Abstract
Reactive oxygen species (ROS) are important signaling molecules, but their overproduction is associated with many cardiovascular diseases, including cardiomyopathy. ROS induce various oxidative modifications, among which glutathionylation is one of the significant protein oxidations that occur under oxidative stress. Despite previous efforts, direct and site-specific identification of glutathionylated proteins in cardiomyocytes has been limited. In this report, we used a clickable glutathione approach in a HL-1 mouse cardiomyocyte cell line under exposure to hydrogen peroxide, finding 1,763 glutathionylated peptides with specific Cys modification sites, which include many muscle-specific proteins. Bioinformatic and cluster analyses found 125 glutathionylated proteins, whose mutations or dysfunctions are associated with cardiomyopathy, many of which include sarcomeric structural and contractile proteins, chaperone, and other signaling or regulatory proteins. We further provide functional implication of glutathionylation for several identified proteins, including CSRP3/MLP and complex I, II, and III, by analyzing glutathionylated sites in their structures. Our report establishes a chemoselective method for direct identification of glutathionylated proteins and provides potential target proteins whose glutathionylation may contribute to muscle diseases.
Keywords: glutathionylation, site-specific identification, clickable glutathione, reactive oxygen species, cardiomyocytes
Graphical Abstract
INTRODUCTION
Reactive oxygen species (ROS) have emerged as important regulatory signaling molecules while their excess production contributes to various diseases, especially in the cardiovascular system.1–2 Myocardial ischemic reperfusion, which has been investigated for several decades, is well-known to induce excess ROS production from various sources, including NADPH oxidase and mitochondria.3 A high level of ROS production in the heart is associated with dysregulation of calcium signaling, reduction of myofilament contractile force, and mitochondrial dysfunction,1 all of which contribute to organ or tissue damages.3–4 In addition, more recent studies have demonstrated that oxidative stress resulting from ischemic reperfusion induces activation of several proteases, including MMP-2, with subsequent degradation of several sarcomeric proteins, such as titin and α-actinin, that are essential for maintenance of contractile sarcomeres,5–7 suggesting significance of ROS in cardiac and skeletal myocytes.
One of the major consequences of ROS in cells includes protein oxidation,8 especially at redox-sensitive Cys residues, such as sulfenylation, disulfide formation, and glutathionylation.9 Glutathionylation involves formation of a disulfide bond between a protein Cys residue and glutathione in response to various redox stimuli, thus causing structural and functional changes of numerous proteins.10 Several examples, especially in muscle cells, include glutathionylation of SERCA and ryanodine receptors that regulate calcium flux.11–12 Glutathionylation of complex I in the electron transport chain correlates with decrease in cellular energy production.10, 13–14 Glutathionylation of myofilament and sarcomeric proteins, such as titin,15 actin,16 and troponin I,17 alters contractile functions of myofilaments. We recently showed that glutathionylation of sarcomere-associated SMYD2 contributes to degradation of sarcomeric proteins.18 In addition, glutathionylation of myosin-binding protein C (MyBP-C) was shown to correlate with diastolic dysfunction in a hypertensive stress model.19
With the growing importance of glutathionylation in cardiac function, several strategies have been developed for proteomic identification of glutathionylated proteins.20 Biotinylated glutathione disulfide (biotin-GSSG) has been used to induce glutathionylation in the heart and other tissue homogenates, finding ~33 proteins.21 Gel-based diagonal electrophoresis has been applied to cardiomyocytes, finding protein disulfide and glutathionylation in ~93 proteins.22 Nitric oxide-induced S-oxidations in cardiomyocytes have been studied, finding over 260 Cys sites of glutathionylation.23 More recently, exercise-induced glutathionylation in mouse skeletal muscle tissue has been identified and quantified, finding over 2,200 Cys sites of glutathionylation.24 The latter two approaches23–24 provide an efficient means to identify glutathionylated proteins, but rely on indirect detection of glutathionylated residues, such as a resin-capture or isobaric tagging after reduction of glutathionylated proteins. Despite these significant advance, direct detection of specifically glutathionylated proteins at a proteomic level has been limited, especially in cardiomyocytes.
We recently developed a clickable glutathione method to detect glutathionylated proteins.25–26 In our approach, a mutant of glutathione synthetase (GS M4) leads to in situ biosynthesis of clickable glutathione (γGlu-Cys-azido-Ala, azido-glutathione, N3-GSH) upon incubation of azido-Ala to cells (Figure 1a).25 A clickable functionality on glutathione serves as a chemical tag for direct identification of glutathionylated proteins after click reaction with biotin-alkyne or rhodamine-alkyne (Figure 1a).20 A modified azido-glutathione is a good substrate of glutathione disulfide reductase (GR), glutathione transferase omega (GSTO) and glutaredoxin, without significant alteration of the redox system.18, 27 We have previously used a clickable glutathione approach for proteomic analysis of glutathionylated proteins in HEK293 cells, without identification of specific Cys residues.27 In this report, we have established our clickable glutathione method for direct identification of glutathionylated peptides with specific Cys sites, especially by adopting silyl ether-based cleavable biotin-alkyne28 for chemoselective elution and subsequent tandem mass analyses. We have applied this approach to HL-1 mouse cardiomyocytes, finding over 1,700 glutathionylated peptides with specific Cys sites. Bioinformatic analyses suggest that various biological processes could be affected by glutathionylation, including metabolism, translation, and protein folding. Further STRING and clustering analyses with cardiomyopathy-relevant genes found 125 glutathionylated proteins whose mutation or functional alterations are associated with cardiomyopathy. We also highlight several glutathionylated proteins in the sarcomere and the electron transport chain to suggest the potential functional effects of their glutathionylation based on the Cys sites.
Figure 1. A clickable glutathione approach to identify glutathionylated peptides with specific Cys residues.
