triangle a convolutional neural network (CNN) was trained to estimate the distances between the mesh triangles and organ boundary, which were then combined into a single network, as described by [1]. We evaluate both methods using a 5-fold cross- validation on both T1w and T2w brain MRI data from sixteen primary and metastatic brain cancer patients (some post-surgical). Using CNN-based boundary detectors improved the results for all structures in both T1w and T2w data. The improvements were statistically significant (p < 0.05) for all segmented structures in the T1w images and only for the auditory system in the T2w images.
Keywords: organ-at-risk brain segmentation, model-based segmentation, deep learning
1. Introduction
Organ-at-risk (OAR) segmentation is a key step for radiotherapy treatment planning. However, efficient and automated segmentation is still an unmet need. Manual delineation of these organs is a tedious process, time consuming, and prone to errors due to intra- and inter-observer variations.
Many automatic organ segmentation approaches work on a voxel-by-voxel basis by inspecting the local neighborhood of a voxel (e.g. region growing, front propagation, level sets etc., but also classification methods like decision forests or neural networks). However, those approaches are often prone to segmentation errors due to imaging artifacts. Another popular segmentation technique is model-based segmentation (MBS) [2]. With this technique a triangulated mesh representing the organ boundary is adapted to the medical image in a controlled way so that the general organ shape is preserved, thus regularizing the segmentation with prior anatomical knowledge. This makes the technique quite robust against many imaging artifacts. MBS usually uses simple features to detect organ boundaries, such as strong intensity gradients and a set of additional intensity-based constraints [4] or scale invariant feature transforms [5]. These features were proven to be reliable when used to segment well-defined grey values. Recent developments in deep learning have allowed the use of neural networks for boundary detection feature learning [1].
In this work, we propose to use an automatic model-based OAR segmentation to support MR-based treatment planning. We train our model features using two different boundary detection approaches: classic strong gradients as described in [4] and as a locally adaptive regression task, where for each triangle a convolutional neural network is trained to estimate the distance between the mesh and organ boundary, which are then combined into a single network, as described by [1]. We evaluate both methods using a 5-fold cross-validation on both T1-weighted (T1w) and T2-weighted (T2w) brain MRI data from sixteen primary and metastatic brain cancer patients (some post- surgical). CNN-based boundary detectors improve the results for all structures in both T1w and T2w data. The improvements were statistically significant (p < 0.05) for all segmented structures in the T1w images results and for the auditory system in the T2w images. Combining the strength and robustness of neural networks with the prior anatomical shape knowledge and shape regularisation of the model-based segmentation is a promising and powerful tool for organ segmentation.
2. Methods
In this section, we describe the basics of model-based segmentation and model development, as well as the data used and the two different trainings we performed.
2.1. Model-based Segmentation
Model-based segmentation (MBS) is a popular segmentation framework. MBS was originally developed as a generic algorithm, but firstly applied mainly to cardiac applications [2], but has been widely used for a variety of other applications and modalities. The main idea of MBS is to adapt a triangulated surface mesh with a fixed number of vertices V and triangles T to an image.
This adaption is commonly performed in three steps: Step 1: Shapefinder. The first step places the surface mesh within the image to be segmented, using for example a Generalised Hough Transform. Step 2: Parametric adaptation. In the second step, global or local parametric transformations (rigid or a ne) are applied to improve the alignment of the mesh to the image. Step 3: Deformable adaption. In the last step, the alignment of the mesh to the image is further improved by applying local mesh deformations.
The latter two steps are both based on triangle-specific features trained on reference populations. During segmentation, each mesh triangle detects an image boundary point along a search profile which is perpendicular to the triangle surface. Image segmentation is then achieved by aligning the mesh with the detected points. The adaptation of the image to the mesh is thus achieved by minimising the objective function E defined as:
| (1) |
where Eint is the internal energy which attempts to minimise large deformations with respect to the mean mesh shape, Eext is the external energy that attracts the mesh to the image boundary points and is a parameter that balances the contribution of both Eint and Eext [2].
2.2. Segmentation Model
The direct segmentation of the organs-at-risk is challenging due to variations in image orientation and due to image quality issues. Hence, we first segment the skull and hemispheres to provide an estimate of the initial location of OARs with respect to the other organs. The proposed head segmentation model is composed of triangle-based meshes for each individual structure to be segmented: skull; hemispheres; brainstem; optic nerves, globes, lenses, chiasm (optic system) and auditory system. To improve segmentation accuracy, we apply a hierarchical segmentation strategy, by segmenting larger-to-smaller structures. Hence, the skull is first segmented, providing a robust localisation of the head, after which the hemispheres are activated and segmented inside of the skull. Finally, the brainstem, and the remaining OARs are segmented.
