Abstract
Purpose
Neurosarcoidosis and primary central nervous system lymphomas, although distinct disease entities, can both have overlapping neuroimaging findings. The purpose of our preliminary study was to assess if magnetic resonance texture analysis can differentiate parenchymal mass-like neurosarcoidosis granulomas from primary central nervous system lymphomas.
Methods
A total of nine patients was evaluated, four with parenchymal neurosarcoidosis granulomas and five with primary central nervous system lymphomas. Magnetic resonance texture analysis was performed with commercial software using a filtration histogram technique. Texture features of different sizes and variations in signal intensity were extracted at six different spatial scale filters, followed by feature quantification using statistical and histogram parameters and 36 features were analysed for each sequence (T1-weighted, T2-weighted, fluid-attenuated inversion recovery, diffusion-weighted, apparent diffusion coefficient, T1-post contrast). The non-parametric Mann–Whitney test was used to evaluate the differences between different texture parameters.
Results
The differences in distribution of entropy on T2-weighted imaging, apparent diffusion coefficient and T1-weighted post-contrast images were statistically significant on all spatial scale filters. Magnetic resonance texture analysis using medium and coarse spatial scale filters was especially useful in discriminating neurosarcoidosis from primary central nervous system lymphomas for mean, mean positive pixels, kurtosis, and skewness on diffusion-weighted imaging (P < 0.004–0.030). At spatial scale filter 5, entropy on T2-weighted imaging (P = 0.001) was the most useful discriminator with a cut-off value of 6.12 (P = 0.001, area under the curve (AUC)-1, sensitivity (Sn)-100%, specificity (Sp)-100%), followed by kurtosis and skewness on diffusion-weighted imaging with a cut-off value of −0.565 (P = 0.011, AUC-0.97, Sn-100%, Sp-83%) and–0.365 (P = 0.008, AUC-0.98, Sn-100%, Sp-100%) respectively.
Conclusion
Filtration histogram-based magnetic resonance texture analysis appears to be a promising modality to distinguish parenchymal neurosarcoidosis granulomas from primary central nervous system lymphomas.
Keywords: Texture analysis, MRI, neurosarcoidosis, primary central nervous system lymphoma, histogram
Introduction
Sarcoidosis is a well-described idiopathic inflammatory disease, with a global incidence of 10–20 cases per 100,000. It is more common in certain parts of the world and is about four times more frequent in African-Americans.1,2 Involvement of the central nervous system (CNS), also referred to as neurosarcoidosis (NS), is seen in about 5% of cases clinically.3 Primary central nervous system lymphoma (PCNSL), on the other hand, accounts for about 5% of all brain tumours.4 Although pathologically distinct, NS and PCNSL share multiple imaging findings such as parenchymal lesions, perivascular enhancement, leptomeningeal involvement, etc. and are often difficult to distinguish on neuroimaging.3,4 This is of obvious diagnostic significance because both entities can have overlapping clinical presentations and both respond to steroids, but overall they have different clinical courses and therapeutic implications.
Texture analysis encompasses a number of mathematical methods to analyse the distribution of pixels within an image to derive texture features.5 It is based on the premise that mathematical image processing can provide quantifiable information about biological behaviour and heterogeneity. Texture analysis can therefore indirectly provide additional insights into the microstructural heterogeneity beyond human visual perception.6 Texture analysis may be performed using different methods such as statistical, model, and transform-based methods. Although not a new technique per se, it has re-emerged as a potential problem-solving modality over recent years, not just in magnetic resonance imaging (MRI), but also in computed tomography (CT) and positron emission tomography-CT studies.6,7 Recent studies using magnetic resonance texture analysis (MRTA) have shown promise in differentiating between PCNSL and glioblastoma.8–10 We hypothesised that MRTA could differentiate PCNSL from mass-like NS granulomas, as these are biologically different processes.
Purpose
The purpose of our preliminary study was to determine the feasibility of MRTA in differentiating parenchymal mass-like NS granulomas from PCNSL.