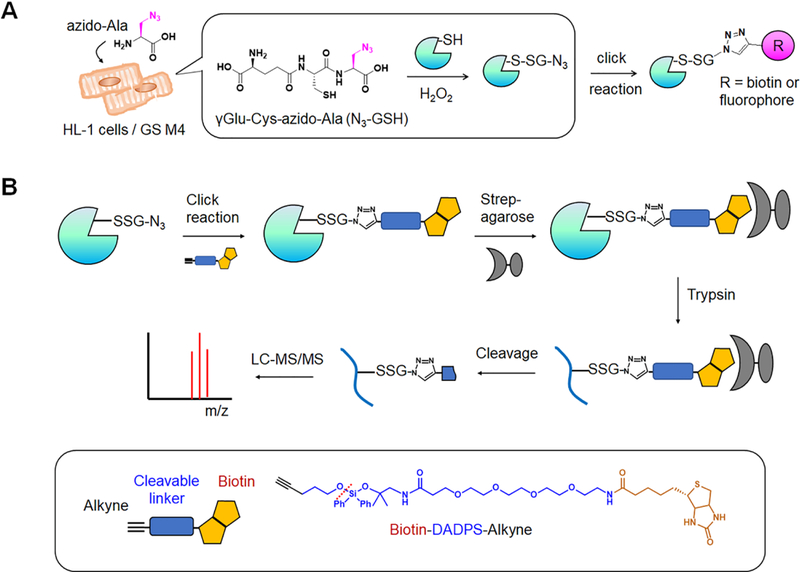
(A) A scheme for a clickable glutathione approach. HL-1 cells were transfected with a mutant of glutathione synthetase (GS M4), which uses azido-Ala to synthesize azido-glutathione (N3-GSH). After addition of ROS stimulus, glutathionylated proteins were identified after click reaction. (B) A scheme for isolation and elution of glutathionylated peptides by using biotin-DADPS-alkyne. After click reaction, biotinylated glutathionylated proteins were bound to streptavidin-agarose and digested by trypsin. Glutathionylated peptides were eluted by an acidic cleavage of a DADPS linker and analyzed by LC-MS/MS.
EXPERIMENTAL PROCEDURES
Material
Azido-alanine was synthesized as previously described.29 Other materials were purchased from the following vendors: biotin-DADPS-alkyne, biotin-DDE-alkyne, biotin-PC-alkyne and THPTA (Click Chemistry Tools), glucose oxidase (G2133), HL-1 cell line (SCC065), Claycomb medium (51800C), norepinephrine (A0937) (Sigma), Polybrene (Millipore), fetal bovine serum (HyClone), penicillin and streptomycin (Gibco), streptavidin-agarose (20359, Pierce), protease-inhibitor cocktail (A32955, Pierce), and Trypsin/Lys-C Mass spec grade (V5072, Promega). Adenovirus expressing GS M4 (Ad/GS M4) was prepared by Vector Biolabs.
Cell culture and induction of glutathionylation.
HL-1 cells were maintained at 37 °C in a 5% CO2 humidified atmosphere and were cultured in Claycomb medium supplemented with 10% FBS, penicillin (100 units/mL), streptomycin (100 μg/mL), norepinephrine (0.1 mM), and L-glutamine (2 mM) in fibronectin-gelatin-coated flasks. HL-1 cells were tested negative in mycoplasma detection test. HL-1 cells (80% confluency) in DMEM with 2% FBS were infected by incubating Ad-GS M4 along with polybrene. After 6 h, cells were incubated in Claycomb medium with 10% FBS for additional 18 h. Cells were then incubated with azido-Ala (0.6 mM) in Claycomb medium with 10% FBS for 24 h and treated with glucose oxidase (6 units) for 10 min to induce glutathionylation. Glucose oxidase (6 units) produces 6 μmole of hydrogen peroxide (H2O2) per min at pH 5.1, which equals to 1.5 mM per min (4 mL volume in our study). Glucose oxidase activity decreases to ~33% at pH 7.5 versus pH 5,30 thus the activity was presumed to be ~0.5 mM of H2O2 per min. Cells were then lysed using a lysis buffer [100 mM HEPES pH 7.6, 1% SDS, 100 mM LiCl, protease inhibitor cocktail, and 50 mM N-ethylmaleimide (NEM)]. Lysates were incubated at 4 °C for 30 min and passed through a 26-gauge needle (15 times) on ice. Protein concentration was determined by the Bradford assay.
Click chemistry and proteomic sample preparation.
Proteins (10 mg) were precipitated with ice-cold acetone (4 volumes of lysates). After centrifugation at 13,000 RPM for 15 min, the supernatant was removed. The pellet was air-dried for 5 min and resuspended in Solution A (1.44 mL for 10 mg proteins, PBS, pH 7.4, 0.1% SDS) with sonication. The resuspended solution was mixed and incubated with biotin-alkyne (final 0.4 mM, 160 μL of 5 mM stock in DMSO), CuBr [final 2 mM, 200 μL of 20 mM stock in DMSO/tBuOH (3/1)], and THPTA (final 2 mM, 200 μL of 20 mM stock in water) for 2 h at room temperature. Proteins were then precipitated with ice-cold acetone and the pellet was dissolved in Solution B (1 mL, PBS, 1.2% SDS) with sonication. Resuspended proteins were added to streptavidin-agarose (100 μL bead volume) suspended in PBS (5 mL) and incubated overnight at 4 °C. Proteins on beads were washed with PBS containing 0.2% SDS (3 × 5 mL) and PBS only (3 × 5 mL). Proteins on beads were incubated in a denaturation solution (0.5 mL, PBS, 6 M urea) at 37 °C for 45 min. After centrifugation, proteins on beads were incubated in a digestion buffer (0.2 mL, PBS, pH 7.4, 2 M urea, 1 mM CaCl2, 5 μg Trypsin/Lys-C) overnight at 37 °C. Beads were then washed with PBS containing 0.2% SDS (3 × 1 mL), PBS only (3 × 1 mL), and water (3 × 1 mL). After digestion, peptides on beads were eluted by incubating beads in 10% aq. formic acid (100 μL x 2) for 30 min for biotin-DADPS-alkyne, followed by a wash (100 μL). Similarly, peptides or proteins were eluted by 2% hydrazine in PBS for biotin-DDE-alkyne or UV-irradiation (365 nm wavelength) for biotin-PC-alkyne. Eluted sample (300 μL) was lyophilized and analyzed by LC-MS/MS.