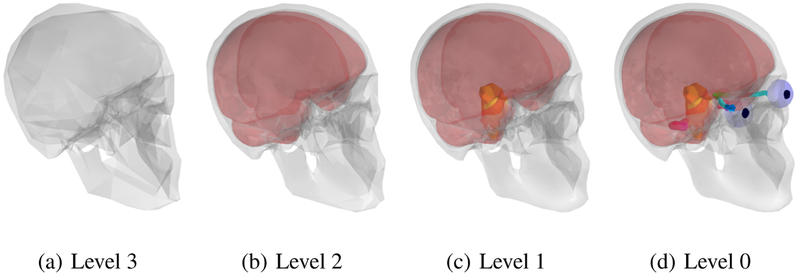
Image segmentation starts by adapting the model from the initial coarsest level to the image. This level is represented by a coarse mesh topology, i.e. only a few triangles are forming the mesh as shown in Figure 1a. Then segmentation is refined by adapting the model of an intermediate level comprising more triangles (Figure 1b). The next segmentation step includes an even finer representation of the skull being adapted, after which the hemispheres are also segmented (Figure 1c). Lastly, the final and highest level of the segmentation is obtained of all structures present in the model: skull, hemispheres and organs-at-risk (Figure 1d). The anatomical correspondence between the different level meshes is ensured by vertex correspondence look-up tables.
Fig. 1.
Head segmentation model with 4 different segmentation levels. Structures segmented include the skull, hemispheres and organs-at-risk
2.3. Boundary Detection
For each triangle, a boundary point is defined by searching along the triangle normal line. In the classical MBS, candidate points on the search line are evaluated using triangle-specific feature functions [4]. The candidate point with the strongest feature response is then selected as the boundary point [4].
Brosch et al. (2018) recently proposed that, instead of looking at discrete points along the search line, one can directly predict the signed distance of the centre of the triangle to the organ boundary using convolutional neural networks (CNN) [1]. This method was used in [1] for the segmentation of the prostate on T2w MRI data.
3. Experiments
3.1. Data and ground truth generation
Sixteen primary and metastatic brain cancer patients (some post-surgical) underwent both CT-SIM and 1.0T MR-SIM (usual clinical protocol including T1-w and T2-w scans with resolutions of 0.9 × 0.9 × 1.25 mm3 and 0.7 × 0.7 × 2.5 mm3 respectively) within 1 week.
Patients were consented to an IRB-approved prospective protocol for imaging on a 1.0 T Panorama High Field Open Magnetic Resonance System (Philips Medical Systems, Cleveland, OH) equipped with flat table top (Civco, Orange City, IA) and external laser system (MR-simulation, or MR-SIM) as described previously. All brain MRI scans were acquired using an 8-channel head coil with no immobilization devices. Brain CT images were acquired using a Brilliance Big Bore (Philips Health Care, Cleveland, OH) scanner with the following settings: 120 kVp, 284 mAs, 512×512 in-plane image dimension, 0.8 − 0.9 × 0.8 − 0.9mm2 in-plane spatial resolution, and 1–3 mm slice thickness.
Ground truth OAR contours were delineated by physicians based on hybrid MR- SIM and CT-SIM information. The frame of reference for all delineation was the CT- SIM dataset, thus we resampled both MR T1w and T2w images into the CT space after elastic registration [3]. The references meshes were generated by adapting triangle-based model meshes to the physician-delineated contours. The reference meshes were further checked to assure consistency, especially for the optic lenses, which exhibit different locations between scans due to eye movement.
3.2. Model training
We trained our model on both the T1-weighted and T2-weighted MRIs of the 16 subjects using a 5-fold cross-validation, and using two different trainings for the boundary detectors: classic gradient-based features (”classic” MBS) as described by [4] and the MBS with deep learning-based boundary detection (DL-MBS) as described by [1]. Both trainings of the boundary detectors are based on the method described in [4] used for selecting optimal boundary detectors from a large set of candidates, called Simulated Search. This method was further adapted for the CNN boundary detectors as described by [1]. Specifically, for each training iteration, mesh triangles were randomly and independently transformed using random translations along the triangle normal, small translations orthogonal to the triangle normal and small random rotations. Subvolumes were then extracted for each newly transformed triangle and the distance to the original reference mesh was computed. The network parameters in this case were optimised using stochastic gradient descent by minimising the root mean square error between the simulated distance and the predicted distance [1].