Materials and methods
Patients
Our departmental radiology neuroimaging MRI database was retrospectively searched, examining a period of 15 years (2002–2017), using the keywords ‘sarcoidosis’ and ‘neurosarcoidosis’ anywhere in the provided history or the final report. The study was approved by the institutional review board. This revealed a total of 49 patients who eventually met the criteria for NS. Enhancing parenchymal granulomas were seen in about 20% (10/49) patients. Patients with lesions less than 1 cm in maximal dimension were excluded for the purpose of MRTA (to ensure sufficient pixels for analysis and better statistical accuracy). A total of six lesions in four patients (three with a solitary lesion, and one with three lesions) were eventually selected for analysis.
For the PCNSL patients, the radiology neuroimaging database was searched using the keywords ‘lymphoma’ and ‘PCNSL’ among patients presenting to our institute from 2008 to 2017. The inclusion criteria were the presence of an index contrast-enhanced magnetic resonance study that was performed at our institution, the presence of a parenchymal mass lesion greater than 1 cm in the shortest dimension, and the absence of motion artifacts. From among this population, five consecutive cases of PCNSL were selected. The clinical, epidemiological and imaging details were collected for these nine patients.
Image acquisition
All patients were examined on a 1.5T system using a standard head coil (Siemens, Erlangen, Germany). The routine protocol for clinical brain MRI study at our institute includes axial T1-weighted (T1W), T2-weighted (T2W), fluid-attenuated inversion recovery (FLAIR), diffusion-weighted imaging (DWI), GRE and tri-planar post-contrast images. For the purpose of MRTA, we used the T1WI (TR/TE/TI: 1950/10/840, NEX: 2, slice thickness: 5 mm, matrix: 320 × 256, field of vision (FOV) 240 mm, pixel size 0.75 mm), T2WI (TR/TE: 4000/89, NEX: 2, slice thickness: 5 mm, matrix: 512 × 408, FOV 240 mm, pixel size 0.5 mm); FLAIR (TR/TE/TI: 9000/109/2500, NEX: 1, slice thickness: 5 mm, matrix: 384 × 308, FOV 240 mm, pixel size 0.6 mm), and DWI (TR/TE: 2900/79, NEX:1, slice thickness: 5 mm, matrix: 128 × 128, FOV 240 mm, pixel size 1.8 mm). Post-contrast images (TR/TE: 462/11, NEX: 2, slice thickness: 5 mm, matrix: 320 × 256, FOV 240 mm, pixel size 0.75 mm) were acquired after administration of gadobutrol contrast (Gadavist; Bayer Healthcare Pharma, Berlin, Germany) for eight lesions and after administration of gadobentate dimeglumine (Multihance; Bayer Healthcare Pharma) for three lesions. The contrast dose was 0.1 mL/kg body weight for the former and 0.2 mL/kg for the latter.
Magnetic resonance texture analysis
The contrast-enhanced image with the largest axial cross-section of the lesion, and the corresponding T1WI, T2WI, FLAIR, DWI (b value 1000) and apparent diffusion coefficient (ADC) images, were selected. MRTA was performed using commercially available software (TexRAD version 3.3, www.texrad.com; Feedback Medical Ltd., Cambridge, UK). Lesion segmentation on individual images was performed by utilising the software’s free-hand drawing tool. The regions of interest were drawn by one user (RE) under the supervision of a fellowship-trained neuroradiologist with 6 years post-fellowship experience, and an MRTA expert (BG) with 10 years of experience, without knowledge of the final histopathological diagnosis. The surrounding oedema was not included in the MRTA.