LC-MS/MS analysis.
Peptides were separated by UHPLC reverse phase chromatography with an EASY-nLC 1000 liquid chromatography system (Thermo) and introduced into an Orbitrap FUSION mass spectrometer (Thermo). MS1 scans were between 350–1600 m/z and at 240,000 orbitrap resolution. Abundant peptides with +2 or +3 charges were fragmented by collision-induced dissociation (CID) at 35% collision energy, and peptides with charges between +3 and +7 were fragmented by electron-transfer dissociation (ETD) using calibrated charge-dependent ETD parameters. All MS2 fragmentations were acquired with ion traps at 0.60 Da resolution.
Database searching.
All MS/MS samples were analyzed using PEAKS Studio [Bioinformatics Solutions, version 8.5 (2018–05-07), Waterloo, Canada]. Charge state deconvolution and deisotoping were not performed. PEAKS Studio was set up to search the Uniprot_Mus_Compl_20170714 database, assuming the digestion enzyme as trypsin. PEAKS Studio was searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10.0 ppm. Glu to pyro-Glu of the N-terminus, Gln to pyro-Glu of the N-terminus, deamidation of Asn and Gln, oxidation of Met, acetylation of the N-terminus, NEM modification of cysteine, and glutathionylation of cysteine (+444 Da) were specified in PEAKS Studio as variable modifications.
Protein identification.
Scaffold (version Scaffold_4.8.6, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 99.0% probability by the peptide prophet algorithm31 with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the protein prophet algorithm.32 A protein prophet false discovery rate (FDR) was 0.4% and a peptide prophet FDR was 0.06%. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins were annotated with GO terms from NCBI (downloaded Oct 25, 2018).33
Bioinformatics.
Proteins that appeared N≥2 times in the positive samples were submitted to DAVID GO analysis (https://david.ncifcrf.gov/). For STRING analysis using the Cytoscape software, the identified mouse proteins for glutathionylation were converted to the human equivalents, which were loaded into STRING program as a ‘glutathionylation’ network. This ‘glutathionylation’ network was then merged with a ‘cardiomyopathy’ network that consists of cardiomyopathy proteins (cutoff value of 0.4) loaded from the STRING disease search.34 This merged network was then subjected to CLUSTER MAKER analysis using MCL Clustering with a granularity parameter of 4 and array sources from score. Similarly, 125 proteins that belong to both ‘cardiomyopathy’ and ‘glutathionylation’ networks were selected and clustered as described above.
RESULT
Detection of glutathionylated proteins in HL-1 cells
We previously used a clickable glutathione approach in HEK293 and H9c2 cells.18, 27 To extend our work to cardiomyocytes, we used a HL-1 mouse cardiomyocyte cell line, which is derived from AT-1 mouse atrial cardiomyocyte tumor lineage, maintains striated myofibrils with contractility, and retains a similar gene expression profile to adult atrial myocytes,35 thus suitable to find cardiac proteins in proteomic analyses. A glutathione synthetase mutant (GS M4) was expressed to HL-1 cells by infecting with adenovirus expressing GS M4 (Ad-GS M4). The level of overexpressed GS M4 was significantly higher than that of endogenous glutathione synthetase (GS) (Figure S1A). GS M4 expression did not alter expression of γ-glutamyl-cysteine-ligase (γGCLC), a rate-limiting enzyme in glutathione biosynthesis, nor levels of Trx1, Grx1, and cellular thiols (Figure S1), suggesting no significant perturbation of redox systems in cells.18, 27 After incubation of azido-Ala, HL-1 cells were treated with glucose oxidase (GluOx), which produces hydrogen peroxide (H2O2) by consuming glucose.36 The subsequent click reaction with Cy5-alkyne and in-gel fluorescence analysis showed strong signals of glutathionylated proteins upon addition of GluOx in dose- and time-dependent manners (Figure 2A and Figure S2). To identify a high number of potential glutathionylated proteins, we selected a relatively high concentration of GluOx, while incubating a short time period (10 min), which is expected to produce a relatively high amount of H2O2 based on catalytic activity of GluOx (~0.5 mM H2O2/min).
Figure 2. Identification of glutathionylated proteins in HL-1 cells.
