Abstract
The Hyperactivated Wnt/β-catenin signaling acts as a switch to induce EMT and promote colorectal cancer. However, due to its essential role in gut homeostasis, therapeutic targeting of this pathway has proven challenging. Additionally, IL-6/Stat-3 signaling, activated by microbial translocation through the dysregulated mucosal barrier in colon adenomas, facilitates the adenoma to adenocarcinomas transition. However, inter-dependence between these signaling pathways and key mucosal barrier components in regulating colon tumorigenesis and cancer progression remains unclear. In current study, we have discovered, using a comprehensive investigative regimen, a novel and tissue specific role of claudin-3, a tight junction integral protein, in inhibiting colon cancer progression by serving as the common rheostat of Stat-3 and Wnt-signaling activation. Loss of claudin-3 also predicted poor patient survival. These findings however contrasted an upregulated claudin-3 expression in other cancer types and implicated role of the epigenetic regulation. Claudin-3−/− mice revealed dedifferentiated and leaky colonic epithelium, and developed invasive adenocarcinoma when subjected to colon cancer. Wnt-signaling hyperactivation, albeit in GSK-3β independent manner, differentiated colon cancer in claudin-3−/− mice versus WT-mice. Claudin-3 loss also upregulated the gp130/IL6/Stat3 signaling in colonic epithelium potentially assisted by infiltrating immune components. Genetic and pharmacological studies confirmed that claudin-3 loss induces Wnt/β-catenin activation, which is further exacerbated by Stat-3-activation and help promote colon cancer. Overall, these novel findings identify claudin-3 as a therapeutic target for inhibiting overactivation of Wnt-signaling to prevent CRC malignancy.
Keywords: Colon cancer, Barrier Dysfunction, Dedifferentiation, Proliferation, Invasion, Tight Junction
Introduction
Genetic and epigenetic aberrations drive the formation of the benign colon adenoma and its progression to adenocarcinoma. However, recent studies have also demonstrated key role of the impaired mucosal barrier in adenoma to carcinoma transition by enabling cancer permissive and inflammatory signaling (1). In this regard, causal role of IL-6/Stat-3 signaling in CRC progression was recently reported (2). Moreover, deregulated epithelial barrier can also allow growth factor infiltration into the mucosa to induce receptor tyrosine kinase signaling known to promote neoplastic transformation and growth (3–5). Notably, TJ help maintain mucosal barrier properties. Additionally, TJ helps regulate epithelial cell differentiation and polarity, and are accordingly deregulated during neoplastic transformation of colonic epithelial cells (6). Furthermore, Wnt/β-catenin signaling has gained particular attention, in regulating the epithelial to mesenchymal transition (EMT) to facilitate the adenoma to carcinoma transition and CRC-progression (7, 8). However, key regulatory mechanism/s governing the de-differentiated colonic epithelial phenotype in correlation with cancer promoting inflammatory microenvironment and its causal association with Wnt/β-catenin signaling remain largely unclear. Thus, improved understanding of the molecular and genetic routes that help regulate inter-dependence between Wnt/β-catenin and Stat-3 signaling to facilitate CRC-progression can help enable therapeutic mechanisms for lessening cancer aggressiveness and patient death.
In current studies, we have identified claudin-3 as the highest expressed cell-adhesion protein, among prominent cell-adhesion proteins tested, in the normal colon and positive association between claudin-3 expression and colonocyte differentiation. Similar observation by other published reports renders support to our findings (9). Therefore, we postulated loss of colonic claudin-3 expression to deregulate mucosal barrier function and colonocyte differentiation, and therefore promote CRC-malignancy. However, limited published studies implicate claudin-3 in CRC-progression similar to prostate and ovarian cancers (10, 11). Due to these inconsistencies and our hypothesis that claudin-3 loss may promote CRC-malignancy, we performed a comprehensive investigation. Using CRC cell lines of differing tumorigenic and metastatic potential, mouse model of colon cancer and CRC patient samples, we here demonstrate a novel colon cancer suppressive role of claudin-3. Using genetic and pharmacological manipulations, we further confirm a role for claudin-3 by regulating intratumoral Wnt/β-catenin- and IL6/gp130/Stat3-signaling pathways. Overall, we here provide first biochemically and clinically validated evidence to support a CRC-suppressive function of claudin-3 by serving as a conjoint rheostat for regulating Stat-3 and Wnt-signaling.
Results
Claudin-3 is the most abundant cell-adhesion moiety in normal colon, expressed predominantly by differentiated colonocytes:
A key role of TJ in regulating epithelial barrier properties, cell differentiation and polarity is established (12, 13). Considering the newly emerging nexus between epithelial and immune homeostasis in regulating colon carcinogenesis, we first determined relative abundance of the key cell-adhesion proteins in normal colon. Immunoblot analysis using mice colon lysate demonstrated claudin-3 expression to be the highest compared to claudin-1, claudin-2, claudin-7, Occludin, ZO-1, E-cadherin, β-catenin and β1-integrin (p<0.01; Fig-1A and Supplementary Fig-S1A). Immunohistochemical (IHC) analysis further demonstrated an expression gradient with predominant claudin-3 expression at the crypt top, among terminally differentiated colonocytes (Fig-1B).
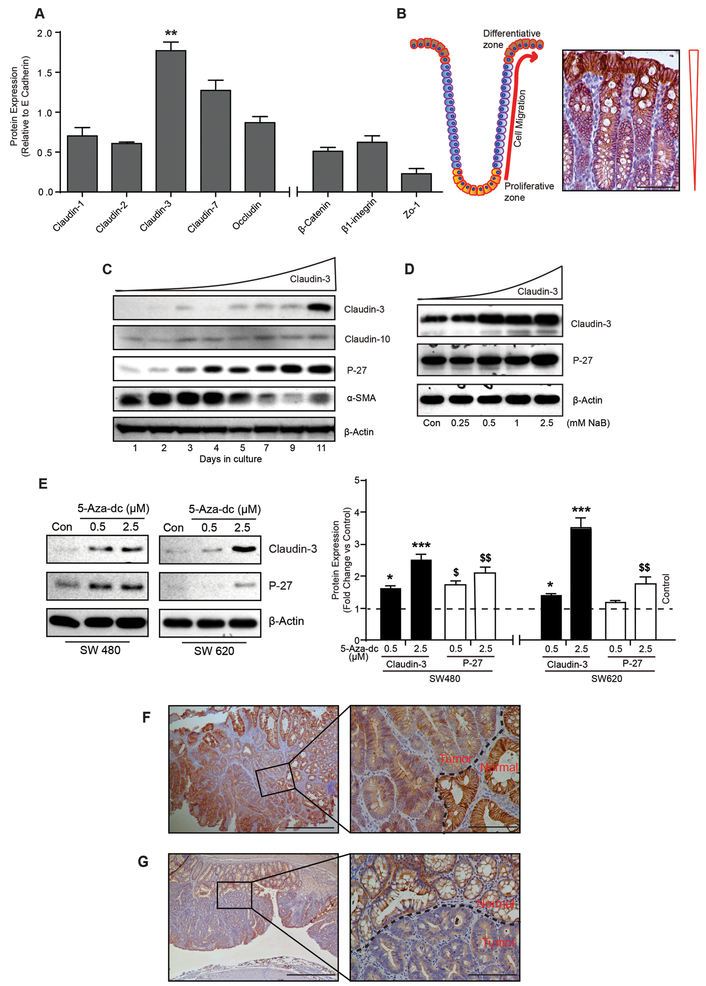
Figure 1: Claudin-3 expression associates positively with colonic epithelial differentiation and decreases sharply in colon tumors.