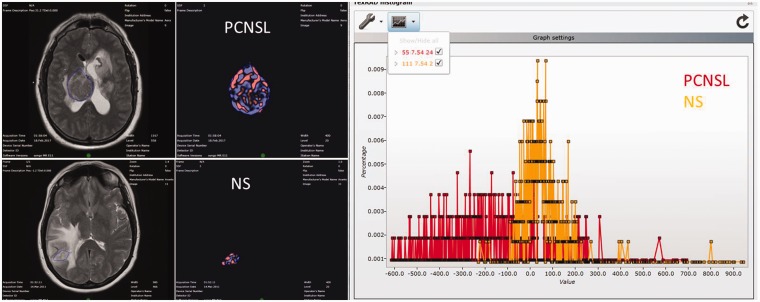
The MRTA methodology consisted of a filtration histogram technique, which extracts and enhances texture features of different sizes and signal intensity within the region of interest, followed by histogram quantification (Figure 1). The initial filtration step uses a Laplacian of Gaussian (LoG) band-pass filtration (similar to non-orthogonal wavelet) to extract and enhance image features of different size and variation in signal intensity, corresponding to spatial scale filter (SSF) values of 0, 2, 3, 4, 5 and 6 mm in width, where SSF = 0 corresponds to conventional image with no filtration. The different SSF values correspond to different texture scale such as fine (SSF = 2), medium (SSF = 3–5), and coarse (SSF = 6). SSF values lower than 2 tend to represent image noise and were not considered. Following feature extraction, quantification of texture was performed using statistical and histogram shape and distribution parameters (mean, standard deviation, entropy, mean positive pixels (MPPs), skewness and kurtosis) for each sequence in both groups of patients.
Figure 1.
Illustrative example of the magnetic resonance texture analysis (MRTA) methodology. The far-left column demonstrates the axial T2-weighted image (T2WI) with the region of interest (ROI) outlined, based on the post-contrast images. The middle column demonstrates the filtered texture images. The right-sided image shows the histogram distribution within the ROI.
Statistical analysis
Non-parametric Mann–Whitney test was performed to evaluate for statistical significance for various MRTA parameters. Receiver operating characteristic (ROC) analysis was performed and area under the curve (AUC) calculated to seek the best cut-off values of MRTA variables to differentiate PCNSL from NS. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) and its 95% confidence intervals (CIs) were calculated for the cut-off values. A P value less than 0.05 was considered statistically significant. Statistical analysis was performed using SPSS (IBM SPSS Statistics for Windows, version 24; IBM Corp., Armonk, NY, USA).
Results
The average age of the NS patients was 37.8 years (range 28–55 years) and for the PCNSL patients it was 64.8 years (range 44–83 years). The clinical and imaging details of all patients are presented in Table 1. The distribution of entropy on T2WI, ADC, and T1W post-contrast images were statistically significant on all spatial scale filters (fine, medium, and coarse). The statistically significant differences are presented in Table 2.
Table 1.
Clinical and epidemiological details of the analysed patients with primary central nervous system lymphoma (PCNSL) and neurosarcoidosis (NS).
| Age (yrs) | Gender | Location | Immuno-compromised | Contrast | |
|---|---|---|---|---|---|
| PCNSL | |||||
| P1 | 82.6 | F | R temporal | N | MH |
| P2 | 63.6 | M | R middle cerebral peduncle | N | GV |
| P3 | 66 | F | Bil-parietal lobes [R > L] | N | GV |
| P4 | 44.6 | M | Multiple periventricular | N | GV |
| P5 | 67.2 | F | L frontal | N | MH |
| Neurosarcoidosis | |||||
| P1 | 35.2 | F | R parietal | N | MH |
| P2 | 32 | M | Hypothalamus & L temporal | N | GV |
| P3 | 28.7 | M | N | GV | |
| L1 | 28.7 | M | R basal ganglia | N | GV |
| L2 | L basal ganglia | GV | L basal ganglia | GV | |
| L3 | Midbrain | GV | Mid brain | GV | |
| P4 | 55.4 | F | L frontal | N | GV |
F: female; M: male; N: normal; GV: gadavist; L: left; MH: multihance; R: right.
Table 2.
