Key Points
Question
Can crowd innovation be used to rapidly prototype artificial intelligence (AI) solutions that automatically segment lung tumors for radiation therapy targeting, and can AI performance match expert radiation oncologists for this time- and training-intensive task?
Findings
A 3-phase, prize-based crowd innovation challenge over 10 weeks, including 34 contestants who submitted 45 algorithms, identified multiple AI solutions that replicated the accuracy of an expert radiation oncologist in targeting lung tumors and performed the task more rapidly.
Meaning
On-demand, crowdsourcing methods can be used to rapidly prototype AI algorithms to replicate and transfer expert skill and knowledge to underresourced health care settings and improve the quality of radiation therapy globally.
This study compares a crowdsourced artificial intelligence solution for segmenting lung tumors for radiation therapy with expert radiation oncologists.
Abstract
Importance
Radiation therapy (RT) is a critical cancer treatment, but the existing radiation oncologist work force does not meet growing global demand. One key physician task in RT planning involves tumor segmentation for targeting, which requires substantial training and is subject to significant interobserver variation.
Objective
To determine whether crowd innovation could be used to rapidly produce artificial intelligence (AI) solutions that replicate the accuracy of an expert radiation oncologist in segmenting lung tumors for RT targeting.
Design, Setting, and Participants
We conducted a 10-week, prize-based, online, 3-phase challenge (prizes totaled $55 000). A well-curated data set, including computed tomographic (CT) scans and lung tumor segmentations generated by an expert for clinical care, was used for the contest (CT scans from 461 patients; median 157 images per scan; 77 942 images in total; 8144 images with tumor present). Contestants were provided a training set of 229 CT scans with accompanying expert contours to develop their algorithms and given feedback on their performance throughout the contest, including from the expert clinician.
Main Outcomes and Measures
The AI algorithms generated by contestants were automatically scored on an independent data set that was withheld from contestants, and performance ranked using quantitative metrics that evaluated overlap of each algorithm’s automated segmentations with the expert’s segmentations. Performance was further benchmarked against human expert interobserver and intraobserver variation.
Results
A total of 564 contestants from 62 countries registered for this challenge, and 34 (6%) submitted algorithms. The automated segmentations produced by the top 5 AI algorithms, when combined using an ensemble model, had an accuracy (Dice coefficient = 0.79) that was within the benchmark of mean interobserver variation measured between 6 human experts. For phase 1, the top 7 algorithms had average custom segmentation scores (S scores) on the holdout data set ranging from 0.15 to 0.38, and suboptimal performance using relative measures of error. The average S scores for phase 2 increased to 0.53 to 0.57, with a similar improvement in other performance metrics. In phase 3, performance of the top algorithm increased by an additional 9%. Combining the top 5 algorithms from phase 2 and phase 3 using an ensemble model, yielded an additional 9% to 12% improvement in performance with a final S score reaching 0.68.
Conclusions and Relevance
A combined crowd innovation and AI approach rapidly produced automated algorithms that replicated the skills of a highly trained physician for a critical task in radiation therapy. These AI algorithms could improve cancer care globally by transferring the skills of expert clinicians to under-resourced health care settings.
Introduction
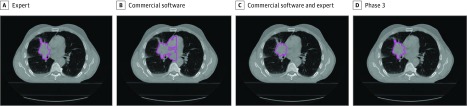
Lung cancer remains the second most common cancer, and leading cause of cancer mortality, in the United States1 with approximately 150 000 deaths estimated in 2018. Radiation therapy (RT) plays a critical role in the treatment of this disease, and 20% of early stage (I-II) and 50% of advanced stage (III-IV) lung cancer patients receive RT2 with projections of approximately 96 000 patients requiring this treatment modality in 2020.2,3 The precise and accurate volumetric segmentation of tumors, which determines where the radiation dose is delivered into the patient, is a critical part of RT targeting and planning, and has a direct impact on tumor control and radiation-induced toxic effects. Typically, tumor segmentation is performed manually, slice-by-slice on computed tomography (CT) scans by trained radiation oncologists (Figure 1) (eFigure 1 in the Supplement) and can be extremely time consuming. However, there is significant interobserver variation even among experts (eg, 7-fold variation among 5 experts in 1 study),4,5 and the quality of segmentation may directly impact clinical outcomes.6,7,8 Even in prospective clinical trials with prespecified RT parameters, major RT planning deviations occur in 8% to 71% of patients and are associated with increased mortality and treatment failure.9
Figure 1. Lung Tumor Segmentations.