A. Endogenous expression of prominent cell adhesion proteins in mouse colon (n=6). Data presented in the graph represent values after normalizing the band intensity for each protein to E-cadherin expression; B. Cartoon depicting normal organization of a colonic crypt and differentiation zone, and representative immunohistochemistry using anti-claudin-3 antibody; C. Immunoblot analysis using cell lysate from Caco-2 cells subjected to spontaneous differentiation, and D. NaB treatment; E. Immunoblot and quantitative analysis showing effect of global demethylation using 5-aza-2’-deoxycytidine upon claudin-3 and P-27 expressions in colon cancer cells. F. Representative immunohistochemical analysis using APCmin mice colon (n=4) and anti-claudin-3 antibody; and G. Representative immunohistochemical analysis using AOM-DSS treated mice colon (n=4) and anti-claudin-3 antibody. Tumor area is marked by black dotted line. For immunoblot analysis, β-actin was used to confirm equal protein loading. **P>0.01. Values presented are mean ± S.E.M.
We further evaluated a causal link between claudin-3 expression and colonocyte differentiation using in vitro models of colonic epithelial cell (CEC) differentiation. In this regard, Caco-2 cells undergo spontaneous differentiation in culture or when subjected to HDACs (histone deacetylase) inhibitors (14, 15). Samples from both models of differentiation were examined. We used P-27(Kip1) and α-SMA (Smooth Muscle Actin), as positive and negative markers of CEC differentiation, respectively as controls (16) (17). A sustained increase in P-27(Kip1) expression and simultaneous decrease in α-SMA levels supported the validity of differentiation models. Moreover, claudin-3 expression followed suit with P-27(Kip1) expression and contrasted with α-SMA levels. In same samples, claudin-10 expression remained largely unaltered (Fig-1C–D).
Importantly, de-differentiated phenotype characterizes colon cancer cells where poorly differentiated CRC cells are highly aggressive. Accordingly, immunoblot analysis using a panel of 14 CRC cell lines demonstrated markedly suppressed claudin-3 levels in 10 of 14 cell lines and almost undetectable expression in HCT116 and SW620 cells (Supplementary Fig-S1B). Of note, both cell lines are poorly differentiated and highly aggressive in vivo (18, 19). Taken together, our data suggested a positive association of claudin-3 expression with CEC differentiation.
Colonic claudin-3 expression is epigenetically regulated:
Cancer cells use epigenetic regulation to exert inhibitory effect upon tumor suppressive genes and/or induce synthesis of oncogenic factors (20). Claudin-3 gene contains multiple CPG islands in its promoter (21) and epigenetic regulation of claudin-3 expression, in other cancers, has been demonstrated. We therefore asked if loss of claudin-3 expression in CRC cells is epigenetically regulated. To determine, we subjected SW480 and SW620 cells to global demethylation using 5-aza-2’-deoxycytidine (2.5μm). Samples were collected 72-hours post-treatment. Notably, in our screening, SW480 cells demonstrated low and SW620 cells demonstrated almost undetectable claudin-3 expression (Supplementary Fig-S1B). Indeed, in both cell types 5-aza treatment induced robust claudin-3 expression in dose-dependent manner (Figure-1E). These data supported epigenetic silencing as potential mechanism underlying suppressed claudin-3 expression in colon cancer.
Claudin-3 loss is an early event and associates positively with CRC progression and poor patient survival:
To further determine if claudin-3 expression is similarly suppressed in vivo, we examined the colon of APCmin mice (Supplementary Fig-S2A–B) (22). Since normal colon harbors robust claudin-3 levels, we reasoned that immunoblotting may not suffice, with reasonable confidence, to differentiate claudin-3 expression between APCmin mice and wild type (WT) mice colon lysate. In lieu, we performed IHC which showed chronic decrease in claudin-3 expression in colon tumor specific manner (Fig-1F). In adjacent normal crypts, claudin-3 expression was intense, membrane localized and apicolateral (versus expected apical localization). Interestingly, the loss of lateral claudin-3 immunoreactivity preceded the loss of the apical claudin-3 in these colon tumors (Supplementary Fig-S1C–D). Moreover, tumors deficient in claudin-3 expression still retained E-cadherin expression and thus supported potential of claudin-3 to serve as a sensitive prognostic biomarker to assess colonocyte transformation than E-cadherin (Supplementary Fig-S2F–G). We found similar tumor specific loss of claudin-3 in mice subjected to colitis-associated cancer (CAC; AOM-DSS-induced colon cancer (Fig-1G, Supplementary Fig-S1D, –S2C–E and –G) (23). Overall, our data demonstrated early loss of claudin-3 expression in colon cancer.
To further determine clinical significance of our observations, we determined claudin-3 expression in CRC patients and correlation with cancer progression. Initial assessment of claudin-3 mRNA expression in a large patient cohort (>250 CRC patient samples; demographics previously described (19) indeed demonstrated significant decrease in claudin-3 mRNA expression in colon adenomas (P<0.05). However, this decrease was progressive as loss of claudin-3 in all stages of CRC (versus normal mucosa) was highly significant (p<0.001) (Fig-2A). Subsequent IHC analysis of a CRC TMA (tissue microarray; 157 cores) demonstrated, similar to the mouse colon, robust apicolateral claudin-3 expression in normal colon. However, expression and membrane localization of claudin-3 were drastically compromised in the adenocarcinomas and metastatic samples (Fig-2B and Supplementary Fig-S3A–B). Blinded evaluation of staining intensity in normal, adenocarcinoma and metastatic CRC samples by a pathologist further confirmed significant decreases in claudin-3 expression in CRC-samples (versus normal colon, P<0.0001 (Fig-2C). We found similar decreases in claudin-3 expression in specimen from colitis-associated cancer patients (Fig-2D). Parallel use of the same antibody in normal prostate and prostate cancer demonstrated robust increase in adenocacrcinoma versus normal tissue, and thus supported antibody specificity (Supplementary Fig-S3C). We further examined if claudin-3 levels may also be of prognostic significance by evaluating association of claudin-3 expression with overall survival among CRC-patients. Based upon median claudin-3 expression values, the Kaplan-Meier survival analysis indeed demonstrated a significant and positive correlation of greater claudin-3 expression with better patient survival (p<0.0491) (Fig-2E). Taken together, we found a positive correlation between claudin-3 expression loss, CRC-progression, and poor patient survival.
Figure 2: Claudin-3 expression decreases in colon cancer in stage specific manner.
A. Claudin-3 transcriptome analysis using RNA isolated from colon cancer patients. *P>0.05 (normal versus adenoma) and ***P>0.001 (normal versus all stages) (n=250); B and C. Representative immunohistochemical staining of spontaneous colon cancer [CRC; normal mucosa (n=10) (a), adenoma (n=5) (b), adenocarcinoma (n=20) (c) and lymph node metastasis (n=16) (d)] using anti-claudin-3 antibody and intensity score analysis of immunohistochemical staining of CRC samples ****P>0.0001 (versus normal mucosa); D. Representative immunohistochemical staining of colitis associated cancer (CAC) using anti-claudin-3 antibody; E. Kaplan-Meir analysis to determine overall survival where red and blue colors indicate high and low claudin-3 expression, respectively.