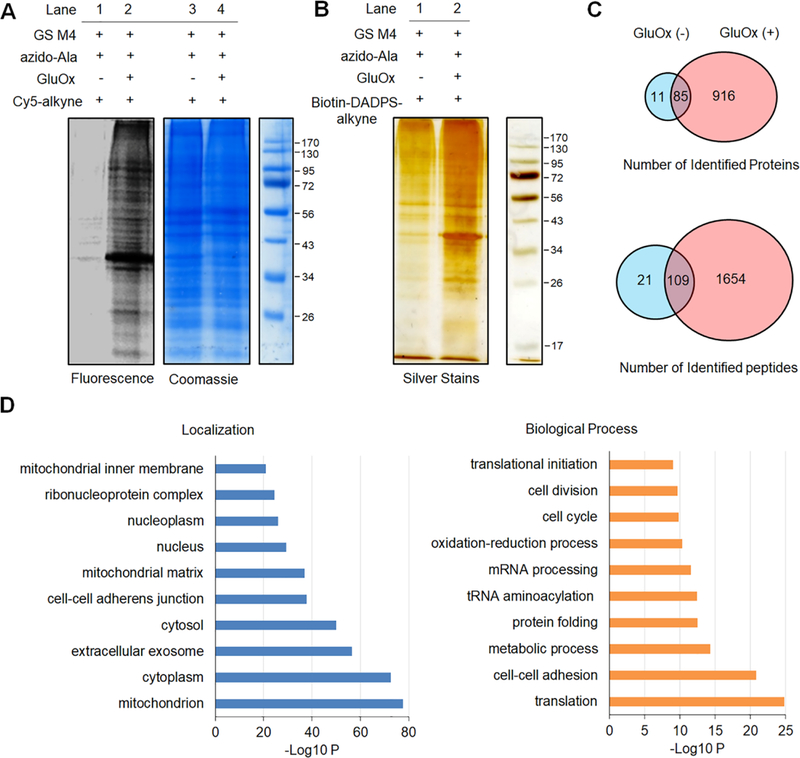
HL-1 cells expressing GS M4 were incubated with azido-Ala (0.6 mM) for 24 h and treated with glucose oxidase (GluOx, 6 units) for 10 min. (A) In-gel fluorescence detection of glutathionylated proteins. Collected lysates were subjected to click reaction with Cy5-alkyne for fluorescence detection. (B) Enrichment and elution of glutathionylated proteins. After click reaction of lysates with biotin-DADPS-alkyne, biotinylated glutathionylated proteins were bound to streptavidin-agarose, washed, eluted in an acidic cleavage condition (10% formic acid), and analyzed by silver staining. (C) The number of glutathionylated proteins and peptides under indicated conditions by LC-MS. Biotinylated glutathionylated proteins bound on streptavidin-agarose were digested by trypsin/Lys-C, eluted in an acidic cleavage solution, and analyzed by LC-MS/MS. (D) DAVID gene ontology (GO) analysis of identified glutathionylated proteins.
Optimization for mass spectrometry identification of glutathionylated proteins
Our clickable glutathione approach uses azido-glutathione in situ synthesized in cells expressing GS M4.25 After glutathionylation, all glutathionylated proteins can be enriched by binding to streptavidin-agarose after click reaction with biotin-alkyne. After on-bead trypsin and Lys-C digestion, glutathionylated peptides can be eluted and analyzed by LC-MS (Figure 1B). To detect the specific Cys sites, we used cleavable biotin-alkyne,28 which allows for elution of glutathionylated peptides without biotin, thus leaving a relatively small fragment of modification to the glutathione derivative conjugated to a Cys residue (Figure 1B and Figure S3). To maintain disulfide bonds in glutathionylation, we selected biotin-alkyne derivatives that include three different types of cleavable linkers compatible with non-reducing conditions, such as [1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-ethyl, (DDE)],37 dialkoxydiphenylsilane (DADPS),28 or photocleavable group (PC)28 (Figure 1B and Figure S3). Biotin-DDE-alkyne, biotin-DADPS-alkyne, and biotin-PC-alkyne can be cleaved by hydrazine at neutral pH,37 an acidic condition (10% formic acid), or irradiation at 365 nm, respectively.28
After confirming detection of glutathionylation in HL-1 cells (Figure 2A), we investigated enrichment of glutathionylated proteins and the efficiency of their elution by use of three biotin-alkyne derivatives. After click reaction with three different biotin-alkyne derivatives, all biotinylated glutathionylated proteins were captured on streptavidin beads and eluted at elution conditions specific to individual linkers, which were analyzed by silver staining in gels. With both DDE and DADPS linkers, stronger signals of eluted proteins were detected with samples treated with GluOx as opposed to without GluOx (Figure 2B and Figures S4A), suggesting selective elution of glutathionylated proteins by cleavable linkers. However, the PC linker did not show efficient elution upon irradiation while there is significant enrichment with biotin-PC-alkyne comparable to other linkers (Figure S4A).
Due to low efficiency of elution by the PC linker, we next proceeded to digestion and elution of peptides by two linkers (DDE and DADPS). The enriched glutathionylated proteins on beads were subjected to on-bead digestion with trypsin and Lys-C overnight. Our gel analysis showed apparently complete digestion of all proteins by trypsin and Lys-C under non-reducing conditions (Figure S4B). After digestion, the cleavage of a DDE linker by hydrazine in a neutral buffer or water resulted in a high amount of salts, which needed a significant desalting step before or during LC-MS analyses. Despite highly efficient elution of glutathionylated proteins with a DDE linker in gel analyses, the number of identified peptides in LC-MS/MS with a DDE linker was relatively small (<100). We also observed the fragile nature of γ-glutamyl-cysteine bond in glutathione during LC-MS analyses (Figure S5), as seen previously,13 suggesting the challenge of direct detection of glutathionylated peptides in mass analyses. Further optimizations, such as different collision energy in LC-MS, removal of desalting steps, or in-solution digestion, did not improve the result significantly. Despite the successful use of a DDE-linker in previous proteomic studies,37–38 we concluded that this linker may not be suitable in our proteomic analyses that involve disulfide bonds in glutathionylation. In contrast to a DDE-linker, the same procedure of enrichment and on-bead digestion, while eluting in an acidic condition from the DADPS linker, resulted in about 10 times higher number of glutathionylated peptides in LC-MS analyses, suggesting that biotin-DADPS-alkyne is more suitable to find glutathionylated peptides in our proteomic analyses than other cleavable linkers.
