Abstract
Glomerular biology is dependent on tightly controlled signal transduction networks that control phosphorylation of signaling proteins such as cytoskeletal regulators or slit diaphragm proteins of kidney podocytes. Cross-species comparison of phosphorylation events is a powerful mean to functionally prioritize and identify physiologically meaningful phosphorylation sites. Here, we present the result of phosphoproteomic analyses of cow and rat glomeruli to allow cross-species comparisons. We discovered several phosphorylation sites with potentially high biological relevance, e.g. tyrosine phosphorylation of the cytoskeletal regulator synaptopodin and the slit diaphragm protein neph-1 (Kirrel). Moreover, cross-species comparisons revealed conserved phosphorylation of the slit diaphragm protein nephrin on an acidic cluster at the intracellular terminus and conserved podocin phosphorylation on the very carboxyl terminus of the protein. We studied a highly conserved podocin phosphorylation site in greater detail and show that phosphorylation regulates affinity of the interaction with nephrin and CD2AP. Taken together, these results suggest that species comparisons of phosphoproteomic data may reveal regulatory principles in glomerular biology. All MS data have been deposited in the ProteomeXchange with identifier PXD001005 (http://proteomecentral.proteomexchange.org/dataset/PXD001005).
Keywords: Animal proteomics, Glomerulus, Kidney, Phosphoproteomics, Podocin, Slit diaphragm complex
Maintenance of the kidney filtration barrier is one of the most important functions of kidney podocytes, highly specialized epithelial cells. Podocyte integrity and function largely depends on the physiological organization of a specialized protein complex, the slit diaphragm [1]. The slit diaphragm protein complex is not only part of a mechanical filter, but is also appreciated as a site of active signaling events [1]. However, the signaling pathways, which directly regulate these proteins are not fully understood and only partially identified. Previous studies have identified phosphorylation dependent regulation of protein-protein interactions at the slit diaphragm with focus on nephrin tyrosine phosphorylation [2, 3]. To pioneer phosphoproteomics in glomerular biology, we have published an initial map of the glomerular phosphoproteome applying an unbiased MS/MS-based approach in mice where we discovered >2000 phosphorylated proteins corresponding to >4000 high-confidence phosphorylated residues [4]. This large number of phosphorylated proteins and residues warrants further assessment to identify biologically relevant phosphorylation sites.
The mere amount of phosphorylation sites in large-scale phosphoproteomic studies, also at nonconserved sites, led to the hypothesis that some phosphorylation occurs due to undirected kinase activity and might be biologically nonfunctional [5]. Moreover, the stochastic nature of shotgun phosphoproteomic analyses adds another layer of variability to the picture. To overcome these limitations, comparative phosphoproteomic analyses allow functional prioritization of posttranslational modifications to identify phosphorylation-enriched regions within protein families and domains [6]. This approach is more powerful than a mere in silico analysis using multiple sequence alignments across multiple species [6, 7].
To prioritize phosphorylation, we compiled and analyzed the phosphoproteome of glomeruli in three mammalian species. We isolated rat (8 week old male Fisher rats) and cow glomeruli (ca 1 year old male cows) as previously described [8]. We next proceeded with phosphoproteomic analysis using an LTQ orbitrap machine as previously described [4] with each experiment repeated twice (biological replicates). The methods are deposited in the supplement.
The analysis of the rat glomerular phosphoproteome revealed 1417 class-I phosphorylation sites (localization score >0.75). The majority of these sites were serines, followed by threonines and tyrosines (Fig. 1A). The analysis also revealed a bias of phosphorylation sites toward basophilic and proline-directed phosphorylation motifs. The analysis of the bovine glomerular phosphoproteome revealed 1700 identified class-I phosphorylation sites. Serines were more frequent than threonines and tyrosines, a finding consistent with other mammalian species (Fig. 1B). Only 13 of the discovered phosphorylation sites were previously reposited in the phosphosite.org or phosphoELM repository [9, 10]. Prediction of kinase substrates revealed similar properties of the phosphoproteomic datasets (Fig. 1C). The raw data containing information on all high-confident phosphorylation sites (“class I”) are made publically available at an online repository at https://helixweb.nih.gov/ESBL/Database/GloPhos/RGloPhos.htm (Rat glomerular phosphoproteomic database) and https://helixweb.nih.gov/ESBL/Database/GloPhos/BGloPhos.htm (Bovine glomerular phosphoproteomic database). The MS proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) [11] via the PRIDE partner repository with the dataset identifier PXD001005.