Claudin-3 null (Cldn-3KO) mice demonstrate de-differentiated colonic epithelium, infiltratory immune microenvironment and aggressive tumor burden:
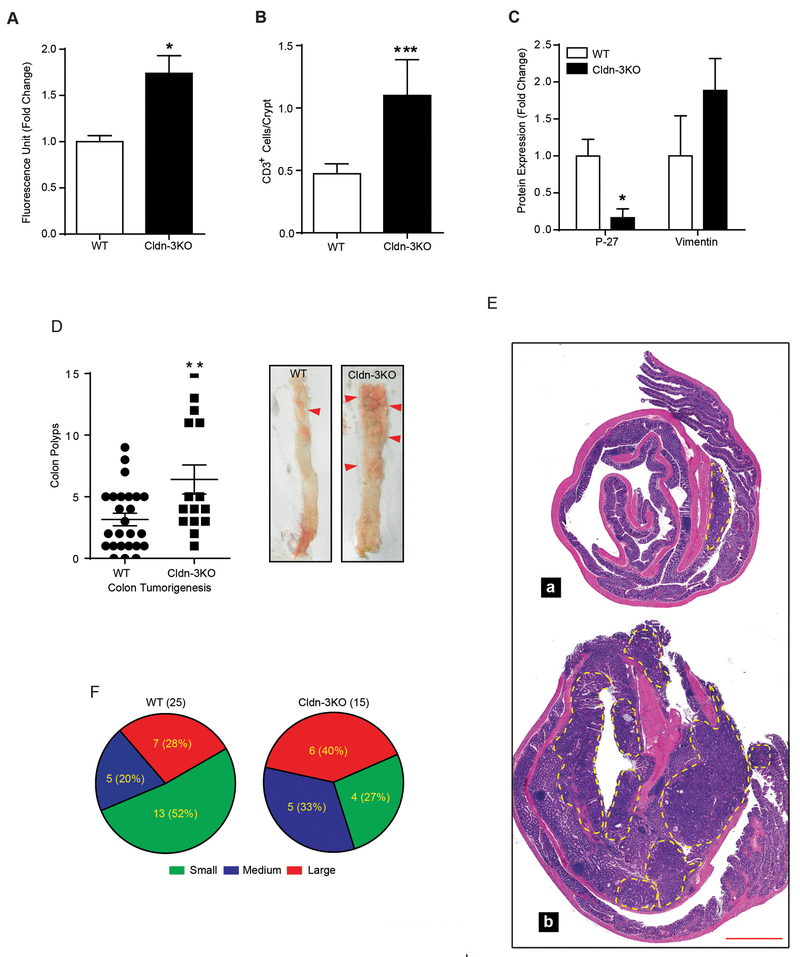
To further evaluate if claudin-3 loss is causally associated with colon tumorigenesis, we used Cldn-3KO mice (see methods). Immunoblot analysis using mice colon lysate confirmed inhibition of claudin-3 protein synthesis in Cldn-3KO mice. Interestingly, claudin-3 depletion did not affect colonic expression or cellular localization of claudin-1, −2 or −7 proteins, known to be altered in CRC (19, 24, 25) (Supplementary Fig-S4A and –S4B). Irrespective, analysis using rectal administration of FITC-Dextran (4kDa) to determine mucosal barrier dysfunction demonstrated a significant increase in mucosal leakage in Cldn-3KO mice (versus WT-mice; p<0.05) (Fig-3A). Additionally, IHC analysis using anti-CD3 antibody demonstrated marked upregulation in T-cell infiltration in naïve Cldn-3KO mice (versus WT-mice, p<0.001; Fig-3B and Supplementary Fig-S4D). Most notably, immunoblot analysis demonstrated suppressed P-27(Kip-1) expression (p<0.05) and contrasting upregulation of Vimentin, a mesenchymal cell marker, in Cldn-3KO mice colon (versus WT mice; Fig-3C and Supplementary Fig-S4C). Taken together, our data supported an important role of claudin-3 in maintaining differentiated CEC phenotype and mucosal barrier function, and potential susceptibility of Cldn-3KO mice to colon tumorigenesis.
Figure 3: Deletion (Genetic) of colonic claudin-3 expression induces colonic epithelial de-differentiation and leaky gut, and promotes colon cancer burden.
A. FITC-dextran passes (4kDa) to determine mucosal permeability (at least 4 mice/group); B. Quantitative analysis of immune infiltration (CD3+ cells) into the mucosa (3 mice/group with at least 100 crypts/mice examined); C. Quantitative analysis of the immunoblot outcome using colon lysates from naïve Cldn-3KO and WT mice (4 mice/ group); D. Polyps numbers in Cldn-3KO (n#15) and littermate WT-mice (n#25) subjected to colon tumorigenesis and representative images of the colon from Cldn-3KO and WT-mice subjected to colon tumorigenesis; E. H&E staining of the swiss-rolled colons; F. Graphical representation of colonic tumor distribution according to size. (**p<0.01); Values presented are mean ± S.E.M.
We further examined if loss of colonic claudin-3 expression can promote colon cancer progression. Cldn-3KO mice were subjected to the AOM-DSS induced colon carcinogenesis. Remarkably, Cldn-3KO mice subjected to colon cancer not only demonstrated significant body weight loss during the DSSIII cycle (p>0.05; versus WT-mice; Supplementary Fig-S5A). Also, some AOM-DSS treated Cldn-3KO mice died before the study termination (data not included). Moreover, we observed significant increase in the colon weight (gm/cm length; p<0.05) of AOM-DSS treated Cldn-3KO mice (versus WT mice; Supplementary Fig-S5B). In corroboration, gross tumor numbers, at the study end, demonstrated significant increase (p<0.01) in colon tumor burden in Cldn-3KO mice (versus WT mice; Fig-3D–E). Further analysis revealed that the tumors in Cldn-3KO mice were not only more in number but also in size (large, intermediate and small tumors: 40%, 33% and 27% in Cldn-3KO mice versus 28%, 20% and 52% in WT mice; Fig-3F). In accordance, Cldn-3KO mice showed significantly higher tumor score (p<0.5), a recently described measure of cancer progression (26) (Fig-4A). Most notably, 33% of Cldn-3KO mice developed invasive adenocarcinomas (p<0.01) compared to none (0%) in WT mice (Fig-4B–C). Overall, these results uncovered a novel, and tissue specific cancer suppressing role for claudin-3 in colon cancer.
Figure 4: Loss of colonic claudin-3 induces cancer invasiveness and promotes vimentin expression.
A. Colonic tumor scores in Cldn-3KO (n=15) and WT (n=25) mice (**p<0.05); B. A majority of the colon tumors in Cldn-3KO mice were invasive (Fisher exact test **p<0.01 versus WT mice) and C. Representative H&E staining showing invasive neoplasm in Cldn-3KO mice; D. Silencing of claudin-3 expression in colonic epithelial cells induces de-differentiation: immunoblot analysis using cell lysates from control or claudin-3 siRNA transfected Caco-2 cells; E. Effect of siRNA silencing of claudin-3 expression upon cell mobility and cell invasion; F. Spearman’s correlation analysis of colon cancer patient microarray data to determine association between claudin-3 and vimentin expressions; and G. Representative co-immunofluorescence analysis using AOM/DSS-treated mice colon, anti-claudin-3 (red) and vimentin (green) antibody (n=3 and 15 high power field (20x)/mice was used for determination. Values are mean ± S.E.M. *p<0.05, **p<0.01 versus control.
Claudin-3 depletion in CRC cells induces de-differentiated phenotype and invasive mobility:
To further elucidate the causal role of claudin-3 in regulating CEC differentiation to inhibit cancer cell phenotype, we silenced claudin-3 expression in CRC cells using anti-human claudin-3 or control siRNA. Immunoblotting to determine altered protein expression, wound healing and cell invasion assays were performed. As shown in Fig-4D, immunoblot analysis demonstrated that claudin-3 depletion, similar to in vivo conditions, inhibited P-27(Kip1) expression however upregulated vimentin and cyclin-D1 expression. Moreover, claudin-3 silenced cells demonstrated significant increases in migration (wound-healing assay) and invasion (Boyden chamber assay) (p<0.05 and p<0.01 respectively; versus control cells) (Fig-4E).