4. Results
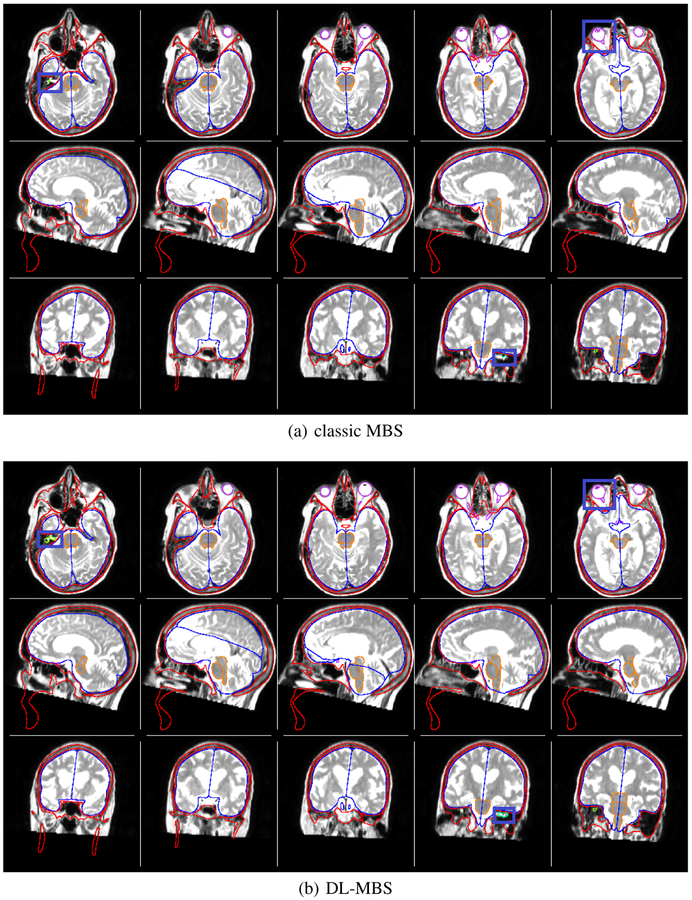
We segmented the organs-at-risk in T1w and T2w brain MRI using the 2 different features on all 16 patients using a 5-fold cross validation. An example segmentation of both classic and DL-MBS on a T2w image is shown in Figure 2.
Fig. 2.
Organs-at-risk segmentation of skull (red), hemispheres (blue), brainstem (orange), optic system (magenta) and auditory system (green). on T2w MR brain data using classic MBS and DL-MBS. It can be observed that DL-MBS provides a more robust segmentation, especially for smaller structures such as the optic lenses and auditory system, thus outperforming classic MBS.
We computed the mean distance in mm between the segmentation and ground truth meshes as well as the maximum dista. Since the boundary between the optic nerves and chiasm is not clearly defined in the MR images, we combined the two for the error analysis. The results are presented in Tables 1 and 2. The DL-MBS outperforms the classic MBS, giving smaller mean distances for both T1w and T2w over all structures of interest (organs-at-risk) investigated.
Table 1.
Mean and maximum distance (mm) per substructure over 16 subjects using 5-fold cross- validation on the T1w images using classic MBS and DL features. Note that 2 subjects presenting cataracts were excluded from both training and testing of the optic lenses.
| Structure | Number of Triangles | Mean Distance (mm) | Maximum Distance (mm) | ||
|---|---|---|---|---|---|
| Classic MBS | DL-MBS | Classic MBS | DL-MBS | ||
| Brainstem | 6054 | 0.729 | 0.608 | 1.967 | 1.586 |
| Optic Globes | 1002 | 0.853 | 0.563 | 2.133 | 1.447 |
| Optic Lenses | 428 | 0.640 | 0.268 | 1.666 | 0.667 |
| Chiasm and Optic Nerves | 1263 | 0.729 | 0.410 | 1.990 | 1.089 |
| Auditory System | 2720 | 1.230 | 0.882 | 2.808 | 2.081 |
Table 2.
Mean and maximum distance (mm) per substructure over 16 subjects using 5-fold cross- validation on the T2w images using classic MBS and DL features. Note that 2 subjects presenting cataracts were excluded from both training and testing of the optic lenses.
| Structure | Number of Triangles | Mean Distance (mm) | Maximum Distance (mm) | ||
|---|---|---|---|---|---|
| Classic MBS | DL-MBS | Classic MBS | DL-MBS | ||
| Brainstem | 6054 | 0.564 | 0.557 | 1.542 | 1.507 |
| Optic Globes | 1002 | 0.593 | 0.526 | 1.388 | 1.252 |
| Optic Lenses | 428 | 0.752 | 0.674 | 1.554 | 1.568 |
| Chiasm and Optic Nerves | 1263 | 0.939 | 0.796 | 2.328 | 1.984 |
| Auditory System | 2720 | 1.163 | 0.639 | 2.764 | 1.640 |
We performed a t-test to investigate if the differences between the classic MBS and DL-MBS are statistically significant. The difference T1w segmentation results between classic MBS and DL-MBS are statistically significant (p < 0.05) for all structures segmented, while for the T2w segmentation the results are statistically significant different (p < 0.05) only for the auditory system.
