Abstract
In a high-throughput screening campaign, we recently discovered the rRNA-binding tetracyclines, methacycline and meclocycline, as inhibitors of Dicer-mediated processing of microRNAs. Herein, we describe our biophysical and biochemical characterization of these compounds. Interestingly, although direct, albeit weak, binding to the pre-microRNA hairpins was observed, the inhibitory activity of these compounds was not due to RNA binding. Through additional biochemical and chemical studies, we revealed that metal chelation likely plays a principle role in their mechanism of inhibition. By exploring the activity of other known RNA-binding scaffolds, we identified additional disconnections between direct RNA interaction and inhibition of Dicer processing. Thus, the results presented within provide a valuable case study in the complexities of targeting RNA with small molecules, particularly with weak binding and potentially promiscuous scaffolds.
Keywords: MicroRNA, Dicer, high-throughput screening, tetracyclines
RNA-targeted probe and drug discovery have become important areas of chemical biology and medicinal chemistry research.1,2 From the promise of personalized medicine to the hugely significant, yet largely understudied role of RNA in human biology and disease, targeting RNA with small molecules has emerged as a sought-after approach in biomedical research. While the complexities of RNA, particularly its net electronegative charge and high degree of structural plasticity, coupled with a lack of high-resolution structural information,3 have historically made such efforts difficult; a rarely discussed challenge associated with targeting RNA are the often-discovered disconnections between RNA binding and inhibition of biological function.
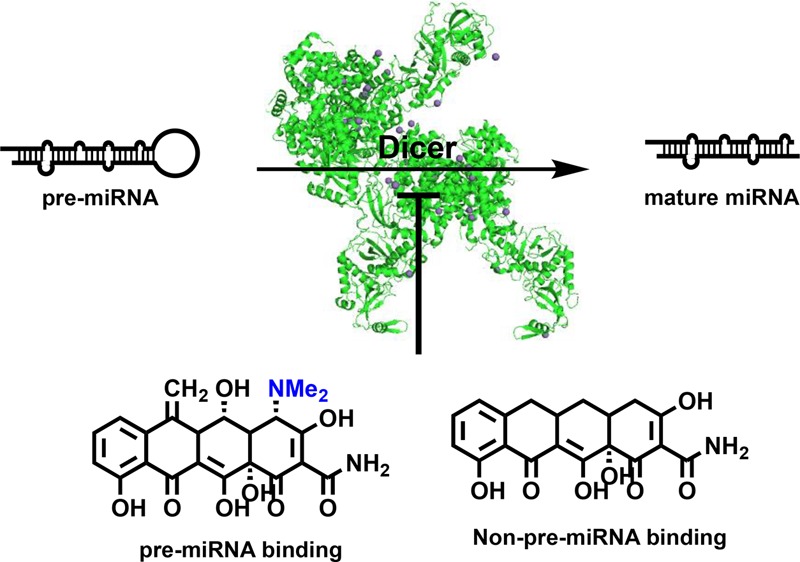
MicroRNAs (miRNAs or miRs) are a family of small noncoding RNAs that play critical roles in the fine-tuning of gene expression.4 Dicer-mediated processing of pre-microRNAs (pre-miRNA or pre-miR) is the penultimate step of miRNA maturation, which occurs prior to loading of the mature miRNA into the RNA-induced silencing complex (RISC).5 As many miRNAs become aberrantly regulated in disease,6−8 we became interested in developing a strategy by which to discover small molecule modulators of this process, with the goal of identifying those that could do so in a RNA-selective manner. Using our catalytic enzyme-linked click chemistry assay (cat-ELCCA) for Dicer,9−11 we performed a high-throughput screen (HTS) targeting pre-miR-21,12 an oncogenic miRNA overexpressed in the majority of human tumors,13 and a target of interest in the field.14−27 Herein, we disclose the hits from our prior HTS12 and reveal tetracyclines as the only RNA-binding hits from our small molecule screen. Interestingly, through additional studies, we have revealed that RNA binding is not required for this activity, highlighting the necessity for careful analysis when characterizing inhibitors of RNA biology.