Median values and distribution parameters for the statistically significant discriminators between primary central nervous system lymphoma (PCNSL) and neurosarcoidosis (NS) at various spatial scale filters (SSF) values.
| Parameter | Median value PCNSL | Median value NS | P value |
|---|---|---|---|
| SSF = 0 | |||
| Entropy_ADC | 4.8 | 4.2 | 0.017 |
| Mean-T2 | 449 | 677 | 0.030 |
| Entropy-T2 | 5.6 | 5.1 | 0.017 |
| MPP-T2 | 449 | 677 | 0.030 |
| SSF = 2 | |||
| Mean-T1PC | 148.6 | 249.3 | 0.030 |
| Entropy-TIPC | 6.1 | 5.8 | 0.009 |
| Entropy-ADC | 4.9 | 4.4 | 0.017 |
| Entropy-T2 | 6.2 | 5.7 | 0.004 |
| SSF = 3 | |||
| Entropy-T1PC | 6.2 | 5.8 | 0.030 |
| MPP-T1PC | 522.7 | 646.9 | 0.030 |
| Entropy-ADC | 4.9 | 4.3 | 0.017 |
| Entropy-T2 | 6.3 | 5.7 | 0.004 |
| Kurtosis-DWI | 1.2 | –0.2 | 0.030 |
| SSF = 4 | |||
| Entropy-ADC | 4.9 | 4.3 | 0.017 |
| Entropy-T2 | 6.4 | 5.6 | 0.004 |
| Skewness-T1 | 0.5 | –0.6 | 0.017 |
| Kurtosis-DWI | 0.6 | –0.5 | 0.017 |
| SSF = 5 | |||
| Entropy-T1PC | 6.2 | 5.8 | 0.030 |
| Entropy-ADC | 4.9 | 4.3 | 0.017 |
| Entropy-T2 | 6.3 | 5.6 | 0.004 |
| Skewness-T1 | 0.7 | –0.6 | 0.030 |
| MPP-DWI | 49.1 | 31.2 | 0.017 |
| Skewness-DWI | 0.9 | –0.5 | 0.004 |
| Kurtosis-DWI | 0.1 | –0.7 | 0.009 |
| SSF = 6 | |||
| Entropy-T1PC | 6.2 | 5.8 | 0.017 |
| Entropy-ADC | 4.9 | 4.3 | 0.017 |
| Entropy-T2 | 6.3 | 5.7 | 0.004 |
| Mean-DWI | 61 | –17.1 | 0.030 |
| MPP-DWI | 79.7 | 37.8 | 0.017 |
| Kurtosis-DWI | –0.1 | –0.9 | 0.030 |
ADC: apparent diffusion coefficient; MPP: mean positive pixels; TIPC: T1 post-contrast image; DWI: diffusion weighted imaging; SSF: spatial scale filters.
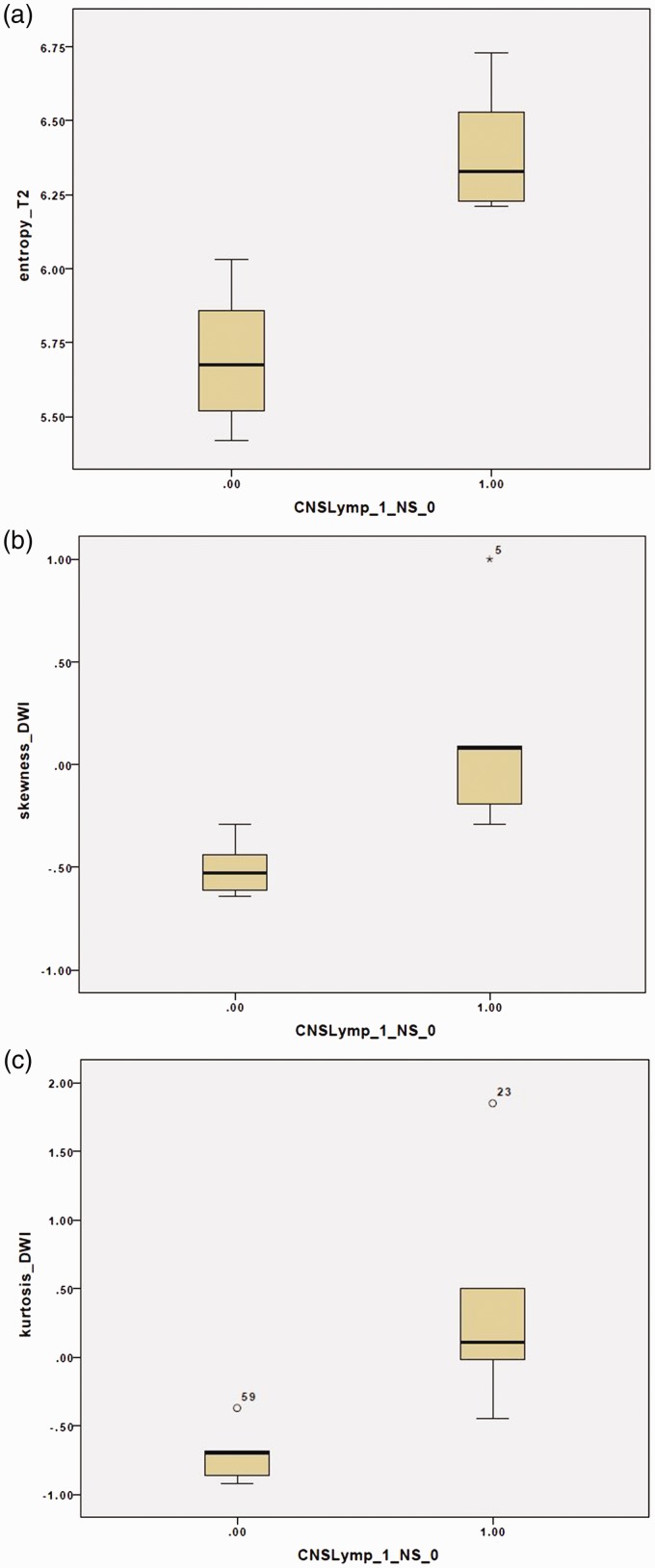
For medium and coarse filters (SSF 5, 6), significant differences were also noted in the kurtosis and MPP on DWI (P values between 0.009 and 0.030). In addition, SSF 5 also showed significant difference in the skewness on DWI (P = 0.015). The discrimination between NS and PCNSL lesions was statistically more robust on higher SSF. In fact, the texture analysis at SSF 5 showed the maximal number of discriminant parameters, followed by coarse texture (SSF 6). Figure 2 shows the box plots for distribution of entropy on T2WI and skewness and kurtosis on DWI at SSF 5.