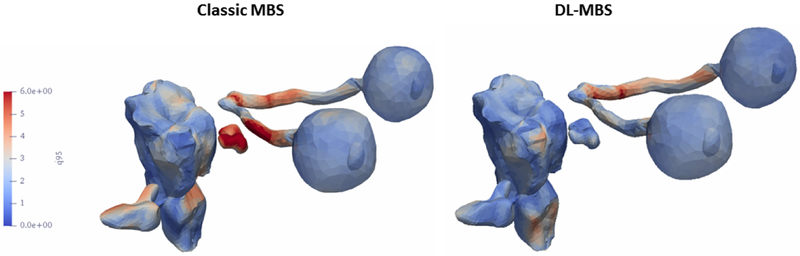
We plotted the maximum distance error for both feature training methods for th T1w (Figure 3) and T2w (Figure 4)) on the organs-at-risk surfaces to investigate the regions were the segmentation presents the most differences. It can be observed that classic MBS is less consistent in segmenting the optical nerves and cochlea, especially in the T2w images with a lower resolution in the z-direction.
Fig. 3.
Maximum segmentation error of the T1w MR images plotted on the organs-at-risk surfaces.
Fig. 4.
Maximum segmentation error of the T2w MR images plotted on the organs-at-risk surfaces.
5. Discussion
In this paper, we investigated the advantages of combining neural networks with the prior anatomical shape knowledge of the model-based segmentation of organs-at-risk for brain RT on MRI. To that end, we trained and compared two boundary detectors for MBS using two different approaches: first using classic gradient-based features (”classic” MBS) [2] and then by using a convolutional neural network that predicts the organ boundary [1]. We validated the results on data from 16 brain cancer patients using a 5-fold cross-validation. The latter provided better segmentation results, with a smaller average distance between the ground truth and segmentation meshes for all organs-at- risk (brainstem, optic nerves, globes, lenses, chiasm, auditory system) on both T1w and T2w MR images, although statistically significant improvements were achieved only on the T1w data.
This work illustrates the strength of using neural networks for segmentation to sup- port MR-based treatment planning in the brain. The CNN-based boundary detection was trained as it was done in [1], where the algorithm was developed and tested for prostate segmentation in T2-weighted images. Nonetheless, the method is also succesful outside of the scope of the prostate segmentation, with its good results being translated also to brain segmentation, where it outperformed classic MBS segmentation. Results could be further improved in the future with small modifications to the training and/or network, in order to adapt it to the specific case of brain organs-at-risk segmentation, where the structures are small and their size is comparable to the image resolution.
We offer solutions to segment the skull, hemispheres and OARs (brainstem, optic nerves, globes, lenses, chiasm and auditory system). The skull segmentation currently has the main purpose of improving the robustness of the segmentation of the other structures, but it may also to be for the generation synthetic CT, the computed tomography substitute data set derived from MRI data and essential for treatment planning.
Furthermore, this study demonstrated the feasibility of developing automated OAR brain segmentation for MR-based treatment planning. Since the segmentation accuracy is below the voxel resolution, our results highlight the importance of acquiring data with high resolution to improve the accuracy of autosegmentation. One limitation of the current work might be the incompatibility of the immobilization masks with the MRI head coil, which may increase image registration uncertainty. Although in this work the images were acquired with dedicated MR-head coils, we hypothesis that the robustness of the CNN and regularising property of the model-based segmentation would provide an accurate segmentation even with an RT-compatible coil setup, with lower signal- to-noise ration. However, this hypothesis remains to be tested. Future work will also include using a broader database and to explore other body regions such as the head and neck.
References
- 1.Brosch T, Peters J, Groth A, Stehle T, Weese J: Deep learning-based boundary detection for model-based segmentation with application to mr prostate segmentation (2018)
- 2.Ecabert O, Peters J, Schramm H, Lorenz C, von Berg J, Walker M, Vembar M, Olszewski M, Subramanyan K, Lavi G, Weese J: Automatic Model-Based Segmentation of the Heart in CT Images. IEEE Transactions on Medical Imaging 27(9), 1189–1201 (September 2008). 10.1109/TMI.2008.918330 [DOI] [PubMed] [Google Scholar]
- 3.Kabus S, Lorenz C: Fast elastic image registration Medical Image Analysis for the Clinic: A Grand Challenge pp. 81–89 (2010) [Google Scholar]
- 4.Peters J, Ecabert O, Meyer C, Kneser R, Weese J: Optimizing boundary detection via simulated search with applications to multi-modal heart segmentation. Medical Image Analysis 14(1), 70 (2010), http://perso.telecom-paristech.fr/ãngelini/MIMED/papers_2010/peters_media_2010.pdf [DOI] [PubMed] [Google Scholar]
- 5.Yang M, Yuan Y, Li X, Yan P: Medical image segmentation using descriptive image features BMVC pp. 1–11 (2011) [Google Scholar]