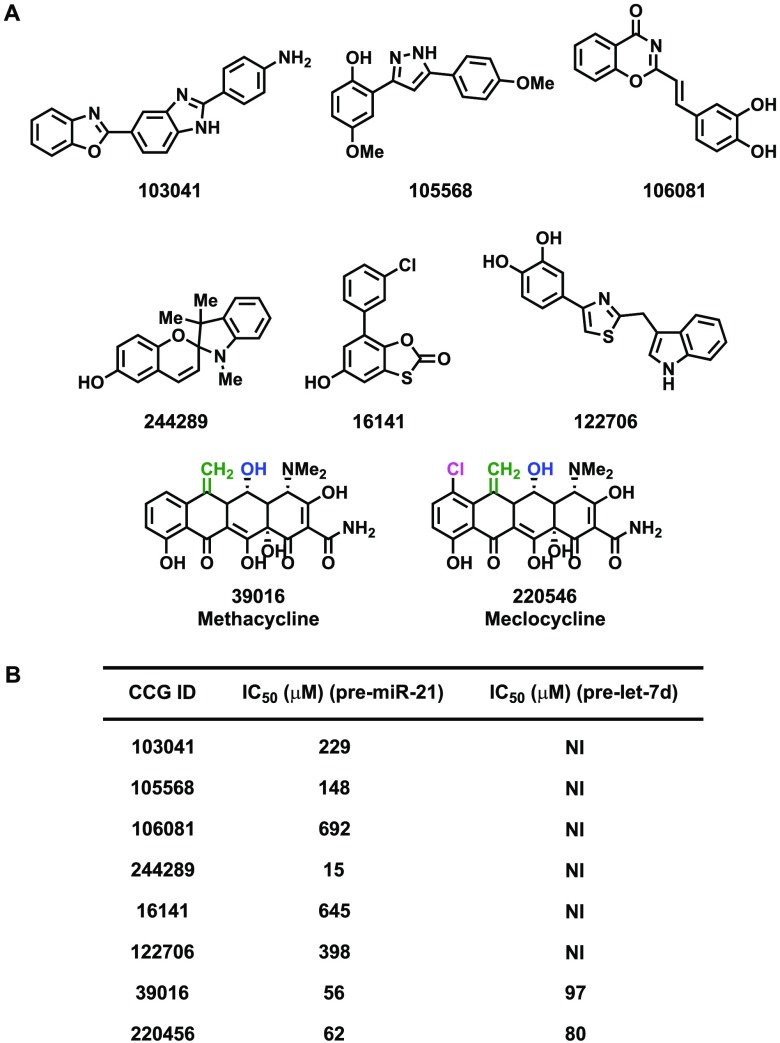
As reported previously, our screening campaign using commercial small molecule libraries resulted in very few hit compounds with demonstrated selectivity for pre-miR-21 over pre-let-7d, which was used in our counterscreen.12 To determine if this apparent selectivity was due to RNA binding properties, six compounds that showed modest, yet specific inhibition of pre-miR-21 processing were purchased and analyzed (Figure 1A). Methacycline and meclocycline, members of the tetracycline family of antibiotics,28,29 which were also identified from the screen and showed nonspecific inhibition of Dicer processing, were also examined (Figure 1A).
Figure 1.
HTS results. (A) Structures of hit compounds from the primary screen that showed selectivity for pre-miR-21 and identified tetracyclines, methacycline, and meclocycline. (B) Table of measured IC50 values (NI = no inhibition).
Direct binding to pre-miR-21 and pre-let-7d was measured via surface plasmon resonance spectroscopy (SPR) using biotinylated pre-miRs immobilized to streptavidin chips. From this analysis, only methacycline and meclocycline were found to exhibit affinity for the pre-miRNA hairpins. Measured Kd values for methacycline were 9.9 and 17 μM, and those for meclocycline were 2.1 and 5.8 μM, for pre-miR-21 and pre-let-7d, respectively (sensorgrams can be found in Figure S1; Figure 2B), which are similar to the reported affinities for ribosomal RNA (rRNA).29 The IC50 values for these compounds from fresh powders were subsequently measured as 383 and 189 μM against pre-miR-21 and 800 and 225 μM against pre-let-7d (Figure 2). We attribute the observed differences in potency from our HTS results (Figure 1B)12 to tetracycline instability during long-term storage in DMSO (see below).30 In sum, these results indicate that the identification of low molecular weight small molecules that target RNA hairpins is difficult, likely due to a bias in the chemical space of these collections toward protein binders.
Figure 2.
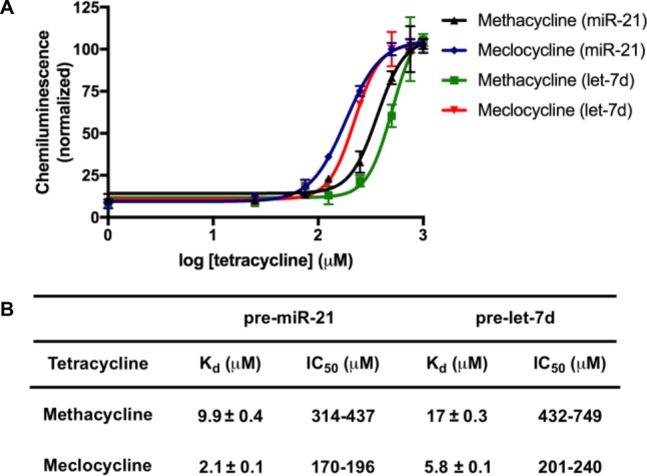
Inhibition of Dicer processing using freshly dissolved methacycline and meclocycline. (A) Dose-dependent inhibition as measured via cat-ELCCA. (B) Table of binding affinities and inhibition constants. IC50 values are represented as 95% confidence intervals.