Claudin-3 expression associates inversely with vimentin expression:
One of our key findings was a consistent increase in vimentin expression, a type III intermediate filament and EMT marker, under claudin-3 suppressed conditions. In colon cancer, vimentin expression associates positively with high malignant potential and poor prognosis of colorectal patients, and is highly expressed in cancer cells at the invading front (27, 28). We therefore examined potential association between claudin-3 and vimentin expression in CRC patients. Spearman’s correlation analysis, using the same patient cohort utilized in the analysis described in Fig-2A, indeed demonstrated a significant inverse correlation between claudin-3 and vimentin expression in patient samples (p<0.0002) (Fig-4F). We further examined whether this correlation is simply byproduct of the end point changes in gene expressions in cancer cells or dynamic association. Co-immunolocalization for claudin-3 and vimentin was performed using the colon of AOM/DSS-treated WT mice. Specific emergence of vimentin expression in tumor areas where claudin-3 expression is chronically suppressed suggested that the observed inverse correlation is dynamic and may regulate emergence of cancer invasive front (Fig-4G).
Hyperactivated Wnt/β-catenin signaling, intratumoral proliferation and cancer cell survival facilitate colon cancer progression in Cldn-3KO mice:
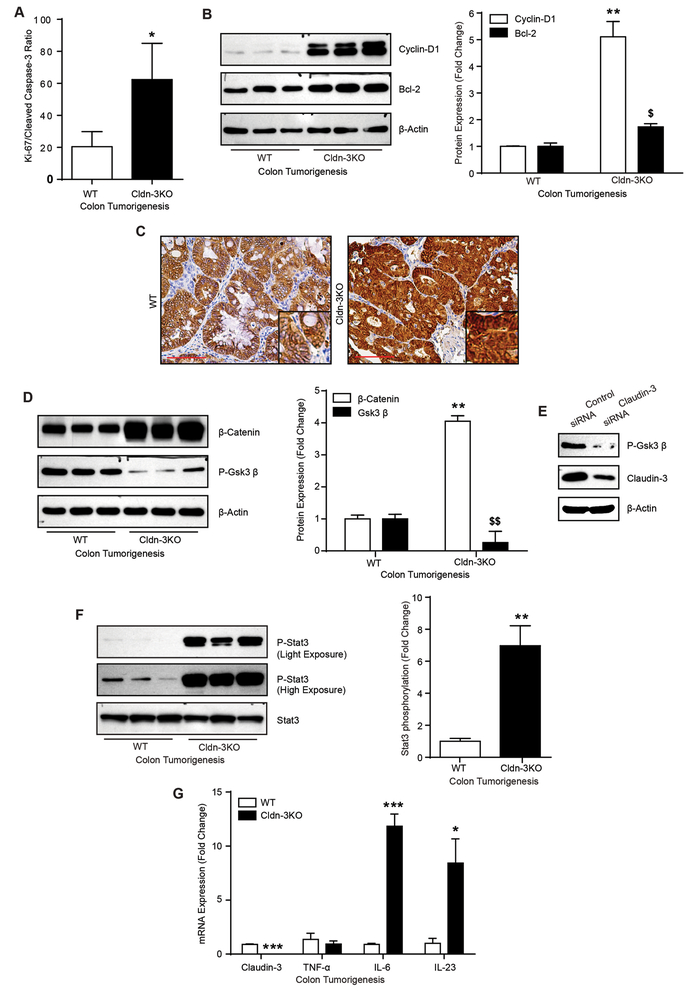
To further ascertain whether increased tumor burden in Cldn-3KO mice is result of an increase in proliferation, cancer cell survival, decreased apoptosis or all combined, we determined intratumoral proliferation and apoptosis by IHC using anti-Ki-67 and -cleaved caspase-3 antibodies. Analysis demonstrated a significant increase in proliferation/apoptosis ratio between Cldn-3KO and WT-mice mice tumors (p<0.05; Fig-5A and Supplementary Fig-S5C). Immunoblot analysis using total colon lysate from mice subjected to colon cancer further demonstrated a significant increase in cyclin-D1 expression (p<0.01), a proliferation inducer and Wnt/β-catenin-signaling target, and Bcl-2 (p<0.05), anti-apoptotic protein, in Cldn-3KO mice (versus WT-mice; Fig-5B). These findings suggested role of the increased tumor cell proliferation/survival in CRC progression fueled by the loss of claudin-3 expression.
Figure 5: Increased intratumoral proliferation and hyperactivation of the Wnt/β-catenin and IL-6/Stat-3 signaling characterize tumors in Cldn-3KO mice.
A. Quantitative representation of the proliferation/apoptosis ratio from immunohistochemical analysis of the serial sections of colon from WT and Cldn-3KO mice subjected to colon tumorigenesis, using Ki67 and cleaved caspase-3 expression as markers; B. Immunoblot analysis using colon lysate from WT and Cldn-3KO mice subjected to colon cancer and quantitative analysis of cyclin D1 and Bcl2 protein expression; C. Immunohistochemical analysis using anti β-catenin antibody; D. Immunoblot analysis using colon lysates from mice subjected to colon cancer and quantitative analysis of P-GSK-3β and β-catenin expression; E. Immunoblot analysis using control and Caco-2siRNA cells. F. Immunoblot analysis using colon lysates from Cldn-3KO and WT-mice subjected to colon tumorigenesis and quantitative analysis of Phospho-stat-3 expression relative to stat-3 levels; G. qRT-PCR using mRNA isolated from mice colon subjected to colon carcinogenesis (n=3 mice/group). Values are mean ± S.E.M. *p<0.05, **p<0.01 versus WT mice.
Notably, colon cancer regulation is a multifaceted process and involves dysregulation of multiple unique and/or overlapping pathways. However, a recent study suggest that claudin-3 loss may promote Wnt/β-catenin-signaling, key regulator of CRC progression, to promote hepatocellular cancer (29). Upregulation of cyclin-D1 in Cldn-3KO mice tumors (Fig-5B) or Caco-2siRNA cells (Fig-4D) further supported potential role of Wnt/β-catenin-signaling in claudin-3 dependent CRC-progression. Since, cytoplasmic/nuclear β-catenin expression signifies Wnt/β-catenin-signaling activation, IHC using anti-β-catenin antibody was done. As expected, we found cytoplasmic anti-β-catenin immunoreactivity in WT-mice tumors. However, the delocalized β-catenin expression was markedly upregulated and predominantly nuclear localized in Cldn-3KO mice colon tumors and thus suggested Wnt-β-catenin-signaling hyperactivation (Fig-5C). Immunoblot analysis using colon lysate further revealed a significant upregulation of β-catenin levels in AOM-DSS treated Cldn-3KO mice (versus WT-mice; p<0.01; Fig-5D) suggesting potential inhibition of protein degradation. Notably, GSK-3β regulates β-catenin expression through the regulation of its protein stability (30) and inactivation of GSK-3β (by phosphorylation) reduces β-catenin ubiquitination resulting in its nuclear accumulation (31). However, contrasting our expectation, P-GSK3β (Ser9) expression in Cldn-3KO mice was sharply decreased (versus WT-mice; p<0.001), (Fig-5D). We found similar decrease in P-GSK3β expression in Caco-2siRNA cells (Fig-5E) suggesting a GSK-3β independent role of claudin-3 in hyperactivating Wnt-β-catenin-signaling during colon tumorigenesis.