Figure 1.
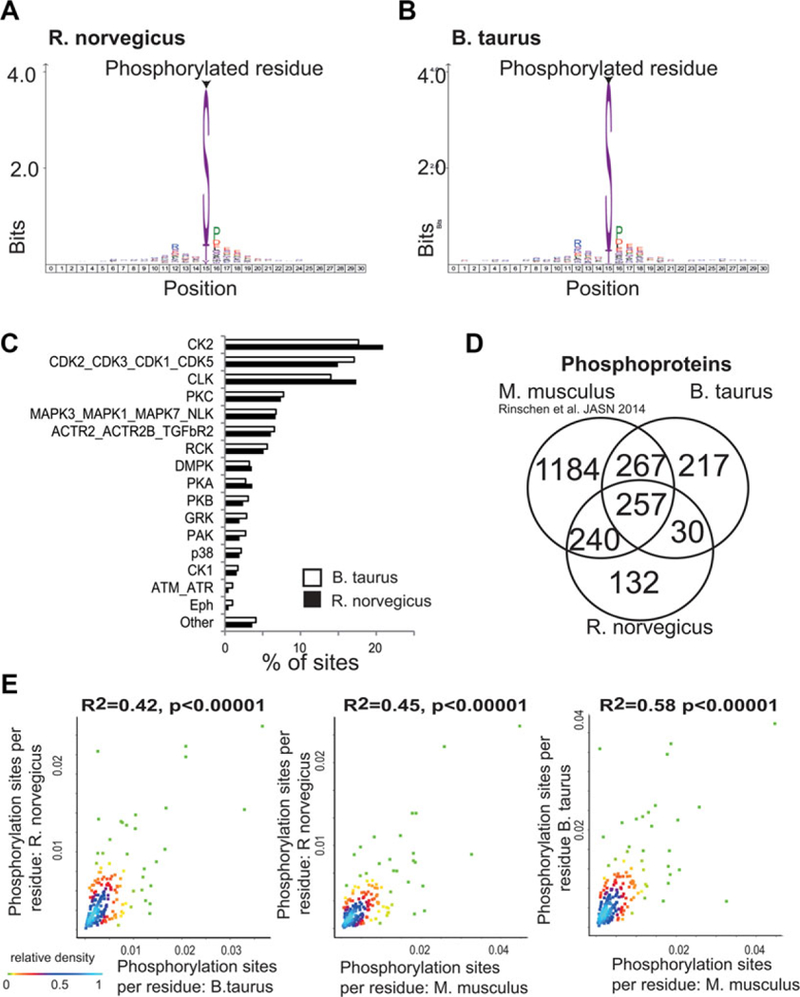
Comparative phosphoproteomic analysis of mammalian glomeruli. (A) Distribution of serine, threonine, and tyrosine phosphorylation in the rat glomerular phosphoproteome. A position weighted matrix of surrounding amino acid windows was created. The matrix shows over-represented residues surrounding the respective phosphorylation site. (B) Distribution of serine, threonine, and tyrosine phosphorylation in the bovine glomerular phosphoproteome. (C) Prediction of kinase substrates. The frequency of each kinase group predicting to phosphorylate the respective sites is shown. (D) Venn diagram indicating the overlap in homologous phosphoproteins. The venn diagram shows only proteins which are matched against the NCBI homologene repository. The mouse dataset was previously published as Rinschen et al., JASN 2014. (E) Correlation between site number per protein (normalized over number of residues) between the rat, mouse, and bovine dataset. The relative contribution of the data points to overall density is color coded.