Identification and STRING analyses of glutathionylated proteins
Mass analysis of glutathionylated peptides from HL-1 cells incubated with GluOx (positive sample, minimum 2 out of 3 hits in triplicates) has found 1,001 glutathionylated proteins and 1,763 peptides (positive sample, minimum 2 out of 3 hits) with specific cysteine sites (99% protein and peptide thresholds) (Figure 2C and Table S1). A large number of glutathionylated peptides were identified with +3 charge states (71.6%), which were equally annotated by both fragmentation methods, CID (49%) and ETD (51%), in MS2 spectrums. Among all identified spectrums, 93% spectrums contain glutathionylation on peptides, while 7% or less spectrums show peptides without glutathionylation, suggesting relatively specific enrichment and cleavage of glutathionylated peptides. The sequence analysis surrounding glutathionylated Cys did not find any apparent conserved sequence motif (Figure S6), which agrees with a previous report that the local environment around Cys, rather than the sequence itself, may be important for glutathionylation.39–40 Interestingly, a relatively small portion of our identified proteins (ca. 27%, 274 out of 1,012 proteins) was found in previous proteomic analysis of glutathionylation from fatigue mouse skeletal muscle (Figure S6), suggesting that identity of glutathionylated proteins may depend on biological contexts. Notably, without treatment of GluOx, we have found 96 glutathionylated proteins (negative sample, minimum 2 out of 3 hits in triplicates) among which 85 proteins were also found in the positive samples, whereas 11 proteins were only found in negative samples (Figure 2C). For example, SERCA was found to be glutathionylated at C998 in the C-terminus even without incubation of GluOx, while its glutathionylation was found at C998 and additional Cys residues after incubation of GluOx (Table S1). However, the number of total spectrum counts of glutathionylated peptides in the negative samples was relatively low, suggesting that they may represent a low level of glutathionylation or result from their high redox-sensitivity prone to oxidation even under a low level of ROS.
Interestingly, DAVID gene ontology (GO) analyses41 showed that the largest number of glutathionylated proteins are localized at mitochondria (Figure 2D), suggesting that glutathionylation would make a significant impact on mitochondrial function. Consistently, many identified proteins are involved in mitochondrial metabolic processes, but also notably in translation, cell-cell adhesion, and protein folding (Figure 2D). Next, we sought to analyze our list of glutathionylated proteins for its relevance to cardiomyopathy by using STRING analysis.34 Because cardiomyopathy-relevant genes are documented in human proteins in STRING analysis, our identified proteins from the mouse HL-1 cell line were converted to corresponding human orthologs (964 proteins, ‘glutathionylation’ network), which were then merged with all proteins (1,359 proteins, ‘cardiomyopathy’ network) linked to cardiomyopathy imported from STRING disease query (Figure 3A). After merging two networks, clustering analyses showed that glutathionylated proteins interact with cardiomyopathy-relevant genes in the process of metabolism, tRNA acylation, chaperone, ubiquitination, sarcomere assembly, electron transport complex, and apoptosis (Figure S7). Importantly, we found that 125 out of 964 glutathionylated proteins belong to the genes whose mutations or dysfunctions are associated with cardiomyopathy (Figure 3A and Table S2). 125 identified proteins are mainly clustered in the process of sarcomere assembly, chaperone, mitochondrial metabolism, tRNA aminoacylation, and calcium signaling (Figure 3B), suggesting significance of glutathionylation in these processes in the etiology of cardiomyopathy.
Figure 3. STRING and cluster analysis of glutathionylated proteins with cardiomyopathy-relevant genes.
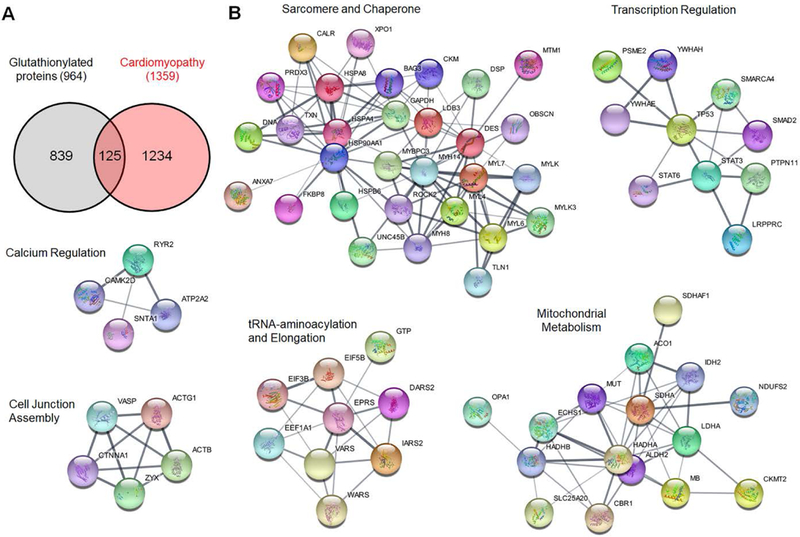
(A) The number of two merged networks that consist of ‘glutathionylated proteins’ and ‘cardiomyopathy-relevant genes’ loaded from STRING disease query in Cytoscape program. (B) Cluster analysis of 125 proteins that belong to both ‘glutathionylated protein’ and ‘cardiomyopathy-relevant gene’. The name of clusters was assigned after classifying proteins in individual clusters by DAVID GO analysis.