Example of a human expert segmentation (A) compared with automated segmentation from commercially-available region growing–based segmentation technique before (B) and after (C) human adjustment of settings, and automated segmentations from the top artificial intelligence algorithm from phase 3 of the contest (D).
Unlike cancer image analysis for diagnostic purposes, which produces a single binary answer (yes or no) to a single question (“Is a mass present?”), therapeutic tumor segmentation involves interpretation of medical imaging on a voxel-by-voxel basis to classify cancer vs normal organ and incorporates an intrinsic risk-benefit assessment of where the radiation dose is to be delivered. For an expert radiation oncologist, this requires both training and intuition, and experience may directly impact lung cancer outcomes.10 However, this critical human resource is not accessible to many underserved patients in both the United States and globally.11,12 Although approximately 58% of lung cancer cases occur in less developed countries,13 these countries have a staggering shortage of radiation oncologists, with an estimated 23 952 radiation oncologists required in 84 low- and middle-income countries by 2020 yet only 11 803 were available in 2012.14
We used a novel combined approach of crowd innovation and artificial intelligence (AI) to address this unmet need in global cancer care. Crowd innovation has been successfully applied to a variety of genomic and computational biology problems by using prize-based competitions to identify extreme value solutions that outperform those developed by conventional academic approaches.15,16,17,18 Online contests expand the pool of potential problem solvers substantially beyond traditional academic expert circles to include individuals with a more diverse set of skills, experience, and perspectives. Artificial intelligence has been successfully applied to diagnostic subspecialties of medicine, such as pathology and radiology. Examples include diagnosis of skin cancers from photographs,19 lung cancer on screening CT images,20,21 retinal diseases using optical coherence tomography,22 and breast cancer using mammograms,23 or pathology specimens.24 However, applying AI techniques to therapeutic processes in medicine has not been equally well explored because of a lack of large data sets that are well curated by medical experts, and the need for AI techniques capable of adjusting to alterations in practice patterns or risk tolerance and style of individual treating physicians for specific diseases.
To address the global shortage of expert radiation oncologists, we designed a crowd innovation contest to challenge an international community of programmers to rapidly produce automated AI algorithms that could replicate the manual lung tumor segmentations of an expert radiation oncologist. To reach this goal, we developed a novel contest design, including:
A well-curated lung tumor data set segmented by an expert clinician for contestants to train and test their algorithms.
An objective scoring system for automatic evaluation of submitted algorithms to provide contestant feedback and final rankings.
Motivating and guiding contestants to produce clinically relevant solutions with a cost-effective prize pool, information sharing, and access to feedback from the expert clinician.
Methods
Data Set Curation
To encompass a range of tumor biology and size, we collected a data set of 461 patients with stage IA to IV non–small cell lung cancer (NSCLC) with planning CT scans obtained prior to RT under a protocol approved by the institutional review board at the Dana-Farber/Harvard Cancer Center. A waiver of informed consent was granted due to the retrospective data collection. The data set comprised 77 942 images (median, 157 images/scan) of which 8144 images had tumor present. All tumors were segmented by a single expert (R.H.M.) with 4 years of RT specialty training and 7 years of subspecialty experience in treating lung cancers. The data set was anonymized and randomly divided by patient into training (n = 229), validation (n = 96), and holdout test sets (n = 136). Additional information is available in the eMethods section of the Supplement.
Tumor Characteristics
The median tumor volume was 16.40 cm3 (range, 0.28-1103.74 cm3). The volume distribution was reflective of patients with known lung cancer undergoing therapy and differed from publicly available data sets such as the LIDC/IDRI lung nodule atlas25 (eFigure 2 in the Supplement). Additional patient and tumor summary statistics are provided in the Table.