Next, we analyzed whether the phosphoproteome analysis covered homologous phosphoproteins. We compared the obtained data with recently published data of the mouse glomerular phosphoproteome [4]. There were 257 homologous proteins found in all three datasets based on NCBI HomoloGene groups (Fig. 1C) and 537 were found in two species. The proteins found in all three species showed a significant overrepresentation of genes involved in RNA binding (G0:0003732; FDR q value (q) = 1.26 × 10−3), organization of cell-cell contacts (G0:0005911; q = 4.59 × 10−2), and structural components ofthe cytoskeleton (G0:0005200; q = 2.36 × 10−2) as compared to the other phosphorylated proteins [12]. We analyzed whether the total numbers of detectable phosphorylation sites per protein correlated across these proteins. The analysis revealed that the number of phosphorylation sites (normalized over protein residue numbers) correlated significantly, but there was a rather high degree of variability (Fig. 1D). We next looked for homologous phosphorylation sites among the analyzed species by mapping the discovered phosphorylation sites on NCBI HomoloGene protein sequences. The analysis revealed 268 homologous phosphorylation sites found in all three species, and 847 were found in two species (Fig. 2A), all of these sites are available in Supporting Information Table 1. We calculated site and motif conservation scores across all eukaryotic species (http://www.ncbi.nlm.nih.gov/homologene/statistics/) in the HomoloGene database using the C-Phos software (Fig. 2B) [13]. This analysis performs multiple sequence alignments in the respective HomoloGene group and calculates scores for conservation of the respective sites. The analysis revealed that the mean scores of motif and site conservation were significantly higher for homologous phosphorylation sites detected on two or more species as compared with those phosphorylation sites which were only detected on one species (Fig. 2B). All conserved sites are deposited in the Supporting Information Table 1.
Figure 2.
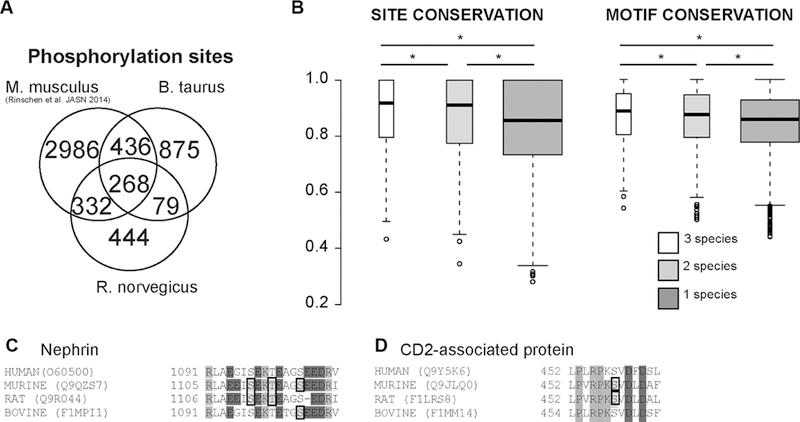
Comparison of phosphorylation site analysis. (A) Venn diagram indicating the overlap of identified phosphoprotein sites. Two hundred and seventy one phosphorylation sites were found at homologous phosphoproteins at homologous sites. Only phosphorylation sites matched to the NCBI homologene repository are shown. (B) Analysis of conservation of sites and sequences by HomoloGene groups. Phosphorylation sites found in more than one organism have significantly higher site and motif conservation (c-Phos) scores within their respective entire HomoloGene group. The statistics were performed using a Wilcoxon-Mann-test (*p < 0.001). Whiskers extend to data points that are less than 1.5 × Interquartile range away from 1st/3rd quartile C), (D) Phosphorylation sites found on slit diaphragm proteins Nephrin and CD2AP in two or more organisms in this and a recent study (Rinschen et al. JASN 2014). Phosphorylated residues are denoted by black boxes.
The phosphoproteomic analysis revealed previously undescribed tyrosine phosphorylation sites on important glomerular proteins, such as Neph1 (Kirrel; Y679 in rats) and Synaptopodin (Synpo; Y870 in rats). Neph1, an important nephrin interactor, has been previously shown to be phosphorylated at the residues Y637/638/716/719 (mouse/rat sequence) by the tyrosine kinase Fyn and at Y637/638 by the tyrosine kinase Src [14, 15] in vitro and in vivo. Serine/threonine phosphorylation of synaptopodin regulates its interaction with the actin-based cytoskeleton [16].