Figure 2.
(a–c) Box plots of the T2-weighted image (T2WI) entropy (a), diffusion-weighted imaging (DWI) skewness (b) and kurtosis (c) showing the differences in these variables at spatial scale filter (SSF) = 5.
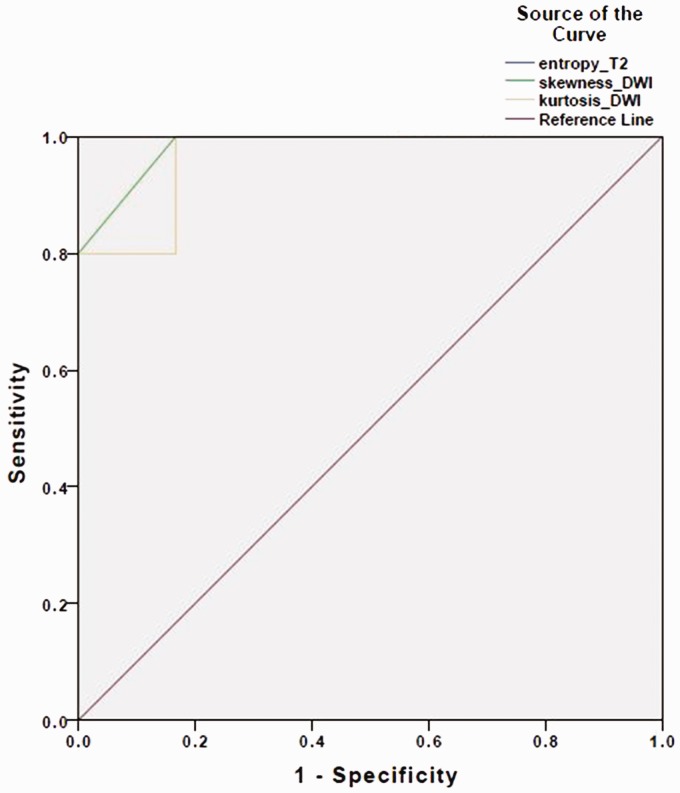
The diagnostic accuracy of the MRTA variables was assessed, and the best possible cut-off value and corresponding diagnostic accuracy were calculated. A ROC analysis for the variables at SSF = 5 revealed that the distribution of entropy on T2WI was the most useful discriminator, followed by kurtosis and skewness on DWI (Figure 3). The mean entropy of PCNSL was significantly higher as compared to NS (6.41 ± 0.22 vs. 5.70 ± 0.23, P = 0.001). Entropy was a perfect diagnostic method to detect the PCNSL (AUC = 1.00, 95% CI 1.00–1.00, P = 0.001). Entropy value 6.12 was the best cut-off value with sensitivity 100% (95% CI 46–100%), specificity 100% (95% CI 52–100%), PPV 100% (95% CI 46–100%), NPV 100% (95% CI 52–100%). Mean skewness of PCNSL was significantly higher than NS (0.14 ± 0.51 vs. −0.51 ± 0.13, P = 0.015). Skewness was identified as a good predictor of the PCNSL (AUC 0.98, 95% CI 0.92–1.00, P = 0.008). Skewness value −0.365 was chosen as the best cut-off value with sensitivity 100% (95% CI 46–100%), specificity 100% (95% CI 52–100%), PPV 100% (95% CI 46–100%), NPV 100% (95% CI 52–100%). The mean kurtosis of PCNSL was significantly higher than NS (0.40 ± 0.88 vs. −0.71 ± 0.19, P = 0.015). Kurtosis was found to be a good predictor (AUC 0.97, 95% CI 0.87–1.00, P = 0.011). Kurtosis −0.565 was considered as the best cut-off value with sensitivity 100% (95% CI 46–100%), specificity 83% (95% CI 36–99%), PPV 83% (95% CI 37–99%), NPV 100% (95% CI 46–100%).
Figure 3.