Table. Summary Statistics of Patient and Tumor Data Set Used in the Competition.
| Categories | No. (%) |
|---|---|
| Patients, No. | 461 |
| Sex | |
| Female | 236 (51) |
| Male | 223 (48) |
| Unspecified | 2 (<1) |
| Median age (range), y | 73 (39 to ≥89a) |
| Median tumor volume (range), cm3 | 16.40 (0.28-1103.74) |
| Clinical stageb | |
| IA/IB | 97 (21)/15 (3) |
| IIA/IIB | 12 (3)/10 (2) |
| IIIA | 171 (37) |
| IIIB | 91 (20) |
| IV | 48 (10) |
| Unspecified | 17 (4) |
| Lobe (categories not exclusive) | |
| No primary lung tumor | 8 (2) |
| Right upper lobe | 169 (37) |
| Right middle lobe | 34 (7) |
| Right lower lobe | 67 (15) |
| Left upper lobe | 115 (25) |
| Left lower lobe | 58 (13) |
| Right endobronchial | 3 (1) |
| Left endobronchial | 3 (1) |
| Unspecified | 20 (4) |
| IV contrast | |
| Yes | 154 (33) |
| No | 307 (67) |
| Histologic type | |
| Adenocarcinoma | 274 (59) |
| Squamous cell carcinoma | 92 (20) |
| Non–small cell lung carcinoma | 54 (12) |
| Other | 41 (9) |
Seventeen patients classified as 89 years or older during anonymization process.
American Joint Committee on Cancer Staging, 7th edition.
Contest Design
We conducted our contest on Topcoder.com (Wipro, Bengalaru, India), a commercial platform that hosts online algorithm challenges for a community of more than 1 000 000 programmers who compete for prizes while solving computational problems. The contest was designed with 3 interconnected phases with the results of each phase informing the design of subsequent phases. Participants were oriented to the underlying medical problem and the contest design through online written materials and a video (https://youtu.be/An-YDBjFDV8) of the clinician expert demonstrating the manual lung tumor segmentation task.
The first 2 phases each ran for 3 weeks over 70 calendar days and offered $35 000 and $15 000 prize pools (eTable 1 in the Supplement), respectively, with entry open to anyone registered on the Topcoder platform. The third invitation-only phase ran for 4 weeks, with an additional approximately $5000 in prizes.
Segmentation Scoring
The contestants’ algorithms were scored by comparing the volumetric segmentation produced by each algorithm on a given patient’s CT scan (including all CT slices) against the expert’s segmentation. The performance of the contestants’ algorithms were assessed using a custom segmentation score (S score) (eTable 2 in the Supplement), that incorporated both the impact of relative and absolute errors. A higher score reflects an automated segmentation for a given patient’s entire tumor that has a high level of both relative and absolute overlap with the expert’s segmentation. Incorporating absolute error was particularly important in this therapeutic RT segmentation task because missing any volume of tumor would lead to an RT miss and increased likelihood of tumor recurrence. We also analyzed performance using traditional measures of relative error, including the Dice coefficient (Dice) and Jaccard index.
Contest Execution
Participants were provided with the full training set (CT scans, expert lung tumor and organ segmentations, and other clinical data), and the CT scans without segmentations from a validation set. During each phase, contestants produced segmentations using their algorithms on the validation set and received real-time evaluation of their algorithm’s performance based on the S score on a public leaderboard. With this feedback, contestants could modify their solution or generate new approaches to improve their scores. At the end of each phase, participants submitted their final algorithms for independent evaluation by the study team on the holdout test data set (which was not available to the contestants), and prizes were distributed to contestants who submitted algorithms generating the highest scores.
Between each phase, the study team including the clinical expert reviewed the winning algorithms’ performance and segmentations on individual patient scans to revise the contest design and objectives in each subsequent phase. In the first phase, the contestants were tasked with producing an algorithm that would both locate the tumor and replicate the expert’s segmentations. After review of phase 1 performance identified deficiencies in tumor localization as the major limiting factor, the contest was redesigned in phase 2 to allow contestants to focus their efforts on the therapeutic task of lung tumor targeting, as opposed to the distinct task of tumor diagnosis. Accordingly, we identified tumor location by providing a randomly generated seed point within each tumor, and asked the contestants to optimize their algorithms with that a priori knowledge.
The top 5 contestants from phase 2 were invited to work with the investigators in a collaborative phase 3 challenge to address deficits observed in the phase 2 solutions. The collaborative model allowed contestants to work together to improve algorithm performance.
Benchmarks
We compared the performance of the top AI algorithms from each phase against benchmarks including: (1) a commercially available, semi-automated segmentation software program (MIM Maestro, MIM Software) with and without human optimization of parameters; (2) the interobserver variation in manual segmentations between R.H.M. and 5 radiation oncologists in a previously published and publicly available,26,27 but similar external data set of 21 patients with lung cancer planned for RT (21 CT scans; median 178 images per scan; 3666 images in total; mean, roughly 226 images with tumor present); (3) intraobserver variation in the human expert performing the same segmentation task twice (independently, 3 months apart).