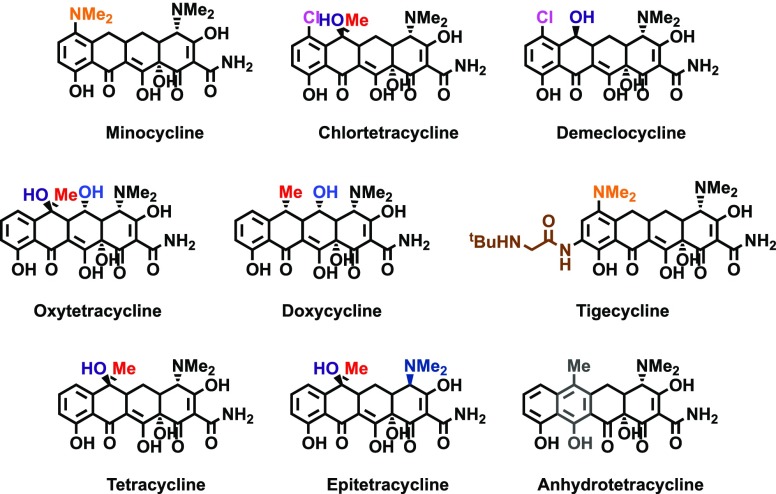
The tetracyclines are natural product antibiotics that inhibit bacterial translation by binding to 16S rRNA and blocking amino acyl-tRNA delivery.31 However, this class of molecules has been linked to a number of other RNA- and protein-related biological processes, including inhibition of the mitochondrial32 and human33 ribosomes, promotion of SMN2 splicing,34 and inhibition of matrix metalloproteinases.35 To determine if this scaffold generally inhibits Dicer processing of pre-miRNAs, we screened a collection of tetracyclines (Figure 3).
Figure 3.
Structures of select tetracyclines analyzed in cat-ELCCA.
As shown in Figure 4A, inhibition ranged from 40–100% after testing at a single concentration of 1.0 mM. We then profiled the dose-responsive inhibition of select tetracyclines, and IC50 values were measured as 194–558 μM against pre-miR-21 and 265–664 μM against pre-let-7d (Figures 4B–D). From these analyses, it was noted that the presence of a chlorine substituent, as found in meclocycline and chlortetracycline, was important for potent inhibition of Dicer-mediated maturation, and 2–4-fold improvements were observed for each over the parent scaffolds of methacycline and tetracycline, respectively (Figures 2 and 4B–D). Additionally, upon analysis of the two most common tetracycline degradation products, epitetracycline and anhydrotetracycline (Figure 3),36 we found that the anhydro product boosted the inhibitory activity of the tetracycline scaffold (Figure 4A). Thus, this or other degradation products could have contributed to our conflicting results between the HTS stocks and fresh powders. It is important to note that although tetracyclines are not potent inhibitors of pre-miRNA maturation by Dicer, inhibition of this process has historically been a challenge due to the complexity of the Dicer enzyme and its interaction with a pre-miRNA.37 In fact, even RNA-binding proteins,38,39 RNA aptamers,40 and peptides18,26 that are known to inhibit Dicer processing in cells, exhibit high IC50 values relative to their binding affinities in in vitro assays. Because of the poor catalytic properties of Dicer in vitro, our assay uses excess enzyme to ensure sufficient signal-to-background for accurate and reproducible detection; and thus, antagonism of processing with a nonoptimized small molecule is a challenge.
Figure 4.
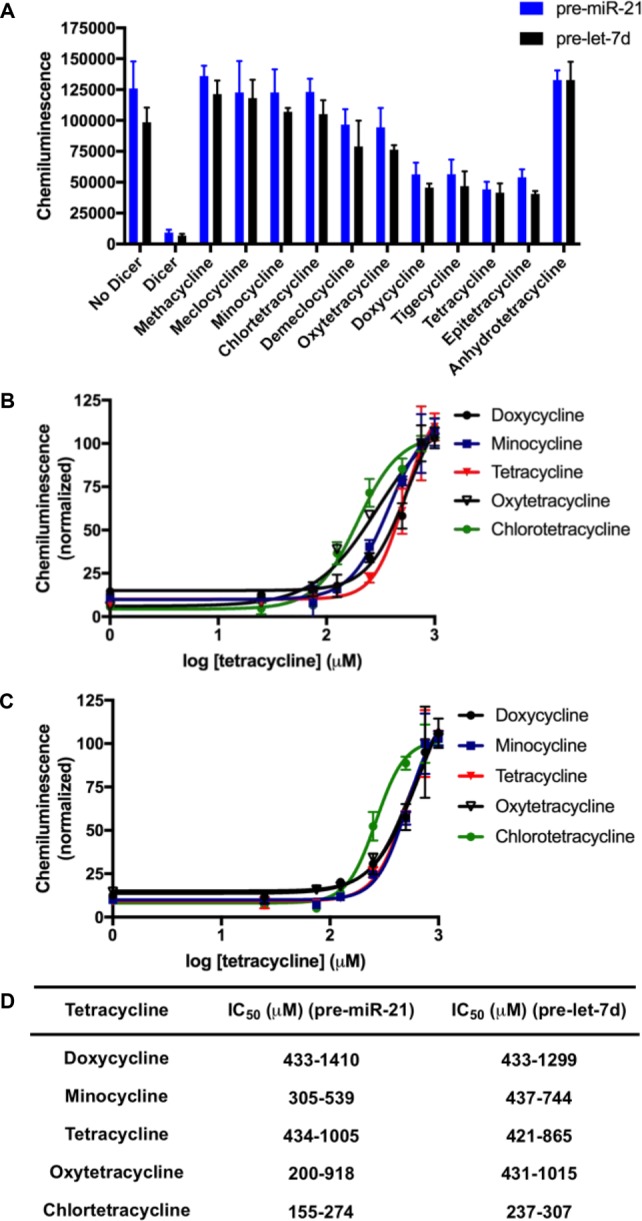
Tetracyclines as inhibitors of Dicer-mediated pre-miRNA maturation. (A) Tetracycline library screening at 1.0 mM. (B,C) Plot of dose-dependent inhibition by select tetracyclines against pre-miR-21 and pre-let-7d processing, respectively, in cat-ELCCA. (D) Table of IC50 values represented as 95% confidence intervals.