Claudin-3 loss induces IL6/gp130/Stat-3 signaling:
As described, we found increased gut permeability (p<0.05; Fig-3A) and exacerbated immune organization (CD3+ immune cell, Fig-3B) in naïve Cldn-3KO mice colon which was further augmented in their tumors (Supplementary Fig-S6A). Emerging studies suggest an intricate role of Stat-3 signaling in colon cancer progression by interlacing epithelial and immune homeostasis (2). Therefore, we examined if claudin-3 loss induces Stat-3 signaling. Immunoblot analysis using colon lysate indeed demonstrated a specific and significant (p<0.001) increase in P-Stat3(Tyr705) expression in Cldn-3KO mice subjected to colon cancer (versus WT mice; Fig-5F). Thereafter, IHC analysis was done to determine the source of the activated Stat-3 as phosphorylation shuttles Stat-3 protein into the nucleus to initiate a transcriptional program (Supplementary Fig-S6B). Of interest, the role of the IL-6/Stat-3 signaling, upregulated by dysregulated barrier function in colon adenomas, in CRC progression was recently reported (2). In concurrence, IHC analysis using anti-IL6 antibody demonstrated markedly higher immunoreactivity in Cldn-3KO versus WT mice tumors which was predominantly epithelial but also of immune cell origin (Supplementary Fig-S6C). qRT-PCR further demonstrated significant increases in IL-6 and IL-23, pro-tumorigenic cytokines but not in TNF-α expression in these mice (Fig-5G). Taken together, our data suggested that claudin-3 loss induces IL-6/IL23/Stat-3 signaling to promote colon cancer.
IL-6/gp130/Stat-3-signaling hyperactivates Wnt-signaling induction by the loss of claudin-3 expression:
A key finding in Caco-2siRNA cells was the marked delocalization of the membrane expressed β-catenin, similar to the in vivo findings (Fig-6A). Moreover, TOP-FLASH reporter activity was significantly increased (p<0.001) in these cells (Fig-6B). Additional studies where control or Caco-2siRNA cells were cultured on transwell filter support, bathed in same culture medium apically and basolaterally, demonstrated similar upregulation of Wnt-signaling suggesting that the effect of claudin-3 loss in inducing Wnt-signaling may be direct and not necessarily dependent on the leaky barrier (Supplementary Fig-S7A–C). We further determined if there is a cross talk between Wnt- and Stat-3 signaling in hyperactivating Wnt-signaling under claudin-3 depleted conditions. Previous studies have demonstrated a cross talk between Wnt signaling and IL-6/STAT3 signaling and cell survival (32, 33). To test, we first determined if Stat-3 signaling is also activated in Caco-2siRNA cells. IL-6 treatment served as positive control. Similar to the Cldn-3KO mice, we found robust P-Stat3(Tyr705) expression in Caco-2siRNA cells which was further augmented by IL-6 treatment (Fig-6C). Further determinations also uncovered marked increases in gp130 expression, the IL-6 receptor, in Caco-2siRNA cells, and also in Cldn-3KO mice (Supplementary Fig-S6D–E), suggesting potential activation of IL-6/gp130/Stat-3 signaling under claudin-3 depleted condition. Interestingly, IL-6 treatment also induced cyclin-D1 expression which again was highly upregulated in Caco-2siRNA cells (Fig-4D and –6C), similar to Cldn-3KO mice (Fig-5B). Cyclin-D1 is a Wnt-signaling target. Therefore, we further determined causal association between Stat-3 signaling and Wnt/β-catenin signaling. In this regard, IL-6 treatment exacerbated the delocalized β-catenin expression in Caco-2siRNA cells. In contrast, inhibiting Stat-3 signaling using Niclosamide (10 uM), a Phopsho-Stat-3 specific inhibitor, rescued the membrane localized β-catenin expression (Fig-6A). Niclosamide treatment also inhibited TOP-FLASH reporter activity in Caco-2siRNA cells and expression of Wnt-target genes cyclin-D1 and c-Myc (Fig-6D–E). Taken together, our studies suggested that the loss of claudin-3 expression promotes colon cancer by modifying epithelial and immune homeostasis by serving as a common rheostat for Stat-3 and Wnt-signaling (Fig-6F).
Figure 6: Claudin-3 loss induces IL6/gp130/Stat-3 dependent activation of the Wnt/β-catenin signaling.
A. Co-immunofluorescence analysis of β-catenin and claudin-3 in control or anti-claudin-3 siRNA transfected cells and in response to IL-6 treatment alone or in combination with Nicosolamide; B. TOP-FLASH reporter activity using control and claudin-3siRNA transfected cells. S33Y transfected cells served as positive control; C. Effect of IL-6 treatment upon Stat-3 phosphorylation and cyclin D1 expression in control and claudin-3siRNA transfected cells; D. TOP-FLASH reporter activity in control and claudin-3 siRNA transfected cells subjected to niclosamide treatment; E. mRNA expression analysis of Wnt-signaling targets cyclin-D1 and C-myc; F. Cartoon depicting how loss of colonic claudin-3 expression can modulate molecular and signaling events to promote CRC progression. Values are mean ± S.E.M. (*p<0.05, **p<0.01, ***p<0.001).
Discussion
A highly prevalent nuclear β-catenin expression and an overactivated Wnt/β-catenin signaling in aggressive colon cancer has fueled the concept that hyperactivated Wnt-signaling promotes CRC-progression, possibly by regulating the EMT switch (34, 35). However, Wnt-signaling is required for normal gut homeostasis and regeneration of adult intestinal epithelium, and therefore direct therapeutic targeting of this pathway to control CRC-malignancy has proven to be challenging. Thus, current findings confirming a novel colon cancer suppressive role of claudin-3 in its capacity to avert intratumoral Wnt-signaling hyperactivation is an important step in this direction. Our finding that the loss of claudin-3 activates Wnt-signaling which is then hyperactivated by the IL-6/gp130/Stat3 signaling further highlights the pivotal role of claudin-3 in regulating colonic epithelial and immune homeostasis. A role for Stat-3 signaling in accelerating CRC-progression is established (1, 2, 36). Also, Stat-3 dependent Wnt-signaling activation is now reported in specific neoplasms (33, 37). Overall, we here describe a novel role of claudin-3 in regulating CRC malignancy by serving as a common rheostat for two critical CRC-promoting signaling pathways.
Our findings however contrast an upregulated claudin-3 expression in many cancer types including the prostate, ovarian and renal cancers and thus suggest tissue specificity (11, 38, 39). Our findings also contradict limited reports suggesting CRC promoting role for claudin-3 (10). These inconsistencies necessitated comprehensive investigation to clarify the status and role of claudin-3 in colon carcinogenesis. Accordingly, our conclusions are based upon studies utilizing a panel of CRC cells with differing tumorigenic abilities, murine models of colorectal cancer, mRNA and protein expression analysis using a large CRC patient cohort, and genetic manipulation of claudin-3 expression in vitro and in vivo in claudin-3KO mice in the context of colon tumorigenesis. Moreover, we have confirmed the specificity of the reagents used to probe claudin-3 levels, using prostate cancer tissue where claudin-3 expression is highly upregulated. A decrease in claudin-3 expression in APCmin mice colon tumors in a recent study probing mechanisms underlying CRC progression supports our findings (1). Additionally, inhibition of PI-3 kinase signaling in poorly differentiated HCT116 cells induced claudin-3 expression along with epithelial differentiation (40). A similar decrease in claudin-3 expression was recently reported to promote hepatocellular carcinoma. Moreover, promoter hypermeythylation, also observed in our studies, suppresses claudin-3 levels in hepatocellular carcinomas (29). Interestingly, epigenetic regulation involving promoter derepression induces claudin-3 upregulation in prostate and ovarian cancers (41). Coincidently, siRNA silencing of DNA methytransferase, DNMT-1, altered claudin-3 expression differentially between PC3 (prostate cancer cells) and Caco-2 cells, suggesting tissue-specific regulation (42). While these data support tissue-specific epigenetic regulation of claudin-3 expression in functional correlation with cancer progression, further investigations are clearly needed for improved understanding of this differential regulation of claudin-3 in different cancer types.