However, we focused on conserved phosphorylation events on proteins whose mutation is responsible for human diseases [4]. The analysis revealed that there was a conserved presence of phosphorylation sites at an acidic phosphorylation cluster on Nephrin (Fig. 2C) as well as on CD2AP (Fig. 2D) across species. In addition, evidence for C-terminal phosphorylation of podocin at S382 was found in all species studied. We decided to further study podocin phosphorylation because of its extraordinary importance for maintenance of the glomerular filter in health and disease [17]. Podocin S382 is highly conserved and localizes in close proximity of a highly positively charged amino acid cluster and phosphorylation of this site would lead to a reversal of the total terminal charge (Fig. 3A). Bioinformatic analysis [18] predicted that the proline-rich C-terminus (residues 355–382) of podocin is likely to be intrinsically disordered. We speculated that this phosphorylation could alter the interaction with relevant proteins of the kidney filtration barrier. As we could detect podocin S382 phosphorylation in HEK293T cells expressing podocin (Fig. 3B) we generated FLAG-tagged phosphomimicking (Podocin S382D) or phosphoablating (Podocin S382A) mutants. We performed these experiments as previously described [19]. Co-immunoprecipitation assays revealed a significant reduction in V5-tagged Nephrin binding by the S382D mutant as compared to the S382A mutant (Fig. 3C). The binding with Trpc6 was not affected by this mutant (Fig. 3D). Interaction with V5-tagged CD2AP, however, was decreased by coexpressing a phosphomimicking mutant (Fig. 3E). We also tested whether phosphorylation may interfere with membrane localization by expressing the proteins in HeLa cells but could not observe any significant alteration in subcellular localization (Fig. 2F) [20]. The FLAG-tagged short isoform of podocin was used as a negative control protein which was not targeted to the plasma membrane as previously described [20]. The hypothesis of a phosphorylation-dependent podocin-nephrin and podocin-CD2AP interaction is consistent with previous findings from our group and others mapping this interaction on the podocin-C-terminus [21, 22]. In addition, our work indicates that podocin-Trpc6 interaction is mediated via the PHB domain [23].
Figure 3.
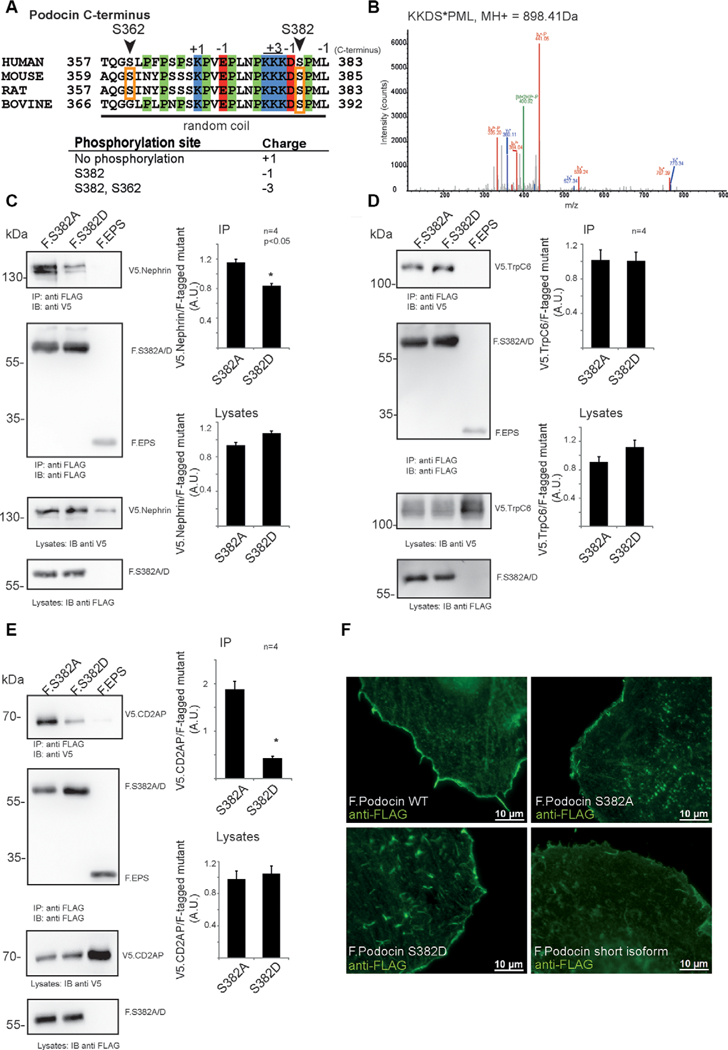
Characterization of Podocin S382 (murine sequence), a phosphorylation site discovered in mouse, rat, and bovine glomeruli. (A) Sequence of the C-terminal part of podocin, indicating a proline-rich random coil at the distal C-terminus. Phosphorylated residues are denoted by orange boxes. Site residue numbering is based on mouse sequence. (B) Evidence for phosphorylation of mouse podocin residue S382 in the protein expressed in HEK293T cells. (C)-(E) Coimmunoprecipitation of members of the slit diaphragm complex with FLAG-tagged phosphoablating (F.S382A) or phosphomimicking (F.S382D) mouse podocin expressed in 293Tcells. Protein complexes were purified using anti-FLAG antibody coupled to sepharose beads. All experiments were carried out in biologically independent quadruplicates. Bands were quantified using densitometry. p < 0.05 in a two-tailed, paired student’s t-test was considered significant. (C) Immunoprecipitation of sV5-tagged nephrin with FLAG-tagged phosphoablating or phosphomimicking mouse podocin mutants. (D) Immunoprecipitation of V5-tagged TrpC6 with FLAG-tagged phosphoablating or phosphomimicking podocin mutants. (E) Immunoprecipitation of sV5-tagged CD2AP with FLAG-tagged phosphoablating or phosphomimicking podocin mutants. (F) Localization of podocin wildtype and phosphoablating (F Podocin S382A) or phosphomimicking (F. Podocin S382D) mutant transiently expressed in HeLa cells. The podocin short isoform was used as a control protein which is not targeted to the membrane. Cells were stained using an anti-FLAG antibody. Appropriate negative controls were performed.