Prior to our work, other researchers have investigated the tetracyclines as inhibitors of Dicer-mediated pre-miRNA maturation;41,42 however, conflicting results were reported. Using a fluorescent intercalator displacement (FID) assay, Herdewijn and co-workers observed no inhibition with select family members.41 This finding is not surprising, as tetracyclines are not known to participate in this type of interaction with nucleic acids. Similar to our results, but using a FRET assay, Duca and colleagues observed inhibition in the presence of doxycycline, oxytetracycline, and minocycline, with IC50 values of 129–380 μM.42 While RNA binding was hypothesized to play a significant role in the mechanism-of-action,42 this was not definitively proven. To probe this possibility, we utilized the chemically modified tetracycline (CMT) derivative, CMT-3, which lacks antibacterial properties and was designed as a metalloproteinase inhibitor.43,44 Although this compound has been recently shown to bind to human 80S ribosome,33 because it lacks the dimethylamino group (Figures 5A), we suspected that its general RNA binding affinity would be weakened due to its absence of a basic amine. Indeed, we observed no binding of CMT-3 to either pre-miR-21 or pre-let-7d via SPR (sensorgrams can be found in Figure S2). Despite this, inhibition, with measured IC50 values very similar to that of methacycline (Figures 2 and 5B,C), was observed, indicating that RNA binding is likely not important for the mechanism of inhibition by the tetracyclines.
Figure 5.
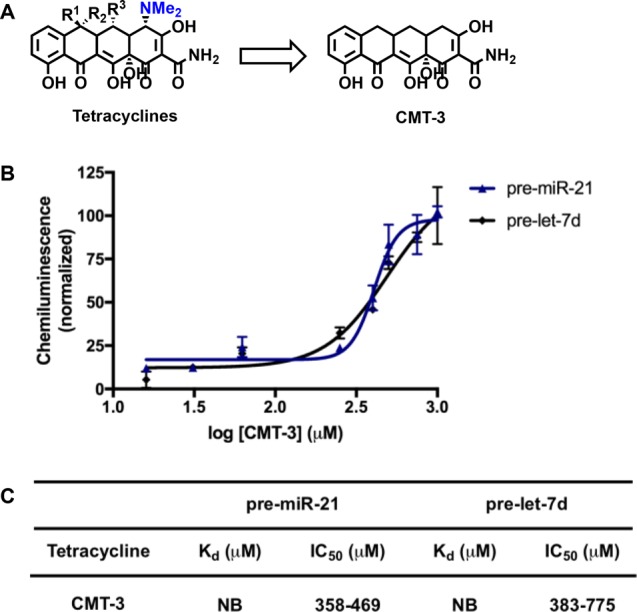
CMT-3. (A) Structure of CMT-3 in comparison to the tetracycline scaffold. (B) Plot of dose-dependent inhibition by CMT-3. (C) Table of binding affinities and inhibition constants (NB = no binding). The IC50 values are represented as 95% confidence intervals.
Aside from rRNA binding, another important activity of the tetracyclines is their ability to coordinate metal ions.35 In fact, the pharmacological actions of the tetracyclines are dependent upon metal complexes, most notably with Ca2+ and Mg2+.35 Because Dicer is a Mg2+-dependent RNase III enzyme,45 we first investigated whether the tetracyclines were scavenging this metal ion from the buffer, thereby preventing formation of an active Dicer enzyme. Even in the presence of lower or higher concentrations of MgCl2 (1–3 mM), methacycline was still able to inhibit Dicer activity (Figure S3A,B). Thus, these results demonstrate that the tetracyclines are not sequestering Mg2+. As many studies have found that Mg2+ and Ca2+ can form differential complexes with the tetracyclines,35,46 we next examined the effect of Ca2+ on methacycline inhibition. While testing in the presence of excess CaCl2 (2:1 CaCl2/MgCl2) was impossible due to the abolishment of Dicer function,47,48 Dicer activity and inhibition by methacycline in a 1:1 mixture of CaCl2 and MgCl2 was retained potentially indicating that both metal complexes are active (Figure S3C,D). Based on the inconclusive nature of these results, we prepared an acetyl protected tetracycline (Figure 6A) to more thoroughly probe the requirement of metal binding for tetracycline inhibitory activity. Using this analogue, Dicer inhibition was weakened (Figures 6B), providing some evidence for the role of metal chelation in tetracycline activity, although hydrogen-bonding interactions could also be disrupted with this analogue. Of note, this is not the first instance of a metal chelating group showing inhibition of Dicer.25
Figure 6.