Furthermore, we validated algorithm performance and assessed for overfitting by applying the winning algorithms to the external, independent data set above. We estimated efficiency gains by comparing speed of expert manual segmentation vs the winning algorithms.
Results
Contest Participation
A total of 564 contestants from 62 countries registered for this challenge, and 34 (6%) submitted algorithms, including 244 unique submissions in phase 1 (mean, 8.4/participant), 164 in phase 2 (mean, 14.9/participant), and 180 in phase 3 (mean, 36/participant). Forty-five of these algorithms were submitted for the final scoring (phase 1, 29; phase 2, 11; and phase 3, 5). From these, we collected 10 independent, winning algorithms (eFigure 4 in the Supplement) developed by 9 unique winners.
Contest Results
Performance results for each phase are provided in eTable 3 in the Supplement; and Figure 2. For phase 1, the top 7 algorithms had average S scores on the holdout data set ranging from 0.15 to 0.38, and suboptimal performance using relative measures of error. The average S scores for phase 2 increased to 0.53 to 0.57, with a similar improvement in other performance metrics. In phase 3, performance of the top algorithm increased by an additional 9%. Combining the top 5 algorithms from phase 2 and phase 3 using an ensemble model (selected based on performance on the validation set; eMethods in the Supplement), yielded an additional 9% to 12% improvement in performance with a final S score reaching 0.68. The algorithms performed well for a variety of clinical situations and tumor locations (eFigures 1 and 5 in the Supplement).
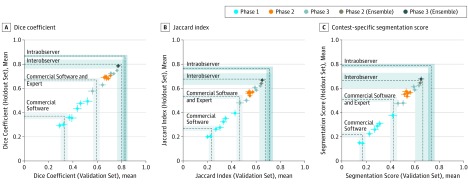
Figure 2. Artificial Intelligence Algorithm Performance in Each Phase of the Contest Compared Against Benchmarks.
A comparison of average performance (point) and standard error (whiskers) based on metrics (A) Dice coefficient, (B) Jaccard Index, and (C) contest-specific segmentation score (S score) of top 7 algorithms from phase 1, top 5 algorithms from phase 2, top 5 algorithms from phase 3, phase 2 ensemble model, and phase 3 ensemble model on the validation (x axis) and holdout data sets (y axis). Performance benchmarks for comparison include commercially available region-growing-based algorithms before (commercial software) and after (commercial software and expert) human intervention, the average of human interobserver comparisons between 6 radiation oncologists, and intraobserver variation in the human expert performing the segmentation task twice independently on the same tumor. Benchmarks are shown as dashed lines (gray bands indicate estimated uncertainties as described in eTable 3 in the Supplement).
Characteristics of the Winning Algorithms
The top contestants used a variety of approaches, including convolutional neural networks (CNNs), cluster growth, and random forest algorithms (eTable 4 in the Supplement). Solutions based on CNNs involved both custom and published architectures and frameworks to perform the tasks of object detection and localization (eg, Overfeat28), and/or segmentation (eg, SegNet29,30,31 and U-Net32). The latter were originally developed for the purpose of facial detection, biomedical image segmentation and road scene segmentation for autonomous vehicles research and adapted for the present task. The phase 3 algorithms produced segmentations at rates between 15 seconds/scan to 2 minutes/scan.
Comparison With Benchmarks
Examples of segmentations generated by human expert, commercially available software and contest algorithms are shown in Figure 1; and eFigure 1 in the Supplement. The phase 2 algorithms performed better than a commercially available, threshold-based autosegmentation software (eTable 3 in the Supplement) (Figure 2). The winning phase 3 algorithm and the ensemble models achieved scores within the interobserver variation and comparable to the interobserver mean between 5 other radiation oncologists and the expert from this study. Although the ensemble solution did not exceed the performance of the intraobserver benchmark (Figure 2 and Figure 3), approximately 75% of ensemble segmentations exceeded an S score of 0.60 (the lower threshold of intraobserver performance) which suggests that the contest produced algorithms capable of matching expert performance.
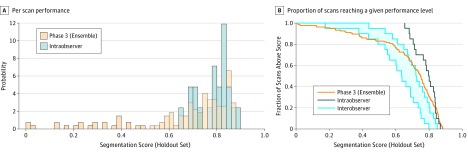
Figure 3. Performance of Best AI Algorithm Against Human Expert Intraobserver and Interobserver Variation Benchmarks .