Ischemic reperfusion causes a production of ROS, which makes a significant impact on the muscle contractile units, sarcomeres and myofibrils, by direct oxidative modifications on sarcomeric proteins.8 Our cluster analyses of 125 proteins also indicated the significance of sarcomeric proteins for cardiomyopathy (Figure 3B). Therefore, we searched for a list of glutathionylated proteins associated with the sarcomere. We found 31 glutathionylated sarcomeric proteins (selected from NCBI GO annotation database in Scaffold) in the positive samples (Table 1), which include myofilament proteins [e.g. myosin heavy and light chains (MYH and MYL), actin, and myosin-binding protein C (MYPC3)], sarcomere-associated structural proteins [e.g. α-actinin (ACTN4), capping protein (CAPZB), desmin (DESM), and filamin C (FLNC)], and regulatory or adapter proteins [e.g. cysteine and glycine-rich protein 3 (CSRP3) and LIM domain-binding protein 3 (LDB3)]. Interestingly, many identified proteins are localized at the Z-disk of a sarcomere (Table 1), the lateral boundary of a sarcomere, which is important for structural stability of sarcomeres and is a nodal point of mechano-transduction signaling, such as force transmission from and to the sarcolemma.42–43 Identified proteins were provided with a disease score (in a range of 0–5) in STRING analysis (Table 1) with the highest values for several proteins (e.g. CSRP3,44–45 desmin,46–47 LDB3,48–49 and nexilin50) that are important for maintaining the sarcomere integrity. To validate mass spectrometry-based identifications, we have examined glutathionylation of selected sarcomere-related proteins (CSRP3 and α-actinin) by an alternative method. After enrichment of glutathionylated proteins by streptavidin-agarose, eluted proteins were analyzed by Western blotting, which confirmed glutathionylation of CSRP3 and α-actinin upon incubation of GluOx (Figure S2). Overall, these data suggest that glutathionylation of the identified sarcomeric proteins may alter the protein network in the sarcomere, contributing to muscle contractile dysfunction.
Table 1.
A list of glutathionylated sarcomere-associated proteins
| Accession # | Name | Glutathionylated peptide | (#/3)a | Lb | D-sc |
|---|---|---|---|---|---|
| BAG3_MOUSE | BCL2-associated athanogene 3 | SQSPAASDC185*SSSSSSASLPSSGR SGTPVHC295*PSPIR VSSAPIPC378*PSPSPAPSAVPSPPK |
2/3 2/3 3/3 |
Z | 5 |
| CSRP3_MOUSE | cysteine and glycine-rich protein 3 | TVYHAEEIQC25*NGR | 2/3 | Z | 5 |
| DESM_MOUSE | desmin | HQIQSYTC332*EIDALKGTNDSLMR | 2/3 | Z | 5 |
| LDB3_MOUSE | LIM domain binding 3 | TSLADVC584*FVEEQNNVYCER | 3/3 | Z | 5 |
| NEXN_MOUSE | nexilin | MRYEEC149*R GETYC569*LYLPETFPEDGGEYMCK |
2/3 3/3 |
Z | 5 |
| RYR2_MOUSE | ryanodine receptor 2 | AEVC1078*SGYGER FSSPSFVSISNDC1776*YQYSPEFPLDILK C2054*SSLQQLISETMVR |
3/3 3/3 3/3 |
A/Z | 5 |
| MYPC3_MOUSE | myosin binding protein C, cardiac | LTIDDVTPADEADYSFVPEGFAC619*NLSAK LLC715*ETEGR KPC1240*PYDGGVYVCR |
3/3 3/3 3/3 |
A | 5 |
| FLNC_MOUSE | filamin C, gamma | LYAQDADGC713*PIDIK VC1067*AYGPGLK GAGTGGLGLTVEGPC1104*EAK VGVTEGC1349*DPTR TPC2680*EEVYVK |
3/3 3/3 3/3 3/3 3/3 |
Z | 4.3 |
| OBSCN_MOUSE | obscurin | VRVEASGC1468*SR | 3/3 | Z/M | 1.8 |
| ILK_MOUSE | integrin-linked protein kinase | FSFQC346*PGR VALEGLRPTIPPGISPHVC422*K |
3/3 2/3 |
Z | 1.2 |
| MYL4_MOUSE | myosin, light polypeptide 4 | ITYGQC74*GDVLR MSEAEVEQLLSGQEADANGC180*INYEAFVK |
3/3 3/3 |
A | 1 |
| MYH8_MOUSE | myosin, heavy polypeptide 8 | AAYLQC392*LNSADLLK C698*NGVLEGIR |
3/3 3/3 |
M | 0.9 |
| MLRA_MOUSE | myosin, light polypeptide 7, regulatory | EAFSC43*IDQNR EAFSC43IDQNRDGIIC53*K SLC164*YIITHGDEKEE |
3/3 3/3 3/3 |
A | 0.9 |
| ACTN1_MOUSE | actinin, alpha 1 | AETAANRIC263*K C322*QLEINFNTLQTK MVSDINNAWGC370*LEQAEK IC480*DQWDNLGALTQK IDQLEC690*DHQLIQEALIFDNK |
3/3 3/3 3/3 3/3 3/3 |
Z | 0.8 |
| MTM1_MOUSE | myotubularin | C191*YELCETYPALLVVPYR | 2/3 | I | 0.8 |