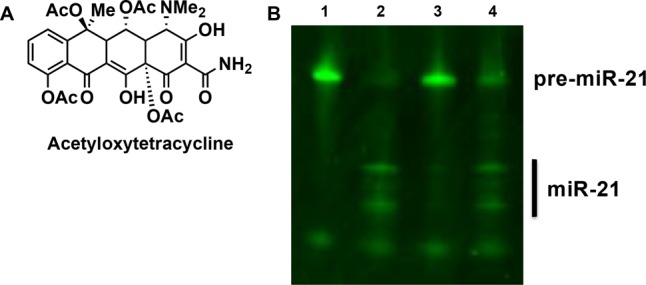
Acetyl-protected oxytetracycline. (A) Structure. (B) Inhibitory activity in an in-solution Dicer assay. Lane 1 = pre-miR-21 (500 nM); Lane 2 = pre-miR-21 + Dicer; Lane 3 = pre-miR-21 + Dicer and oxytetracycline (1.0 mM); Lane 4 = pre-miR-21 + Dicer and the aceylated oxytetracycline (1.0 mM). Two miRNA bands are observed due to the use of a modified pre-miR-21 substrate containing a 5′ biotin connected to the RNA via a PEG linker.10
Human Dicer contains multiple domains outside of its double-stranded RNA-binding domain (dsRBD) and RNase III domains to ensure proper miRNA processing; however, E. coli RNase III functions as a homodimer with only RNase III and dsRBD subunits.45 Thus, to determine which of Dicer’s domains may be the target of the tetracyclines, we tested the activity of methacycline against this homologue. As shown in Figure S4, this enzyme was inhibited by the tetracycline, indicating that the RNase III and dsRBD domains may be the target of these molecules. As the large size of Dicer (200 kDa) and its domains (>100 kDa), which also function as homodimers,49 preclude direct binding studies, future efforts will focus on employing structural biology strategies to decipher the mechanism-of-action of the tetracyclines in inhibiting human Dicer activity. However, because we consistently observed lower IC50 values for pre-miR-21 over pre-let-7d, we are excited about the possibility that the tetracyclines may be functionally inactivating the catalytic activity of the RNase III domains. Previous studies have found that the terminal loop region of a pre-miRNA controls the kinetics of Dicer processing, where hairpins of larger loop size (e.g., pre-let-7) are matured more rapidly than those with smaller loop sizes (e.g., pre-miR-21).50,51 Thus, using this model, inhibition of pre-miR-21 should be observed at lower concentrations due to its slower rate of processing, particularly for weakly potent inhibitors like the tetracyclines. Future explorations in this area will be reported in due course.
Finally, because tetracyclines are members of known RNA-targeted chemical space, we were eager to investigate how their activity compared to other well-established RNA-binding small molecules.3 The aminoglycoside antibiotics, streptomycin, kanamycin, and neomycin, and groove-binding bis-benzimidazole, Hoechst 33258 (Figure 7A), were examined at a single concentration using cat-ELCCA. Interestingly, despite the fact that these molecules displayed nanomolar binding affinity for both pre-miRNAs (Kd values of 100–700 nM; sensorgrams can be found in Figure S5), no inhibition of Dicer maturation was observed (Figure 7B). Thus, we hypothesize that these molecules either do not bind to the competitive Dicer cleavage site or do not induce a structural change in the hairpin structure by which to inhibit Dicer processing. These results, however, should provide caution to those researchers engaged in using binding only assays for targeting RNA with small molecules, and highlight the need for functional assays immediately downstream of such approaches, particularly prior to medicinal chemistry optimization of potency and specificity.
Figure 7.
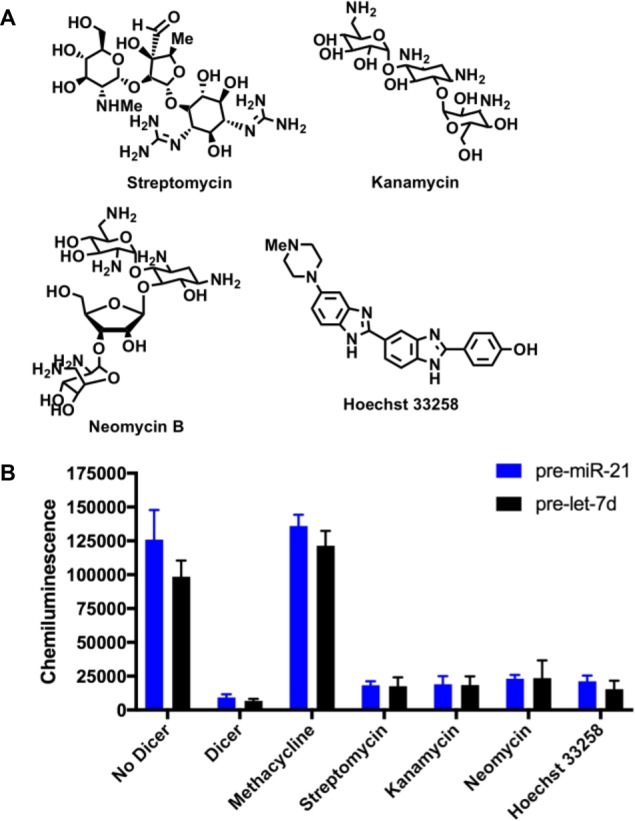
Comparison of the inhibitory activity of methacycline and other known RNA binders (1.0 mM) against Dicer-mediated processing of pre-miR-21 and pre-let-7d. (A) Structures. (B) Activity in cat-ELCCA.