A, Histogram of contest-specific segmentation score (S score) on a per-scan basis on the holdout data set compared with the intraobserver benchmark and (B) fraction of scans with performance greater than a given S score as a function the S score (holdout test set) for the phase 3 ensemble solution compared with the intraobserver benchmark. A comparison with the interobserver benchmark is also shown (gray band indicates first and third quartiles). Seventy-five percent of segmentations produced by the ensemble matched the lower threshold of intraobserver variation (S score, 0.60).
In addition, the performance of the contest’s top algorithms and the ensemble in the independent, external data set matched or exceeded their performance in the contest data set (eTable 3 in the Supplement). The mean time for the expert to perform manual segmentations was 8 minutes (range, 1-23 minutes), substantially longer than even the slowest algorithm.
Discussion
The results of this study show that a combined approach that leverages crowd innovation to access computational expertise to develop AI algorithms coupled with human medical expert feedback can enable rapid development of multiple solutions for a complex medical task with performance comparable to human experts.
Implications for Cancer Care
The ability to rapidly develop high-performing AI algorithms for tumor segmentation via a cost-effective crowd innovation approach has the potential to substantially improve the quality of oncologic care globally. Developing AI solutions for time-intensive tasks such as tumor segmentation can increase productivity and time with patients for busy clinicians by reducing computer-based work, and solve the known oncology workforce crisis (eg, number of trained radiation oncologists) in under-resourced health care systems worldwide.12 Artificially intelligent algorithms can also replicate and transfer expert-level knowledge for education, training, and/or quality assurance to raise the quality of global RT care. Providing quality assurance for RT trials is particularly important because variation in radiation planning even in highly structured protocols may be substantial enough to negatively impact outcomes and drive the results of trials toward the null.9 In addition, the ability to generate automatic tumor segmentations rapidly and accurately could revolutionize therapeutic response assessment in oncology in general, by allowing quantitative assessments of tumor imaging features during and after treatment, which may provide better predictive capability than traditional, manual, linear measurements (eg, RECIST).33,34
Implications for Application of AI to Radiation Therapy
Deep learning methods like CNNs have been increasingly used for visual pattern recognition to automate important diagnostic tasks in medicine.19,21,24,35,36 For the therapeutic task of targeting lung tumors, the top algorithms produced by this challenge (Dice = 0.79 compared with human expert) performed comparably to algorithms used to detect/segment pathologic entities in prior studies, including invasive breast cancer (Dice = 0.76),24 and brain white matter hyperintensities (Dice = 0.79).35 Previous work to apply deep learning to RT by academia and private industry include automated AI segmentation for both tumor targets37,38 and normal organs.39,40,41 Our top algorithms compared favorably against these limited studies as well (eg, Dice = 0.81 for nasopharyngeal tumors).37 Furthermore, this study demonstrated that crowdsourced AI algorithms significantly outperform existing commercially-available, semi-automated segmentation tools, which have historically focused on atlas-based,42 PET-based,43 and single click region grow auto-segmentation44 approaches.
Implications for Crowd Innovation in Oncology
Successful crowd innovation contests start with proper design and methodology. These findings demonstrated that a multiphase challenge design can provide agility, including opportunities for recalibrating objectives to more closely align with the desired clinical output. We opted for phased contests with shorter durations, smaller prizes, and regular feedback because the objective was rapid prototyping of optimal solutions. This methodology helped us to quickly adapt the contest objectives, by obtaining a better understanding of how nondomain expert crowds could perform on this medical task. The timely feedback from the human expert contributed to the iterative improvement in performance. For example, the expert’s input regarding phase 1 algorithm performance was factored into the design of phase 2 to allow a seed point. The clinical expert was able to articulate that this would more closely resemble the clinical situation of RT planning where the clinician’s task is accurate targeting of a known lung tumor. With this input, the phase 2 solution performance improved by approximately 50%. Second, we demonstrated that using different designs for different phases of the contest opened opportunities for further performance gains. With phase 3, we transitioned from a large, crowd innovation competition, to a smaller, invitation-only collaborative contest, involving the top-performing contestants from phase 2. The design encouraged participants to fine-tune the most promising solutions produced in phase 2, with further improvement in performance.