Importantly, claudin family of proteins forms the “backbone” of TJ between epithelial cells (43). While TJ maturation associates with terminal differentiation, the disassembly of TJ can lead to the loss of polarity and increased cell mobility in addition to barrier dysfunction. However, claudin proteins are expressed in cell and tissue specific manner and therefore tissue-specific role of claudin proteins is highly probable (43). Our data suggest that in colonic epithelium claudin-3 protein is essential to uphold the differentiated phenotype. Our findings that claudin-3 loss was sufficient to induce colon cancer permissive microenvironment, without modulating expression of claudin-1, −2 or −7 causally associated with colon tumorigenesis, signifies its key role in colonic epithelial homeostasis (19, 24, 25). Of note, colonic claudin-3 expression increases post-birth and correlates with maturity of the mucosal barrier (44). On the other-hand, sharp decreases in claudin-3 expression associated with impaired barrier function in mice lacking the toll-like receptor adaptor protein MyD88. Moreover, probiotic bacterial colonization in MyD88 null mice colon not only improved the barrier maturity but also colonic increased claudin-3 levels (44). Specific loss of claudin-3 expression, among claudin proteins, also correlates with deregulated mucosal barrier in other pathological conditions of gastrointestinal tract (45–47). Furthermore, a recent study has demonstrated specific loss of claudin-3 expression along with deregulated colonocyte differentiation in mice lacking histone deacytylases HDAC-1 and HDAC-2 in their gut epithelium (48). Coincidentally, these mice also demonstrate increased Stat-3 activity, mucosal inflammation and colonocyte proliferation similar to Cldn-3KO mice (48). Interestingly, claudin-3 expression is also inhibited in the colon of Stat-3 knockout mice (49). Taken together, a complex inter-relationship between claudin-3 expression, mucosal barrier and Stat-3 signaling in regulating colonic homeostasis is plausible. In this regard, we have documented specific increases in the expression of inflammatory cytokine IL-6 in and its receptor gp130 in claudin-3 depleted conditions. Furthermore, upregulated P-Stat3 levels in Caco-2siRNA cells could be upregulated by exogenous administration of IL-6. This observation in association with our observation that P-Stat3 and IL-6 levels increase in both, epithelial and immune components in Cldn-3KO mice suggest collective contribution of epithelial and immune mechanisms in observed increases in Stat-3 signaling in Cldn-3KO mice. The role of the IL-6/Stat-3 signaling by activating IL-17/IL-23 signaling in promoting CRC progression was recently reported (1, 2). As described, impaired mucosal barrier in colon adenomas was found to be responsible for promoting intra-tumoral Stat-3 activation by allowing gut microbe translocation into the mucosa (1). The Cldn-3KO mice gut is significantly leakier than littermate WT-mice. Taken together, we postulate that the loss of claudin-3 expression dysregulates mucosal barrier function and upregulates gp130 expression, possibly in response to the altered epithelial differentiation. Altered mucosal barrier facilitates translocation of the gut antigens into the mucosa. Our data demonstrating significant upregulation of baseline immune cell infiltration into the colonic mucosa in Cldn-3KO versus WT-mice supports such a postulation. Resultant activation of the immune landscape especially IL-6 synthesis accompanied with upregulated gp130 levels results in activated Stat-3 signaling. IL-6 is an established regulator of Stat-3 signaling (50). However, deregulated mucosal barrier can also allow infiltration of the luminal growth factors and thus allow activation of growth permissive signaling cascade, as demonstrated elegantly by Soler et. Al. (3, 51). Clearly, additional studies are needed to understand global changes due to claudin-3 loss in the colonic mucosa and are currently underway using high throughput analysis. The ongoing studies in our laboratory are also aimed at examining the molecular details of claudin-3/IL/Stat-3 regulation and its significance in mucosal inflammatory pathologies.
Another interesting observation of our study, was the finding that Wnt/β-catenin signaling was hyperactivated in Cldn-3KO mice tumors, potentially independent of GSK-3β regulation. This conclusion is based upon multiple parameters associated with activated Wnt/β-catenin and include: 1) upregulated intratumoral nuclear β-catenin expression in Cldn-3KO mice compared to WT-mice; 2) Marked upregulation of total β-catenin expression levels in colon lysate from tumor bearing Cldn-3KO mice (versus WT-mice); 3) increased cyclin-D1 expression and proliferation/apoptosis ratio in tumor bearing Cldn-3KO mice (versus WT-mice); and 4) increased TOP-FLASH reporter activity in claudin-3 silenced CRC cells along with marked increases in Cyclin-D1, C-Myc and Ki-67 expression (versus control cells). Our findings gain strength from similar upregulation of Wnt/β-catenin signaling and suppressed claudin-3 levels in hepatocellular carcinoma (29). Notably, GSK-3β is a key enzyme in regulating Wnt/β-catenin signaling by stabilizing β-catenin expression as observed in our study. Increased phosphorylation (Ser9) of GSK-3β inhibits its ability to phosphorylate β-catenin to target it for ubiquitination and degradation. However, we have found contrasting suppression of P-GSK-3β expression in Cldn-3KO mice suggesting a different mechanism regulating β-catenin in our mouse model. Our data that inhibiting Stat-3 activation in claudin-3 silenced CRC cells inhibited TOP-FLASH reporter activity suggest plausible role of Stat-3 signaling in promoting Wnt-signaling under conditions of the loss of colonic claudin-3 expression in the colon. Notably, Stat-3 activation also promotes intestinal epithelial cell proliferation and Cyclin-D1 expression, a Wnt-signaling target (2, 52, 53). Furthermore, a cross-talk between Stat-3 activation and Wnt-signaling in regulating cancer progression is documented however molecular details remain largely unexplored (37). We predict that the loss of claudin-3 accelerates CRC progression by two distinct mechanisms: 1) by inducing Wnt-signaling and therefore EMT switch due to the loss of epithelial differentiation, and 2) by promoting intratumoral Wnt-signaling hyperactivation by activating Stat-3-signaling. Our findings that the loss of claudin-3 in vitro or in vivo inhibits P-27/Kip-1 expression, upregulates vimentin expression, Wnt- and IL-6/Stat-3-signaling strongly support such a postulation. In accordance, we have found a significant correlation between claudin-3 and vimentin expression in CRC samples and mouse colon tumors. Of interest, vimentin expression marks the invasive front in aggressive CRC samples.
Taken together, we here identify a novel and tissue-specific CRC-suppressive role of caludin-3. The fact that loss of claudin-3 expression not only promoted dedifferentiated colonic epithelial phenotype but also promoted IL-6/Stat-3 signaling suggest its pivotal role in serving as a central regulator of EMT and inflammation, two critical facilitators of CRC malignancy. Moreover, our data that loss of claudin-3 induces hyperactivation of Wnt/β-catenin signaling and its cross talk with Stat-3 signaling identifies this protein as an important therapeutic target in the fight against CRC. Positive association between claudin-3 loss and poor CRC patient survival supports such a postulation.
Materials and Methods
Cell culture and transfection:
The cell lines used in the current study were either purchased from ATCC or acquired from collaborative laboratories and have been described previously (19). Only low cell passages were used, and the identity of cell lines used for detailed studies was routinely verified by DNA fingerprinting. Cells were transfected with human claudin-3 specific or control siRNA or shRNA (Invitrogen, CA and Sigma-Aldrich, MO) using RNAiMAX Reagent (Invitrogen) as per manufacturer instructions. In studies involving determination of reporter activity, cells transfected with anti-claudin-3 siRNA were replated 24 hours post siRNA transfection and re-transfected with the TOPFLASH reporter and/or control constructs. Transfection efficiency was normalized to Renilla luciferase activity of the phRL-TK (Promega Inc.). Results are expressed as the mean relative luciferase activity ± S.E.M. (from at least three independent experiments).