In conclusion, we have characterized tetracycline-based antibiotics as inhibitors of Dicer-mediated pre-miRNA processing. Unexpectedly, despite their ability to directly bind to the RNA hairpins, the inhibitory activity of these compounds was not due to RNA binding capacity. By comparing tetracyclines to other members of known RNA-targeted chemical space, we discovered additional discrepancies between RNA binding and functional inhibitory properties. These combined findings highlight the complexity of targeting RNA with small molecules and the need for thorough characterization, both biochemical and biophysical, of such compounds to provide detailed mechanisms-of-action, particularly with weak binding ligands or potentially promiscuous scaffolds. Although our hit tetracyclines, methacycline and meclocycline, were not active in a cellular miRNA activity assay (Figure S6), further synthetic derivatizations could be carried out to improve activity.52 As tetracyclines have been recently shown to target human rRNA,33 which is much more abundant than miRNAs in a cell, comprising 80–85% and ≪1% of total cellular RNA, respectively,53 this may provide an explanation for this observation. Accordingly, the biological activities of the tetracyclines are likely not due to inhibition of miRNA maturation. Such optimized compounds could be useful probes to interrogate the basic biology of Dicer and its implications in human disease,54,55 in addition to aiding structural biology studies of human Dicer.
Glossary
ABBREVIATIONS
- miRNA or miR
microRNA
- HTS
high-throughput screening
- cat-ELCCA
catalytic enzyme-linked click chemistry assay
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.9b00091.
General materials and methods, assay protocols, synthetic methods, supplementary figures, NMR, and purity spectra (PDF)
Author Contributions
⊥ These authors contributed equally. The manuscript was written through contributions of D.A.L. and A.L.G. All authors have given approval to the final version of the manuscript.
This work was supported by a pilot grant from the University of Michigan Center for the Discovery of New Medicines and the NIH (R01 GM118329 to A.L.G., T32 GM008597 to J.S., and T32 GM007767 to E.E.G.).
The authors declare no competing financial interest.
Supplementary Material
References
- Garner A. L. RNA Therapeutics. Top. Med. Chem. 2018, 10.1007/978-3-319-68091-0. [DOI] [Google Scholar]
- Connelly C. M.; Moon M. H.; Schneekloth J. S. Jr. The emerging role of RNA as a therapeutic target for small molecules. Cell Chem. Biol. 2016, 23, 1077–1090. 10.1016/j.chembiol.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. R.; Hergenrother P. J. Targeting RNA with small molecules. Chem. Rev. 2008, 108, 1171–1224. 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- Vidigal J. A.; Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147. 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M.; Kim V. N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Li Z.; Rana T. M. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discovery 2014, 13, 622–638. 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- Rupaimoole R.; Slack F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discovery 2017, 16, 203–221. 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- Lorenz D. A.; Garner A. L. Approaches for the discovery of small molecule ligands targeting microRNAs. Top. Med. Chem. 2017, 27, 79–110. 10.1007/7355_2017_3. [DOI] [Google Scholar]
- Lorenz D. A.; Song J. M.; Garner A. L. High-throughput platform assay technology for the discovery of pre-microRNA-selective small molecule probes. Bioconjugate Chem. 2015, 26, 19–23. 10.1021/bc500544v. [DOI] [PubMed] [Google Scholar]
- Lorenz D. A.; Garner A. L. A click chemistry-based microRNA maturation assay optimized for high-throughput screening. Chem. Commun. 2016, 52, 8267–8270. 10.1039/C6CC02894B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner A. L. cat-ELCCA: catalyzing drug discovery through click chemistry. Chem. Commun. 2018, 54, 6531–6539. 10.1039/C8CC02332H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz D. A.; Vander Roest S.; Larsen M. J.; Garner A. L. Development and implementation of an HTS-compatible assay for the discovery of selective small-molecule ligands for pre-microRNAs. SLAS Disc. 2018, 23, 47–54. 10.1177/2472555217717944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcuklu S. D.; Donoghue M. T. A.; Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem. Soc. Trans. 2009, 37, 918–925. 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- Gumireddy K.; Young D. D.; Xiong X.; Hogenesch J. B.; Huang Q.; Deiters A. Small-molecule inhibitors of microRNA miR-21 function. Angew. Chem., Int. Ed. 2008, 47, 7482–7484. 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naro Y.; Thomas M.; Stephens M. D.; Connelly C. M.; Deiters A. Aryl amide small-molecule inhibitors of microRNA miR-21 function. Bioorg. Med. Chem. Lett. 2015, 25, 4793–4796. 10.1016/j.bmcl.2015.07.016. [DOI] [PubMed] [Google Scholar]