Receiver operating characteristic (ROC) analysis for the same variables as noted in Figure 2. Entropy on T2-weighted imaging (T2WI) appears to be the most accurate with area under the curve (AUC) of 1.
Discussion
The current proof-of-concept study provides preliminary evidence on the usefulness of MRTA in distinguishing PCNSL from larger NS lesions. Even though these entities are pathologically distinct, NS being inflammatory and PCNSL being a malignant process, they can have similar imaging findings. For example, they both have parenchymal-enhancing lesions, dural-based lesions, and even leptomeningeal and cranial nerve involvement.3,4 In addition, isolated CNS involvement may occur in sarcoidosis in about 20%, rendering distinction from PCNSL very challenging, and often necessitating neural biopsy.1,3 The distinction is, therefore, of obvious therapeutic and prognostic significance.
MRTA has previously shown promise in neuro-oncological applications such as differentiating between low and high-grade gliomas, between tumour infiltration and vasogenic oedema, and also between glioblastoma and PCNSL.8,9,11–13 MRTA has also shown potential in evaluating neuro-inflammatory conditions such as multiple sclerosis.10,14–16 However, the feasibility of MRTA in distinguishing between parenchymal NS granulomas and PCNSL has not been explored.
We noted significant differences in entropy on the ADC and T2W images at all SSF values. In addition, there were significant differences between the entropy values on the post-contrast T1W images at SSF values of 2, 3, 4 and 6. In all cases, PCNSL lesions showed higher entropy when compared to NS. In texture analysis, entropy is a measure of irregularity of gray levels within the matrix.7,15 A higher entropy signifies lesion heterogeneity and is often elevated in malignant lesions. CT-based texture analysis studies have also previously shown elevated entropy in higher-grade gliomas.17 Our findings are consistent with these observations, as PCNSL lesions showed higher entropy.