Although past contests have leveraged crowds to produce AI solutions to problems in diagnostic oncology including in the 2017 Kaggle Bowl (early lung cancer detection in low-dose CT screening scans)45 and the 2016 DREAM Challenge (identifying breast cancer on digital mammograms)46,47 which also used a phased approach, the study reported here applied a multiphase crowd innovation approach with collaborative components to address a therapeutic problem. Whereas diagnostic challenges have a relatively simple gold standard in a pathologic diagnosis, in contrast, solving this therapeutic problem required quality volumetric segmentations of the tumor produced by an expert. Several other differences in contest design included cumulative duration (70 days in 3 phases for ours vs 90 days for Kaggle and 17 weeks for DREAM) and considerably smaller prize pool ($55 000 vs $1 000 000). Although on-demand crowd workers are now widely used in a range of tasks (eg, ride sharing, labor market platforms like UpWork/Freelancer), our research demonstrates that crowds can also augment and complement traditional academic research with considerable cost, time, and performance benefits. As demand for AI talent increases across the economy, and new AI methods rapidly evolve across a range of academic and industrial settings, this study demonstrates that a relatively low-cost, crowd innovation approach can be used to democratize the development of AI solutions in oncology beyond traditional academic and industrial circles, by enabling individual oncologists to access AI expertise, on demand to improve their own clinical and research practices.
Limitations
The data sets used for this competition were relatively small compared with prior diagnostic radiology challenges (the Kaggle and DREAM challenge provided >1000 CTs and >600 000 mammograms, respectively), and the top algorithms likely underperform what would be observed with further training on a larger data set. However, the size of our data sets were comparable to those used in other recent applications of AI to RT, such as Google’s DeepMind’s efforts to automatically segment normal organs in the head and neck.41 Thus, AI can be successfully applied to solve problems in oncology even with limited data resources.
Furthermore, the production of the lung tumor segmentations relied on a single human expert, and the contest’s AI algorithms may have acquired the natural biases of that expert and generated segmentations representative of the training, experience, and judgment of 1 individual rather than objective ground truth (eFigure 6 in the Supplement). Nevertheless, replicating and delivering the manual skillset and experience of a given expert may still provide considerable value in therapeutic oncology.
Conclusions
This multifaceted crowd innovation challenge demonstrated that despite conservative constraints on contest duration and cost, it was possible to produce a diverse set of tumor segmentation AI algorithms that could replicate the abilities of a human expert and were faster. Using crowd innovation to generate clinically-relevant AI algorithms will allow sharing of scarce human expertise to under-resourced health care settings to improve the quality of cancer care globally.
eTable 1. Contest Prize Distributions, with payouts based on ranked performance
eTable 2. Common evaluation metrics for comparing segmentations
eTable 3. Algorithm, ensemble, and benchmark performance averaged over all scans in the validation, holdout and external data set
eTable 4. Primary methods, and utilization of supplemental data for training and/or inference
eTable 5. Algorithm, ensemble, and benchmark performance averaged over all scans in the validation and holdout data sets
eTable 6. Algorithm, ensemble, and benchmark performance averaged over all scans in the external data set
eFigure 1. Additional Examples of Human Expert versus Automated Segmentations
eFigure 2. Lung Tumor Volume Distribution in This Study Versus A Publicly-Available Dataset
eFigure 3. Distribution of Segmentation Score (S-score) as a function of V/V0
eFigure 4. Evolution of Winning AI Algorithms During the Multi-Phase Contest
eFigure 5. Performance of Ensemble AI Algorithms in Clinical Sub-Groups
eFigure 6. Performance of Top Segmentation Algorithms from The Contest on an External Dataset
eReferences.
eAcknowledgements.