- Naro Y.; Ankenbruck N.; Thomas M.; Tivon Y.; Connelly C. M.; Gardner L.; Deiters A. Small molecule inhibition of microRNA miR-21 rescues chemosensitivity of renal-cell carcinoma to topotecan. J. Med. Chem. 2018, 61, 5900–5909. 10.1021/acs.jmedchem.7b01891. [DOI] [PubMed] [Google Scholar]
- Chirayil S.; Chirayil R.; Luebke K. J. Discovering ligands for a microRNA precursor with peptoid microarrays. Nucleic Acids Res. 2009, 37, 5486–5497. 10.1093/nar/gkp549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J. P.; Chirayil R.; Chirayil S.; Tom M.; Head K. J.; Luebke K. J. Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA 2014, 20, 528–539. 10.1261/rna.042911.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose D.; Nahar S.; Rai M. K.; Ray A.; Chakraborty K.; Maiti S. Selective inhibition of miR-21 by phage display screened peptide. Nucleic Acids Res. 2010, 43, 4342–4352. 10.1093/nar/gkv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose D.; Jayaraj G.; Suryawanshi H.; Agarwala P.; Pore S. K.; Banerjee R.; Maiti S. The tuberculosis drug streptomycin as a potential cancer therapeutic: inhibition of miR-21 function by directly targeting its precursor. Angew. Chem., Int. Ed. 2012, 51, 1019–1023. 10.1002/anie.201106455. [DOI] [PubMed] [Google Scholar]
- Ghosh A.; Degyatoreva N.; Kukielski C.; Story S.; Bhaduri S.; Maiti K.; Nahar S.; Ray A.; Arya D. P.; Maiti S. Targeting miRNA by tunable small molecule binders: peptidic aminosugar mediated interference in miR-21 biogenesis reverts epithelial to mesenchymal transition. MedChemComm 2018, 9, 1147–1154. 10.1039/C8MD00092A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z.; Zhang J.; Qian X.; Han L.; Zhang K.; Chen L.; Liu J.; Ren Y.; Yang M.; Zhang A.; Pu P.; Kang C. AC1MMYR2, an inhibitor of Dicer-mediated biogenesis of oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013, 73, 5519–5531. 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- Jiang C.-S.; Wang X.-M.; Zhang S.-Q.; Meng L.-S.; Zhu W.-H.; Xu J.; Lu S.-M. Discovery of 4-benzoylamino-N-(prop-2-yn-1-yl)benzamides as novel microRNA-21 inhibitors. Bioorg. Med. Chem. 2015, 23, 6510–6519. 10.1016/j.bmc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Connelly C. M.; Boer R. E.; Moon M. H.; Gareiss P.; Schneekloth J. S. Jr. Discovery of inhibitors of microRNA-21 processing using small molecule microarrays. ACS Chem. Biol. 2017, 12, 435–443. 10.1021/acschembio.6b00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.; Bhattarai U.; Guo Z.-F.; Liang F.-S. Regulating miRNA-21 biogenesis by bifunctional small molecules. J. Am. Chem. Soc. 2017, 139, 4987–4990. 10.1021/jacs.7b00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge M. D.; Walker M. J.; Pavelitz T.; Chen Y.; Yang W.; Varani G. A macrocyclic peptide ligand binds the oncogenic microRNA-21 precursor and suppresses Dicer processing. ACS Chem. Biol. 2017, 12, 1611–1620. 10.1021/acschembio.7b00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Costales M. G.; Vummidi B. R.; Nakai Y.; Angelbello A. J.; Tran T.; Haniff H. S.; Matsumoto Y.; Wang Z. F.; Chatterjee A. K.; Childs-Disney J. L.; Disney M. D. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem. Biol. 2018, 25, 1086–1094. 10.1016/j.chembiol.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen D. E.; Clemons W. M.; Carter A. P.; Morgan-Warren R. J.; Wimberly B. T.; Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 2000, 103, 1143–1154. 10.1016/S0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Chukwudi C. U. rRNA binding sites and the molecular mechanisms of action of the tetracyclines. Antimicrob. Agents Chemother. 2016, 60, 4433–4441. 10.1128/AAC.00594-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A. C.; Angelino E.; Kishony R. Chemical decay of an antibiotic inverts selection for resistance. Nat. Chem. Biol. 2010, 6, 105–107. 10.1038/nchembio.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014, 12, 35–48. 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- Hong S.; Harris K. A.; Fanning K. D.; Sarachan K. L.; Frohlich K. M.; Agris P. F. Evidence that antibiotics bind to human mitochondrial ribosomal RNA has implications for aminoglycoside toxicity. J. Biol. Chem. 2015, 290, 19273–19286. 10.1074/jbc.M115.655092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortison J. D.; Schenone M.; Myers J. A.; Zhang Z.; Chen L.; Ciarlo C.; Comer E.; Natchiar S. K.; Carr S. A.; Klaholz B. P.; Myers A. G. Tetracyclines modify translation by targeting key human rRNA substructures. Cell Chem. Biol. 2018, 25, 1506–1518. 10.1016/j.chembiol.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M. L.; Berniac J.; Liu Y. H.; Abato P.; Jodelka F. M.; Barthel L.; Kumar S.; Dudley C.; Nelson M.; Larson K.; Edmonds J.; Bowser T.; Draper M.; Higgins P.; Krainer A. R. Tetracyclines that promote SMN2 exon 7 splicing as therapeutics for spinal muscular atrophy. Sci. Transl. Med. 2009, 1, 5ra12. 10.1126/scitranslmed.3000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra W.; Silva-Caldeira P. P.; Terenzi H.; Pereira-Maia E. C. Impact of metal coordination on the antibiotic and non-antibiotic activities of tetracycline-based drugs. Coord. Chem. Rev. 2016, 327–328, 188–199. 10.1016/j.ccr.2016.04.009. [DOI] [Google Scholar]