Similarly, kurtosis is a reflection of the magnitude of pixel distribution, and it reflects the peakedness of the histogram (tissue contrast). We noted higher kurtosis values on the DWI at SSF values of 3–6 (medium and coarse textures). A higher kurtosis reflects tumour heterogeneity and is more in line with a malignant behaviour.7,18
Additional MRTA parameters that were statistically significant at medium SSF values (SSF = 4, 5), pertained to the skewness of the histogram on the T1W and DWI. Skewness is essentially a measure of the asymmetry of a histogram. A zero value signifies even distribution on both sides of the mean. A positive skewness indicates preponderance of bright objects (another component of tumour heterogeneity) and is more in line with a malignant lesion, similar to the findings observed in our cases.7,18–20
It is worth noting here that our methodology employed first-order histogram statistical parameters that are directionally independent, and thus are less complex and easier to reproduce, unlike second-order statistical parameters, which are direction-oriented. In addition, unlike texture algorithms using a machine learning approach, our methodology is not resource-intensive and is not limited by lack of biological intuitiveness because the various quantifiable parameters have been shown by simulation studies to relate to biological heterogeneity, which in turn is related to underlying histology.6,8,12,18 The current methodology therefore is less influenced by issues related to reproducibility between centres.21
Limitations of our proof-of-concept study include a small sample size and its retrospective nature. One reason for the smaller population of NS patients in our study was the exclusion of cases with lesions smaller than 1 cm. However, this was necessary because smaller lesions are often not optimal for MRTA.22 A potential way around the problem of small sample size would be multicentre collaboration and pooling of such cases to generate more robust evidence. Another limitation is the sampling of one slice with the largest lesion area instead of the entire lesion volume. This is a more time and resource-intensive process and was therefore not performed for the purpose of our preliminary analysis. Even though inclusion of the entire lesion volume could improve the detection of lesion heterogeneity, we noted promising results with a single image analysis on different imaging sequences, similar to the experience of previous authors.23,24
Conclusion
Our preliminary study demonstrates that MRTA may be useful to distinguish mass-like NS lesions from PCNSL in immunocompetent patients, thereby implying that the two pathologically distinct entities with multiple overlapping imaging findings can potentially be differentiated non-invasively. This could possibly expand the role of MRTA in clinical neuroimaging and as a potential problem-solving modality in cases with non-specific imaging findings.
Conflict of interest
BG: Shareholder of Feedback Plc and employed by Feedback Medical Ltd (wholly owned by Feedback Plc), which manufactures and commercialises the tumour texture analysis software (TexRAD) described in this papert. GB, RE, NS: None.
Ethical approval
Retrospective study. Institutional review board approved. For this type of study formal consent is not required.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Fritz D, van de Beek D, Brouwer MC. Clinical features, treatment and outcome in neurosarcoidosis: systematic review and meta-analysis. BMC Neurol 2016; 16: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hebel R, Dubaniewicz-Wybieralska M, Dubaniewicz A. Overview of neurosarcoidosis: recent advances. J Neurol 2015; 262: 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bathla G, Singh AK, Policeni B, et al. Imaging of neurosarcoidosis: common, uncommon, and rare. Clin Radiol 2016; 71: 96–106. [DOI] [PubMed] [Google Scholar]
- 4.Bathla G, Hegde A. Lymphomatous involvement of the central nervous system. Clin Radiol 2016; 71: 602–609. [DOI] [PubMed] [Google Scholar]
- 5.Kato H, Kanematsu M, Zhang X, et al. Computer-aided diagnosis of hepatic fibrosis: preliminary evaluation of MRI texture analysis using the finite difference method and an artificial neural network. AJR Am J Roentgenol 2007; 189: 117–122. [DOI] [PubMed] [Google Scholar]