| CHIP_MOUSE | STIP1 homology and U-Box containing protein 1 | AQQAC200*IEAK | 3/3 | Z/I | 0.7 |
| SYNE2_MOUSE | spectrin repeat containing, nuclear envelope 2 | DSASETYC328*NK SSAC1150*LQSK LC3117*AEENSR INNGLHC3691*VEK QLAASVSC4897*PEPEGQIAR TAAC6148*PNSSEVLYTNAK |
2/3 3/3 2/3 2/3 2/3 3/3 |
Z | 0.7 |
| MYH14_MOUSE | myosin, heavy polypeptide 14 | AEDMAELTC111*LNEASVLHNLR C722*NGVLEGIR |
3/3 3/3 |
M | 0.5 |
| MYL6_MOUSE | myosin, light polypeptide 6 | C2*DFTEDQTAEFK | 3/3 | M | 0.5 |
| ACTN4_MOUSE | actinin alpha 4 | AETAANRIC283*K C352*QLEINFNTLQTK IC500*DQWDNLGSLTHSR |
3/3 3/3 3/3 |
Z | N/A |
| ALDOA_MOUSE | aldolase A, fructose-bisphosphate | ALSDHHVYLEGTLLKPNMVTPGHAC240*TQK RALANSLAAC339*QGK |
2/3 2/3 |
Z | N/A |
| CAPZB_MOUSE | capping protein (actin filament) muscle Z-line, beta | NLSDLIDLVPSLC36*EDLLSSVDQPLK GC147*WDSIHVVEVQEK DETVSDC206*SPHIANIGR |
2/3 2/3 3/3 |
Z | N/A |
| DYST_MOUSE | dystonin | AC5144*MQTFLK | 3/3 | Z | N/A |
| FLNB_MOUSE | filamin, beta | GAGTGGLGLTVEGPC1081*EAK VGEPGILC1155*VDCSEAGPGTLGLEAVSDSGAK IAGPGLSSC1434*VR |
2/3 2/3 2/3 |
Z | N/A |
| PDLI5_MOUSE | PDZ and LIM domain 5 | QPTVTSVC213*SESAQELAEGQR | 3/3 | Z | N/A |
| PYGM_MOUSE | muscle glycogen phosphorylase | IC172*GGWQMEEADDWLR | 3/3 | Z | N/A |
| SPTN1_MOUSE | spectrin alpha, non-erythrocytic 1 | ALC315*AEADR TYLLDGSC2233*MVEESGTLESQLEATK |
2/3 3/3 |
Z | N/A |
| SYP2L_MOUSE | synaptopodin 2-like | AGGAESGPEEDALSLGAEAC681*NFMQPLGGR | 2/3 | Z | N/A |
| MYH10_MOUSE | myosin, heavy polypeptide 10 | VEDMAELTC95*LNEASVLHNLK C701*NGVLEGIR QGLETDNKELAC1238*EVK |
3/3 3/3 3/3 |
M | N/A |
| MYH9_MOUSE | myosin, heavy polypeptide 9 | VEDMAELTC91*LNEASVLHNLK C694*NGVLEGIR LEEDQIIMEDQNC988*K KMEDGVGC1379*LETAEEAK |
3/3 3/3 3/3 2/3 |
M | N/A |
| FLNA_MOUSE | filamin, alpha | SPYTVTVGQAC478*NPAACR VGTEC574*GNQK VQVQDNEGC717*SVEATVK YTPC441*GAGSYTIMVLFADQATPTSPIR AHVAPC1157*FDASK LQVEPAVDTSGVQC1260*YGPGIEGQGVFR VANPSGNLTDTYVQDC1312*GDGTYK |
2/3 2/3 3/3 3/3 3/3 2/3 3/3 |
Z | N/A |
The number of identifications of given peptides out of 3 replicates.
Location (L) in the sarcomere: Z, Z-disk; A, A-band; M, M-band; I, I-band.
Disease-score (D-s) from STRING disease query. N/A, not available.
Structural analyses of glutathionylated Cys residues
Next, we sought to locate glutathionylated Cys sites in selected available structural data, which may allow to predict the potential functional effect of glutathionylation. Among sarcomeric proteins, we noted CSRP3 whose human ortholog is known as muscle LIM protein (MLP). CSRP3/MLP plays an important role as a regulatory protein in the sarcomere as well as a co-transcription factor in the nucleus.45, 51 Multiple mutations of MLP are known to cause cardiomyopathy.52–54 MLP/CSRP3 has two zinc-finger LIM domains (LIM1 and LIM2) (Figure 4A), which are involved in self-dimerization and binding interactions with actin filaments, respectively, thus crosslinking actin filaments or blocking actin depolymerization.55–56 More recently, MLP was also reported to be a negative regulator of protein kinase Cα (PKCα) that is a key mediator of hypertrophy and heart failure.45 We identified glutathionylation at C25 in CSRP3/MLP (Figure 4B), which is located at the LIM1 domain (Figure 4C). Although C25 is not involved in zinc-binding, it lies at proximity (6–7 Å) to S54 and E55 (Figure 4C) whose mutations are associated with hypertrophic cardiomyopathy.52 A bulky glutathione (~8 × 10 Å area) modification at C25 is likely to disrupt CSRP3/MLP conformation around S54 and E55, which may contribute to functional alteration of CSRP3/MLP.
Figure 4. Analysis of glutathionylated Cys residues in CSRP3/MLP.
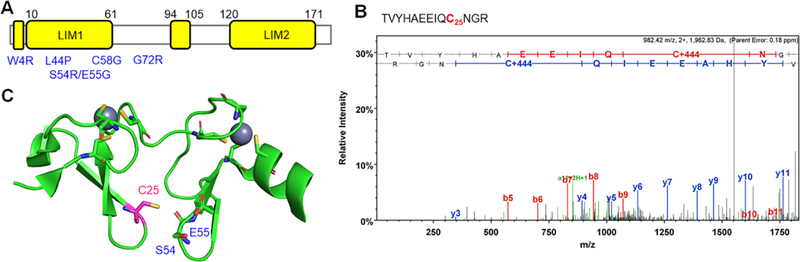
(A) Domains of CSRP3/MLP with mutations (blue) associated with cardiomyopathy. (B) Tandem (MS/MS) spectrum of a glutathionylated peptide at C25 in CSRP3/MLP. (C) A structure of CSRP3/MLP (PDB: 2O13) with a position of C25 (pink) in proximity (~6 Å) to S54 and E55 (blue).