In this dataset brief, we perform phosphoproteomic analyses on rat and bovine glomerular tissue and combined the results with our recently published data set in mouse glomeruli to generate a dataset with conserved phosphorylation sites across species (Supporting Information Table 1, [4]). Little is known about the rat kidney glomerulus phosphoproteome in contrast to distal and proximal tubular proteomes [24, 25]. In bovine tissue, proteomics analyses have been established very recently [26], but no phosphoproteomic studies have been performed to our knowledge. The high percentage of previously undescribed phosphorylation sites is indicative of this finding. The phosphoproteome of rat and bovine tissues reflected typical residue distribution (Fig. 1A, B) [27]. The cross-species comparison between homologous phosphoproteins revealed the expected variabilities in phosphoproteomes (Fig. 1D, E) but still delineated common phosphoproteins and phosphosites in glomeruli that were conserved across species. As expected, phosphorylation sites found in more than one species had significantly higher motif and site conservation scores within their entire HomoloGene groups (Fig. 2B).
We show the potential relevance of such a dataset for the prioritization of phosphorylation sites (Fig. 3), although this and other sites (Kirrel [“Neph1”]; Y679, Synpo; Y870) need to be studied in vivo. In conclusion, this repository of phosphoproteomic data can provide a guide to prioritize phosphorylation sites in glomerular biology and this approach may reveal regulatory principles in glomerular biology.
Supplementary Material
Acknowledgments
The MS proteomics data in this paper have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [11]: dataset identifier PXD001005. This work was supported by the German Research Foundation (Sonderforschungsbereich 635to T.B., SCHE1562–2to B.S. and BR2955 to P.T.B.]) and the German Federal Ministry of Education and Research (GerontoSys2program Sybacol to T.B. and B.S.). T.P. was supported by the Ratchadapiseksomphot Endowment Fund of Chulalongkorn University (RES560530124-HR). M.M.R. was supported by Köln Fortune, by German Research Foundation (DFG) in the framework of the German Excellence Initiative (UoC Postdoc Grant) and by the Fritz-Scheler-Stipendium of the KFH-Stiftung fur Praventivmedizin. The authors thank Ruth Herzog, Astrid Wilbrand-Hennes and Rene Grandjeanfor excellent technical help. The authors thank Andreas Beyer for helpful discussions. The authors thank Fahad Saeed and Boyang Zhao for help with the CPhos software and Jinho Kim and Peter Frommoltfor help with the Netphorest software.
Footnotes
Additional supporting information may be found in the online version ofthis article at the publisher’s web-site
The authors have declared no conflict of interest.