- 6.Kassner A, Thornhill RE. Texture analysis: a review of neurologic MR imaging applications. AJNR Am J Neuroradiol 2010; 31: 809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging 2012; 3: 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcaide-Leon P, Dufort P, Geraldo AF, et al. Differentiation of enhancing glioma and primary central nervous system lymphoma by texture-based machine learning. AJNR Am J Neuroradiol 2017; 38: 1145–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y-h, Muftah M, Das T, et al. Classification of MR tumor images based on Gabor wavelet analysis. J Med Biol Eng 2012; 32: 22–28. [Google Scholar]
- 10.Verma RK, Wiest R, Locher C, et al. Differentiating enhancing multiple sclerosis lesions, glioblastoma, and lymphoma with dynamic texture parameters analysis (DTPA): a feasibility study. Med Phys 2017; 44: 4000–4008. [DOI] [PubMed] [Google Scholar]
- 11.Artzi M, Liberman G, Blumenthal DT, et al. Differentiation between vasogenic edema and infiltrative tumor in patients with high-grade gliomas using texture patch-based analysis. J Magn Reson Imaging 2018; 48: 729–36. [DOI] [PubMed] [Google Scholar]
- 12.Skogen K, Schulz A, Dormagen JB, et al. Diagnostic performance of texture analysis on MRI in grading cerebral gliomas. Eur J Radiol 2016; 85: 824–829. [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki T, Chen T, Hirai T, et al. Classification of cerebral lymphomas and glioblastomas featuring luminance distribution analysis. Comput Math Methods Med 2013; 2013: 619–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbasian Ardakani A, Gharbali A, Saniei Y, et al. Application of texture analysis in diagnosis of multiple sclerosis by magnetic resonance imaging. Glob J Health Sci 2015; 7: 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Michoux N, Guillet A, Rommel D, et al. Texture analysis of T2-weighted MR images to assess acute inflammation in brain MS lesions. PLoS One 2015; 10: e0145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y. MRI texture analysis in multiple sclerosis. Int J Biomed Imaging 2012; 2012: 762804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skogen K, Ganeshan B, Good T, et al. Imaging heterogeneity in gliomas using texture analysis. Cancer Imaging 2011; 11: S113. [Google Scholar]
- 18.Miles KA, Ganeshan B, Hayball MP. CT texture analysis using the filtration-histogram method: what do the measurements mean? Cancer Imaging 2013; 13: 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castellano G, Bonilha L, Li LM, et al. Texture analysis of medical images. Clin Radiol 2004; 59: 1061–1069. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z, Tang LH, Klimstra DS. Effect of tumor heterogeneity on the assessment of Ki67 labeling index in well-differentiated neuroendocrine tumors metastatic to the liver: implications for prognostic stratification. Am J Surg Pathol 2011; 35: 853–860. [DOI] [PubMed] [Google Scholar]
- 21.Gourtsoyianni S, Ljungqvist G, Khan A, et al. Reproducibility of MRI texture analysis in primary rectal cancer, Vienna, Austria: European Society of Radiology, 2013. [Google Scholar]
- 22.Andrés Larroza, Vicente Bodí and David Moratal. Texture Analysis in Magnetic Resonance Imaging: Review and Considerations for Future Applications, Assessment of Cellular and Organ Function and Dysfunction using Direct and Derived MRI Methodologies, Christakis Constantinides, IntechOpen, DOI: 10.5772/64641. Available from: https://www.intechopen.com/books/assessment-of-cellular-and-organ-function-and-dysfunction-using-direct-and-derived-mri-methodologies/texture-analysis-in-magnetic-resonance-imaging-review-and-considerations-for-future-applications (accessed 6 February 2019).
- 23.Ryu YJ, Choi SH, Park SJ, et al. Glioma: application of whole-tumor texture analysis of diffusion-weighted imaging for the evaluation of tumor heterogeneity. PLoS One 2014; 9: e108335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng F, Kozarski R, Ganeshan B, et al. Assessment of tumor heterogeneity by CT texture analysis: can the largest cross-sectional area be used as an alternative to whole tumor analysis? Eur J Radiol 2013; 82: 342–348. [DOI] [PubMed] [Google Scholar]