Antibodies:
The antibodies against claudin-1, −2, −3, −4, and −7, were obtained from Invitrogen. The Bcl-2, E-cadherin and β-Catenin antibodies were obtained from BD Biosciences (San Jose, CA) The P-27 and vimentin antibodies were purchased from Santa Crutz (CA) and the β-actin and anti-α-SMA antibodies were from Sigma-Aldrich (MO). Cyclin-D1, P-Stat3, Stat-3 and GSK3β antibodies purchased from Cell Signaling Inc. (MA).
Immunoblotting and immunohistochemistry:
The methods for immunoblotting and Immunohistochemical analysis have been described previously (54).
Colon cancer patient mRNA analysis:
The protocols and procedures for the procurement of human tissue samples, details of the microarray platforms and statistical analysis have been described previously (19). To compare the expression level of each probe between normal tissue, adenomas and different cancer stages, we employed Mann–Whitney test.
Claudin-3 knockout mice:
Generation of Cldn-3KO mice was recently described (55). Absence of Claudin-3 protein and its mRNA transcript in Cldn-3KO mice was confirmed using Western blotting, immunohistochemistry and real-time quantitative PCR (Supplementary Fig. S4A–B and Fig. 5G). All animal experiments were performed according to institutional guidelines.
Colon permeability measurements:
To determine the potential changes in colonic permeability, FITC-dextran (4 kDa) was used, as described previously (Please see Supplementary Information) (54). Presence of the dye in the blood was assessed in samples collected before (0 h) and 30 min after FITC-dextran administration.
Induction of colonic epithelial differentiation:
Caco-2 cells undergo spontaneous differentiation when cultured over an extended period (14). Accordingly, 1×106 cells were plated in 6-well dishes and culture medium was changed every second day. Cells were harvested to prepare lysate at indicated times. Alternatively, cells were treated with sodium butyrate (NaB), a histone deacetylase inhibitor, using indicated concentrations. Of note, NaB induces differentiation among cancer cells (56).
Statistical analysis:
Data were analyzed with Graph Pad Prism Version 6.0 software and are presented as mean ± SEM. Statistical analysis was performed by Student’s t test, 1-way ANOVA analysis of variance and Fisher’s exact test. The Kaplan-Meier curve was used for survival analysis and Spearman’s correlation analysis was done to analyze the association between variables. P value of less than 0.05 was considered significant. *P < 0.05; **P < 0.01; ***P < 0.005.
Supplementary Material
Acknowlegement:
This work was supported by BX002086 (P.D.), DK088902 and BX002761 (A.B.S.).
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
Supplementary information is available at the Oncogene website.
References
- 1.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soler AP, Miller RD, Laughlin KV, Carp NZ, Klurfeld DM, Mullin JM. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20(8):1425–31. [DOI] [PubMed] [Google Scholar]
- 4.Mullin JM, McGinn MT. Epidermal growth factor-induced mitogenesis in kidney epithelial cells (LLC-PK1). Cancer Res. 1988;48(17):4886–91. [PubMed] [Google Scholar]
- 5.Mullin JM, McGinn MT. Effects of diacylglycerols on LLC-PK1 renal epithelia: similarity to phorbol ester tumor promoters. J Cell Physiol. 1988;134(3):357–66. [DOI] [PubMed] [Google Scholar]
- 6.Singh AB, Dhawan P. Claudins and cancer: Fall of the soldiers entrusted to protect the gate and keep the barrier intact. Seminars in cell & developmental biology. 2015. [DOI] [PubMed] [Google Scholar]
- 7.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–90. [DOI] [PubMed] [Google Scholar]
- 8.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. [DOI] [PubMed] [Google Scholar]
- 9.Holmes JL, Van Itallie CM, Rasmussen JE, Anderson JM. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr Patterns. 2006;6(6):581–8. [DOI] [PubMed] [Google Scholar]
- 10.de Souza WF, Fortunato-Miranda N, Robbs BK, de Araujo WM, de-Freitas-Junior JC, Bastos LG, et al. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: roles of ERK1/2 and PI3K-Akt as modulators of EGFR signaling. PLoS One. 2013;8(9):e74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long H, Crean CD, Lee WH, Cummings OW, Gabig TG. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001;61(21):7878–81. [PubMed] [Google Scholar]
- 12.Royer C, Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18(9):1470–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cereijido M, Contreras RG, Shoshani L, Flores-Benitez D, Larre I. Tight junction and polarity interaction in the transporting epithelial phenotype. Biochim Biophys Acta. 2008;1778(3):770–93. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto H, Erickson RH, Gum JR, Yoshioka M, Gum E, Kim YS. Biosynthesis of alkaline phosphatase during differentiation of the human colon cancer cell line Caco-2. Gastroenterology. 1990;98(5 Pt 1):1199–207. [DOI] [PubMed] [Google Scholar]
- 15.Dzierzewicz Z, Orchel A, Weglarz L, Latocha M, Wilczok T. Changes in the cellular behaviour of human colonic cell line Caco-2 in response to butyrate treatment. Acta Biochim Pol. 2002;49(1):211–20. [PubMed] [Google Scholar]
- 16.Deschenes C, Vezina A, Beaulieu JF, Rivard N. Role of p27(Kip1) in human intestinal cell differentiation. Gastroenterology. 2001;120(2):423–38. [DOI] [PubMed] [Google Scholar]
- 17.Kedinger M, Simon-Assmann P, Bouziges F, Arnold C, Alexandre E, Haffen K. Smooth muscle actin expression during rat gut development and induction in fetal skin fibroblastic cells associated with intestinal embryonic epithelium. Differentiation; research in biological diversity. 1990;43(2):87–97. [DOI] [PubMed] [Google Scholar]
- 18.Rajput A, Dominguez San Martin I, Rose R, Beko A, Levea C, Sharratt E, et al. Characterization of HCT116 human colon cancer cells in an orthotopic model. The Journal of surgical research. 2008;147(2):276–81. [DOI] [PubMed] [Google Scholar]
- 19.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115(7):1765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008;9(2):215–34. [DOI] [PubMed] [Google Scholar]
- 21.Honda H, Pazin MJ, D’Souza T, Ji H, Morin PJ. Regulation of the CLDN3 gene in ovarian cancer cells. Cancer biology & therapy. 2007;6(11):1733–42. [DOI] [PubMed] [Google Scholar]
- 22.Moser AR, Luongo C, Gould KA, McNeley MK, Shoemaker AR, Dove WF. ApcMin: a mouse model for intestinal and mammary tumorigenesis. European journal of cancer. 1995;31A(7–8):1061–4. [DOI] [PubMed] [Google Scholar]
- 23.Thaker AI, Shaker A, Rao MS, Ciorba MA. Modeling colitis-associated cancer with azoxymethane (AOM) and dextran sulfate sodium (DSS). Journal of visualized experiments : JoVE. 2012(67). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhawan P, Ahmad R, Chaturvedi R, Smith JJ, Midha R, Mittal MK, et al. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene. 2011;30(29):3234–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhat AA, Pope JL, Smith JJ, Ahmad R, Chen X, Washington MK, et al. Claudin-7 expression induces mesenchymal to epithelial transformation (MET) to inhibit colon tumorigenesis. Oncogene. 2015;34(35):4570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dube PE, Yan F, Punit S, Girish N, McElroy SJ, Washington MK, et al. Epidermal growth factor receptor inhibits colitis-associated cancer in mice. J Clin Invest. 2012;122(8):2780–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngan CY, Yamamoto H, Seshimo I, Tsujino T, Man-i M, Ikeda JI, et al. Quantitative evaluation of vimentin expression in tumour stroma of colorectal cancer. Br J Cancer. 2007;96(6):986–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaraja GM, Othman M, Fox BP, Alsaber R, Pellegrino CM, Zeng Y, et al. Gene expression signatures and biomarkers of noninvasive and invasive breast cancer cells: comprehensive profiles by representational difference analysis, microarrays and proteomics. Oncogene. 2006;25(16):2328–38. [DOI] [PubMed] [Google Scholar]