References:
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7-30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271-289. doi: 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- 3.Smith BD, Haffty BG, Wilson LD, Smith GL, Patel AN, Buchholz TA. The future of radiation oncology in the United States from 2010 to 2020: will supply keep pace with demand? J Clin Oncol. 2010;28(35):5160-5165. doi: 10.1200/JCO.2010.31.2520 [DOI] [PubMed] [Google Scholar]
- 4.Cui Y, Chen W, Kong F-M, et al. Contouring variations and the role of atlas in non-small cell lung cancer radiation therapy: Analysis of a multi-institutional preclinical trial planning study. Pract Radiat Oncol. 2015;5(2):e67-e75. doi: 10.1016/j.prro.2014.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van de Steene J, Linthout N, de Mey J, et al. Definition of gross tumor volume in lung cancer: inter-observer variability. Radiother Oncol. 2002;62(1):37-49. doi: 10.1016/S0167-8140(01)00453-4 [DOI] [PubMed] [Google Scholar]
- 6.Wuthrick EJ, Zhang Q, Machtay M, et al. Institutional clinical trial accrual volume and survival of patients with head and neck cancer. J Clin Oncol. 2015;33(2):156-164. doi: 10.1200/JCO.2014.56.5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters LJ, O’Sullivan B, Giralt J, et al. Critical impact of radiotherapy protocol compliance and quality in the treatment of advanced head and neck cancer: results from TROG 02.02. J Clin Oncol. 2010;28(18):2996-3001. doi: 10.1200/JCO.2009.27.4498 [DOI] [PubMed] [Google Scholar]
- 8.Eaton BR, Pugh SL, Bradley JD, et al. Institutional enrollment and survival among NSCLC patients receiving chemoradiation: NRG Oncology Radiation Therapy Oncology Group (RTOG) 0617. J Natl Cancer Inst. 2016;108(9):djw034. doi: 10.1093/jnci/djw034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohri N, Shen X, Dicker AP, Doyle LA, Harrison AS, Showalter TN. Radiotherapy protocol deviations and clinical outcomes: a meta-analysis of cooperative group clinical trials. J Natl Cancer Inst. 2013;105(6):387-393. doi: 10.1093/jnci/djt001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang EH, Rutter CE, Corso CD, et al. Patients selected for definitive concurrent chemoradiation at high-volume facilities achieve improved survival in stage III non-small-cell lung cancer. J Thorac Oncol. 2015;10(6):937-943. doi: 10.1097/JTO.0000000000000519 [DOI] [PubMed] [Google Scholar]
- 11.Grover S, Xu MJ, Yeager A, et al. A systematic review of radiotherapy capacity in low- and middle-income countries. Front Oncol. 2015;4(380):380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdel-Wahab M, Zubizarreta E, Polo A, Meghzifene A. Improving quality and access to radiation therapy-an IAEA perspective. Semin Radiat Oncol. 2017;27(2):109-117. doi: 10.1016/j.semradonc.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 13.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 14.Datta NR, Samiei M, Bodis S. Radiation therapy infrastructure and human resources in low- and middle-income countries: present status and projections for 2020. Int J Radiat Biol Phys. 2014;89(3): 448-457. [DOI] [PubMed] [Google Scholar]
- 15.Lakhani KR, Boudreau KJ, Loh PR, et al. Prize-based contests can provide solutions to computational biology problems. Nat Biotechnol. 2013;31(2):108-111. doi: 10.1038/nbt.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costello JC, Heiser LM, Georgii E, et al. ; NCI DREAM Community . A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol. 2014;32(12):1202-1212. doi: 10.1038/nbt.2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill A, Loh P-R, Bharadwaj RB, et al. Stepwise distributed open innovation contests for software development: acceleration of genome-wide association analysis. Gigascience. 2017;6(5):1-10. doi: 10.1093/gigascience/gix009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holland RC, Lynch N. Sequence squeeze: an open contest for sequence compression. Gigascience. 2013;2(1):5. doi: 10.1186/2047-217X-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115-118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Setio AAA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging. 2016;35(5):1160-1169. doi: 10.1109/TMI.2016.2536809 [DOI] [PubMed] [Google Scholar]
- 21.Setio AAA, Traverso A, de Bel T, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Med Image Anal. 2017;42:1-13. doi: 10.1016/j.media.2017.06.015 [DOI] [PubMed] [Google Scholar]
- 22.Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122-1131.e9. doi: 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 23.Kooi T, Litjens G, van Ginneken B, et al. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal. 2017;35:303-312. doi: 10.1016/j.media.2016.07.007 [DOI] [PubMed] [Google Scholar]
- 24.Cruz-Roa A, Gilmore H, Basavanhally A, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: A deep learning approach for quantifying tumor extent. Sci Rep. 2017;7:46450. doi: 10.1038/srep46450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armato SG III, McLennan G, Bidaut L, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys. 2011;38(2):915-931. doi: 10.1118/1.3528204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Baardwijk A, Bosmans G, Boersma L, et al. PET-CT–based auto-contouring in non–small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. Int J Radiat Oncol Biol Phys. 2007;68(3):771-778. [DOI] [PubMed] [Google Scholar]
- 27.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y OverFeat: Integrated Recognition, Localization and Detection using Convolutional Networks. https://arxiv.org/abs/1312.6229. Accessed November 11, 2018.