References
- [1].Benzing T, Signaling at the slit diaphragm. J. Am. Soc. Nephrol. JASN 2004, 15, 1382–1391. [DOI] [PubMed] [Google Scholar]
- [2].Verma R, Wharram B, Kovari I, Kunkel R et al. , Fyn binds to and phosphorylates the kidney slit diaphragm component Nephrin. J. Biol. Chem. 2003, 278, 20716–20723. [DOI] [PubMed] [Google Scholar]
- [3].Jones N, Blasutig IM, Eremina V, Ruston JM et al. , Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature 2006, 440, 818–823. [DOI] [PubMed] [Google Scholar]
- [4].Rinschen MM, Wu X, König T, Pisitkun T et al. , Phosphoproteomic Analysis Reveals Regulatory Mechanisms at the Kidney Filtration Barrier. J. Am. Soc. Nephrol. 2014, 7, 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lienhard GE, Non-functional phosphorylations? Trends Biochem. Sci. 2008, 33, 351–352. [DOI] [PubMed] [Google Scholar]
- [6].Beltrao P, Albanese V, Kenner LR, Swaney DL et al. , Systematic functional prioritization of protein posttranslational modifications. Cell 2012, 150,413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Beltrao P, Trinidad JC, Fiedler D, Roguev A et al. , Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009, 7, e1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gloy J, Henger A, Fischer KG, Nitschke R et al. , Angiotensin II depolarizes podocytes in the intact glomerulus of the Rat. J. Clin. Invest. 1997, 99, 2772–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dinkel H, Chica C, Via A, Gould CM et al. , Phospho.ELM: a database of phosphorylation sites-update 2011. Nucleic Acids Res. 2011, 39, D261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B et al. , PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vizcaino JA, Deutsch EW, Wang R, Csordas A et al. , ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z, GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 2009, 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhao B, Pisitkun T, Hoffert JD, Knepper MA, Saeed F, CPhos: a program to calculate and visualize evolutionarily conserved functional phosphorylation sites. Proteomics 2012, 12, 3299–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Garg P, Verma R, Nihalani D, Johnstone DB, Holzman LB, Nephl cooperates with nephrin to transduce a signal that induces actin polymerization. Mol. Cell. Biol. 2007, 27, 8698–8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Harita Y, Kurihara H, Kosako H, Tezuka T et al. , Nephl, a component of the kidney slit diaphragm, is tyrosine-phosphorylated bythe Src family tyrosine kinase and modulates intracellularsignaling by binding to Grb2. J. Biol. Chem. 2008, 283, 9177–9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Faul C, Donnelly M, Merscher-Gomez S, Chang Y H. et al. , The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat. Med. 2008, 14, 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Boute N, Gribouval O, Roselli S, Benessy F et al. , NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat. Genet. 2000, 24, 349–354. [DOI] [PubMed] [Google Scholar]
- [18].He B, Wang K, Liu Y, Xue B et al. , Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009, 19, 929–949. [DOI] [PubMed] [Google Scholar]
- [19].Kohli P, Bartram MP, Habbig S, Pahmeyer C et al. , Label-free quantitative proteomic analysis of the YAP and TAZ interactome. Am. J. Physiol. Cell Physiol. 2014, 9, 805–818. [DOI] [PubMed] [Google Scholar]
- [20].Völker LA, Schurek E-M, Rinschen MM, Tax J et al. , Characterization of a short isoform of the kidney protein podocin in human kidney. BMC Nephrol. 2013, 14, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huber TB, Kottgen M, Schilling B, Walz G, Benzing T, Interaction with podocin facilitates nephrin signaling. J. Biol. Chem. 2001, 276, 41543–41546. [DOI] [PubMed] [Google Scholar]
- [22].Schwarz K, Simons M, Reiser J, Saleem MA et al. , Podocin, a raft-associated component of the glomerular slit diaphragm, interacts with CD2APand nephrin. J. Clin. Invest. 2001, 108, 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huber TB, Schermer B, Müller RU, Höhne M et al. , Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 17079–17086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Feric M, Zhao B, Hoffert JD, Pisitkun T, Knepper MA, Large-scale phosphoproteomic analysis of membrane proteins in renal proximal and distal tubule. Am. J. Physiol. Cell Physiol. 2011, 300, C755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hoffert JD, Pisitkun T, Saeed F, Song JH et al. , Dynamics of the G protein-coupled vasopressin V2 receptor signaling network revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics MCP 2012, 11, M111.014613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Faulkner S, Elia G, O’Boyle P, Dunn M, Morris D, The composition of the bovine uterine proteome is associated with stage of cycle and concentration of systemic progesterone. Proteomics 2013, 13,3333–3353. [DOI] [PubMed] [Google Scholar]
- [27].Villen J, Beausoleil SA, Gerber SA, Gygi SP, Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 1488–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