- 29.Jiang L, Yang YD, Fu L, Xu W, Liu D, Liang Q, et al. CLDN3 inhibits cancer aggressiveness via Wnt-EMT signaling and is a potential prognostic biomarker for hepatocellular carcinoma. Oncotarget. 2014;5(17):7663–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinoi T, Yamamoto H, Kishida M, Takada S, Kishida S, Kikuchi A. Complex formation of adenomatous polyposis coli gene product and axin facilitates glycogen synthase kinase-3 beta-dependent phosphorylation of beta-catenin and down-regulates beta-catenin. J Biol Chem. 2000;275(44):34399–406. [DOI] [PubMed] [Google Scholar]
- 31.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2003;1653(1):1–24. [DOI] [PubMed] [Google Scholar]
- 32.Pramanik KC, Fofaria NM, Gupta P, Ranjan A, Kim S-H, Srivastava SK. Inhibition of β-Catenin signaling suppresses pancreatic tumor growth by disrupting nuclear β-Catenin/TCF-1 complex: Critical role of STAT-3. Oncotarget. 2015;6(13):11561–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Keng VW, Patmore DM, Kendall JJ, Patel AV, Jousma E, et al. Insertional Mutagenesis Identifies a STAT3/Arid1b/beta-catenin Pathway Driving Neurofibroma Initiation. Cell Rep. 2016;14(8):1979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenbaum SP, Ordonez-Moran P, Puig I, Chicote I, Arques O, Landolfi S, et al. beta-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med. 2012;18(6):892–901. [DOI] [PubMed] [Google Scholar]
- 35.Gavert N, Ben-Ze’ev A. beta-Catenin signaling in biological control and cancer. J Cell Biochem. 2007;102(4):820–8. [DOI] [PubMed] [Google Scholar]
- 36.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pramanik KC, Fofaria NM, Gupta P, Ranjan A, Kim SH, Srivastava SK. Inhibition of beta-catenin signaling suppresses pancreatic tumor growth by disrupting nuclear beta-catenin/TCF-1 complex: critical role of STAT-3. Oncotarget. 2015;6(13):11561–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinzelmann-Schwarz VA, Gardiner-Garden M, Henshall SM, Scurry J, Scolyer RA, Davies MJ, et al. Overexpression of the cell adhesion molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian epithelium and ovarian cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10(13):4427–36. [DOI] [PubMed] [Google Scholar]
- 39.Bartholow TL, Chandran UR, Becich MJ, Parwani AV. Immunohistochemical profiles of claudin-3 in primary and metastatic prostatic adenocarcinoma. Diagnostic pathology. 2011;6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Araujo WM, Vidal FC, de Souza WF, de Freitas JC Jr., de Souza W, Morgado-Diaz JA. PI3K/Akt and GSK-3beta prevents in a differential fashion the malignant phenotype of colorectal cancer cells. J Cancer Res Clin Oncol. 2010;136(11):1773–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon MJ, Kim SS, Choi YL, Jung HS, Balch C, Kim SH, et al. Derepression of CLDN3 and CLDN4 during ovarian tumorigenesis is associated with loss of repressive histone modifications. Carcinogenesis. 2010;31(6):974–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaqinuddin A, Qureshi SA, Qazi R, Farooq S, Abbas F. DNMT1 silencing affects locus specific DNA methylation and increases prostate cancer derived PC3 cell invasiveness. J Urol. 2009;182(2):756–61. [DOI] [PubMed] [Google Scholar]
- 43.Singh AB, Dhawan P. Claudins and cancer: Fall of the soldiers entrusted to protect the gate and keep the barrier intact. Semin Cell Dev Biol. 2015;42:58–65. [DOI] [PubMed] [Google Scholar]
- 44.Patel RM, Myers LS, Kurundkar AR, Maheshwari A, Nusrat A, Lin PW. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am J Pathol. 2012;180(2):626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramalingam A, Wang X, Gabello M, Valenzano MC, Soler AP, Ko A, et al. Dietary methionine restriction improves colon tight junction barrier function and alters claudin expression pattern. Am J Physiol Cell Physiol. 2010;299(5):C1028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gibson DL, Ma C, Rosenberger CM, Bergstrom KS, Valdez Y, Huang JT, et al. Toll-like receptor 2 plays a critical role in maintaining mucosal integrity during Citrobacter rodentium-induced colitis. Cell Microbiol. 2008;10(2):388–403. [DOI] [PubMed] [Google Scholar]
- 47.Clark JA, Doelle SM, Halpern MD, Saunders TA, Holubec H, Dvorak K, et al. Intestinal barrier failure during experimental necrotizing enterocolitis: protective effect of EGF treatment. Am J Physiol Gastrointest Liver Physiol. 2006;291(5):G938–49. [DOI] [PubMed] [Google Scholar]
- 48.Turgeon N, Blais M, Gagne JM, Tardif V, Boudreau F, Perreault N, et al. HDAC1 and HDAC2 restrain the intestinal inflammatory response by regulating intestinal epithelial cell differentiation. PLoS One. 2013;8(9):e73785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J, Kim JC, Lee SE, Quinley C, Kim H, Herdman S, et al. Signal transducer and activator of transcription 3 (STAT3) protein suppresses adenoma-to-carcinoma transition in Apcmin/+ mice via regulation of Snail-1 (SNAI) protein stability. J Biol Chem. 2012;287(22):18182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henttinen T, Levy DE, Silvennoinen O, Hurme M. Activation of the signal transducer and transcription (STAT) signaling pathway in a primary T cell response. Critical role for IL-6. J Immunol. 1995;155(10):4582–7. [PubMed] [Google Scholar]
- 51.Mullin JM. Epithelial barriers, compartmentation, and cancer. Sci STKE. 2004;2004(216):pe2. [DOI] [PubMed] [Google Scholar]
- 52.Ray S, Ju X, Sun H, Finnerty CC, Herndon DN, Brasier AR. The IL-6 trans-signaling-STAT3 pathway mediates ECM and cellular proliferation in fibroblasts from hypertrophic scar. J Invest Dermatol. 2013;133(5):1212–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tadlaoui Hbibi A, Laguillier C, Souissi I, Lesage D, Le Coquil S, Cao A, et al. Efficient killing of SW480 colon carcinoma cells by a signal transducer and activator of transcription (STAT) 3 hairpin decoy oligodeoxynucleotide--interference with interferon-gamma-STAT1-mediated killing. FEBS J. 2009;276(9):2505–15. [DOI] [PubMed] [Google Scholar]
- 54.Ahmad R, Chaturvedi R, Olivares-Villagomez D, Habib T, Asim M, Shivesh P, et al. Targeted colonic claudin-2 expression renders resistance to epithelial injury, induces immune suppression, and protects from colitis. Mucosal Immunol. 2014;7(6):1340–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kooij G, Kopplin K, Blasig R, Stuiver M, Koning N, Goverse G, et al. Disturbed function of the blood-cerebrospinal fluid barrier aggravates neuro-inflammation. Acta Neuropathol. 2014;128(2):267–77. [DOI] [PubMed] [Google Scholar]
- 56.Krishnan M, Singh AB, Smith JJ, Sharma A, Chen X, Eschrich S, et al. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene. 2010;29(2):305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.