- 29.Kendall A, Badrinarayanan V, Cipolla R SegNet: Model Uncertainty in Deep Convolutional Encoder-Decoder Architectures for Scene Understanding. https://arxiv.org/abs/1511.02680. Accessed November 11, 2018.
- 30.Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481-2495. doi: 10.1109/TPAMI.2016.2644615 [DOI] [PubMed] [Google Scholar]
- 31.Badrinarayanan V, Handa A, Cipolla R SegNet: A deep convolutional encoder-decoder architecture for robust semantic pixel-wise labelling. https://arxiv.org/abs/1505.07293. Accessed August 1, 2018.
- 32.Ronneberger O, Fischer P, Brox T U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III. Cham: Springer International Publishing; 2015:234-241. [Google Scholar]
- 33.Agrawal V, Coroller TP, Hou Y, et al. Radiologic-pathologic correlation of response to chemoradiation in resectable locally advanced NSCLC. Lung Cancer. 2016;102:1-8. doi: 10.1016/j.lungcan.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 34.Zhao B, Oxnard GR, Moskowitz CS, et al. A pilot study of volume measurement as a method of tumor response evaluation to aid biomarker development. Clin Cancer Res. 2010;16(18):4647-4653. doi: 10.1158/1078-0432.CCR-10-0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghafoorian M, Karssemeijer N, Heskes T, et al. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci Rep. 2017;7(1):5110. doi: 10.1038/s41598-017-05300-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402-2410. doi: 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 37.Men K, Chen X, Zhang Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017;7(315):315. doi: 10.3389/fonc.2017.00315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cardenas CE, McCarroll RE, Court LE, et al. Deep Learning Algorithm for Auto-Delineation of High-Risk Oropharyngeal Clinical Target Volumes With Built-In Dice Similarity Coefficient Parameter Optimization Function. Int J Radiat Oncol Biol Phys. 2018;101(2):468-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44(2):547-557. doi: 10.1002/mp.12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61(24):8676-8698. doi: 10.1088/1361-6560/61/24/8676 [DOI] [PubMed] [Google Scholar]
- 41.Nikolov S, Blackwell S, Mendes R, et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. https://arxiv.org/abs/1809.04430. Accessed November 11, 2018. [DOI] [PMC free article] [PubMed]
- 42.Delpon G, Escande A, Ruef T, et al. Comparison of automated atlas-based segmentation software for postoperative prostate cancer radiotherapy. Front Oncol. 2016;6:178. doi: 10.3389/fonc.2016.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Markel D, Caldwell C, Alasti H, et al. Automatic segmentation of lung carcinoma using 3D texture features in 18-FDG PET/CT. Int J Mol Imaging. 2013;2013:980769. doi: 10.1155/2013/980769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velazquez ER, Parmar C, Jermoumi M, et al. Volumetric CT-based segmentation of NSCLC using 3D-slicer. Sci Rep. 2013;3:3529. doi: 10.1038/srep03529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aberle DR, Adams AM, Berg CD, et al. ; National Lung Screening Trial Research Team . Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395-409. doi: 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ribli D, Horváth A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with deep learning. Sci Rep. 2018;8(1):4165. doi: 10.1038/s41598-018-22437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trister AD, Buist DSM, Lee CI. Will machine learning tip the balance in breast cancer screening? JAMA Oncol. 2017;3(11):1463-1464. doi: 10.1001/jamaoncol.2017.0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Contest Prize Distributions, with payouts based on ranked performance
eTable 2. Common evaluation metrics for comparing segmentations
eTable 3. Algorithm, ensemble, and benchmark performance averaged over all scans in the validation, holdout and external data set
eTable 4. Primary methods, and utilization of supplemental data for training and/or inference
eTable 5. Algorithm, ensemble, and benchmark performance averaged over all scans in the validation and holdout data sets
eTable 6. Algorithm, ensemble, and benchmark performance averaged over all scans in the external data set
eFigure 1. Additional Examples of Human Expert versus Automated Segmentations
eFigure 2. Lung Tumor Volume Distribution in This Study Versus A Publicly-Available Dataset
eFigure 3. Distribution of Segmentation Score (S-score) as a function of V/V0
eFigure 4. Evolution of Winning AI Algorithms During the Multi-Phase Contest
eFigure 5. Performance of Ensemble AI Algorithms in Clinical Sub-Groups
eFigure 6. Performance of Top Segmentation Algorithms from The Contest on an External Dataset
eReferences.
eAcknowledgements.