- Sokoloski T. D.; Mitscher L. A.; Yuen P. H.; Juvarkar J. V.; Hoener B. Rate and proposed mechanism of anhydrotetracycline epimerization in acid solution. J. Pharm. Sci. 1977, 66, 1159–1165. 10.1002/jps.2600660829. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Wang J.; Cheng H.; Ke X.; Sun L.; Zhang Q. C.; Wang H.-W. Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate. Cell 2018, 173, 1191–1203. 10.1016/j.cell.2018.03.080. [DOI] [PubMed] [Google Scholar]
- Newman M. A.; Thomson J. M.; Hammond S. M. Lin-28 interaction with the let-7 precursor loop mediates regulated microRNA processing. RNA 2008, 14, 1539–1549. 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Yang F.; Zubovic L.; Pavelitz T.; Yang W.; Godin K.; Walker M.; Zheng S.; Macchi P.; Varani G. Targeted inhibition of oncogenic miR-21 maturation with designed RNA-binding proteins. Nat. Chem. Biol. 2016, 12, 717–723. 10.1038/nchembio.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunse C. E.; Michlewski G.; Hopp C. S.; Rentmeister A.; Caceres J. F.; Famulok M.; Mayer G. An aptamer targeting the apical-loop domain modulates pri-miRNA processing. Angew. Chem., Int. Ed. 2010, 49, 4674–4677. 10.1002/anie.200906919. [DOI] [PubMed] [Google Scholar]
- Maiti M.; Nauwelaerts K.; Herdewijn P. Pre-microRNA binding aminoglycosides and antitumor drugs as inhibitors of Dicer catalyzed microRNA processing. Bioorg. Med. Chem. Lett. 2012, 22, 1709–1711. 10.1016/j.bmcl.2011.12.103. [DOI] [PubMed] [Google Scholar]
- Tran T. P. A.; Vo D. D.; Di Giorgio A.; Duca M. Ribosome-targeting antibiotics as inhibitors of oncogenic microRNAs biogenesis: old scaffolds for new perspectives in RNA targeting. Bioorg. Med. Chem. 2015, 23, 5334–5344. 10.1016/j.bmc.2015.07.062. [DOI] [PubMed] [Google Scholar]
- Lokeshwar B. L.; Escatel E.; Zhu B. Cytotoxic activity and inhibition of tumor cell invasion by derivatives of a chemically modified tetracycline CMT-3 (COL-3). Curr. Med. Chem. 2001, 8, 271–279. 10.2174/0929867013373516. [DOI] [PubMed] [Google Scholar]
- Golub L. M.; McNamara T. F.; D’Angelo G. D.; Greenwald R. A.; Ramamurthy N. S. A non-antibacterial chemically-modified tetracycline inhibits mammalian collagenase activity. J. Dent. Res. 1987, 66, 1310–1314. 10.1177/00220345870660080401. [DOI] [PubMed] [Google Scholar]
- Zhang H.; Kolb F. A.; Jaskiewicz L.; Westhof E.; Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Lambs L.; Decock-Le Reverend B.; Kozlowski H.; Berthon G. Metal ion-tetracycline interactions in biological fluids. 9. circular dichroism spectra of calcium and magnesium complexes with tetracycline, oxytetracycline, doxycycline and chlortetracycline and discussion of their binding modes. Inorg. Chem. 1988, 27, 3001–3012. 10.1021/ic00290a022. [DOI] [Google Scholar]
- Provost P.; Dishart D.; Doucet J.; Frendewey D.; Samuelsson B.; Raedmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002, 21, 5864–5874. 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A.; Skakun V. V.; Ziegelmuller P.; Hahn U. A fluorescence correlation spectroscopy-based enzyme assay for human Dicer. Biol. Chem. 2012, 393, 187–193. 10.1515/hsz-2011-0202. [DOI] [PubMed] [Google Scholar]
- Takeshita D.; Zenno S.; Lee W. C.; Nagata K.; Saigo K.; Tanokura M. Homodimeric structure and double-straded RNA cleavage activity of the C-terminal RNase III domain of human Dicer. J. Mol. Biol. 2007, 374, 106–120. 10.1016/j.jmb.2007.08.069. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010, 38, 7689–7697. 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y.; Zhang X.; Graves P.; Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA 2012, 18, 2083–2092. 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.; Myers A. G. Development of a platform for the discovery and practical synthesis of new tetracycline antibiotics. Curr. Opin. Chem. Biol. 2016, 32, 48–57. 10.1016/j.cbpa.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Jankowsky E.; Harris M. E. Specificity and non-specificity in RNA-protein interactions. Nat. Rev. Mol. Cell Biol. 2015, 16, 533–544. 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.-S.; Rossi J. J. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem. J. 2017, 474, 1603–1618. 10.1042/BCJ20160759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes W. D.; Priest J. R.; Duchaine T. F. DICER1: mutations, microRNAs and mechanisms. Nat. Rev. Cancer 2014, 14, 662–672. 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.