In addition to sarcomeric proteins, ATP energy production from mitochondria is essential for muscle function. We have found glutathionylation of complex I, II, and III in the electron transport chains (Table 2), among which glutathionylation of complex I and II has been examined previously.13, 57 Complex I has a large structure (1.7 MDa) with 14 core subunits, which are essential for electron transfer coupled with proton pump, and 31 supernumerary subunits, which are largely absent in prokaryotes.58–59 Among 45 subunits in complex I, we found glutathionylation of 6 Cys residues in two core subunits (NDUFS1 and NDUFS2) and 8 Cys residues in 7 supernumerary subunits (Table 2 and Figure 5A). NDUFS1 (also known as 75 kD subunit, Figure 5A) contains iron-sulfur clusters (ISC), mediating the electron transfer from FMN in NDUFV1 (51 kD) to ubiquinone (UQ) in a binding site surrounded by NDUFS2 (49 kD), ND1, and NDUFS7 (PSST) subunits59 (Figure 5A). Our glutathionylated Cys residues in NDUFS1 include C75, C367, C463, C554, and C727 (Table 2). In the available structure (PDB: 4XTD) of complex I,60 C727 is at the flexible C-terminus, thus not determined in the structure. C367, C463, and C554 are relatively exposed to the protein surface (Figure 5B-C). However, all of them are distant from the electron transfer cofactors (Figure 5A-C). In contrast, C75 in NDUFS1 is relatively buried, but in proximity to iron-sulfur clusters (~5 Å) (Figure 5D), suggesting that glutathionylation at C75 could functionally interfere with the electron transfer through iron-sulfur clusters. In addition to NDUFS1 (75-kD), we also identified glutathionylation of C347 in NDUFS2 (known as 49-kD subunit), which is relatively proximal (~20 Å) to a UQ-binding site (Figure 5E). However, in this structure, C347 is in α-helix while facing away from the protein surface (Figure 5E). Thus, glutathionylation at C347 may not occur significantly, whereas its glutathionylation could distort the conformation around the UQ-binding site.
Table 2.
Complex I, II, and III with identified glutathionylated Cys residues.
| Name | Subunit | Role | Glutathionylated Cys | Note |
|---|---|---|---|---|
| Complex I | NDUFS1 | Core | C75, C367, C463 (S463), C554, C727 | catalytic core |
| NDUFS2 | Core | C347 | catalytic core with UQ site | |
| NDUFB9 | Supernumerary | C4, C31 | Interface with complex III | |
| NDUFA10 | Supernumerary | C183 | At N-terminus of α-helix | |
| NDUFA5 | Supernumerary | C17 | In an exposed loop | |
| NDUFA9 | Supernumerary | C86 | Close to NADPH | |
| NDUFS6 | Supernumerary | C79 (C87) | Close to Cys 115 | |
| NDUFA7 | Supernumerary | C55 | At N-terminus of α-helix | |
| NDUFAB1 | Supernumerary | C140 | Exposed | |
| Complex II | SDHA | C467, C475, C654 | FAD-binding site | |
| Complex III | UQCRC1 | Core | C268, C410, C453 | Close to complex I |
All numbers of Cys residues are based on mouse complex I. If the number is not conserved between mouse and bovine complex I, it is also shown with bovine numbers in parenthesis.
Figure 5. Analysis of glutathionylated Cys residues in Complex I.
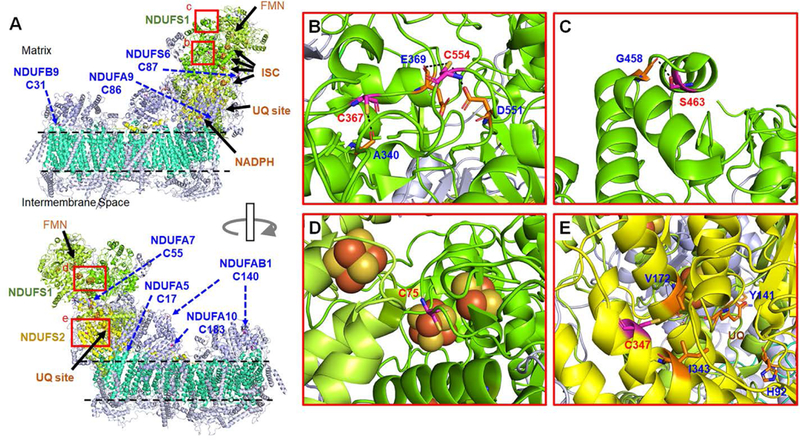
(A) A structure of bovine complex I (PDB: 5XTD) that contains 14 core subunits (7 cores in matrix, shown in lime, green, yellow, and 7 cores in membrane, shown in greencyan) and 31 supernumerary subunits (light blue). Glutathionylated Cys residues in core subunits [NDUFS1 (green) and NDUFS2 (yellow)] and supernumerary subunits are shown in rectangular boxes (b-e, red) and indicated by arrows (blue), respectively. (B-E) The enlarged structures of boxed areas (b-e) in (A), showing the position of glutathionylated Cys residues in core subunits. The residues that have potential interaction with the identified Cys (within 4 Å distance) are labeled with a dotted line. All numberings are based on the residue number in bovine complex I. Note that C463 in mouse NDUFS1 corresponds to S463 in bovine one.
In addition to catalytic core subunits, it is notable to see glutathionylation of 7 supernumerary subunits (Table 2, Figure 5A and Figure S8) where identified Cys residues are largely exposed to surface (Figure 5A and Figure S8). Biological significance of supernumerary subunits are less understood than core subunits, but supernumerary subunits are known to contribute to stability and assembly of the complex among other functions.58, 61 Recently, supernumerary subunits are suggested to be implicated in forming a mega-complex or respirasome containing complex I, II, III, and IV that increases efficiency of electron transfer.60, 62 We have identified glutathionylation of NDUFB9 and NDUFA9 subunits in complex I (Table 2), which are at the interface with complex III and complex II, respectively (Figure 6C, left). Therefore, glutathionylation at NDUFB9 and NDUFA9 in complex I may affect stability of a mega-complex under oxidative stress.
Figure 6. Analysis of glutathionylated Cys residues in Complex II, III, and mega-complex.
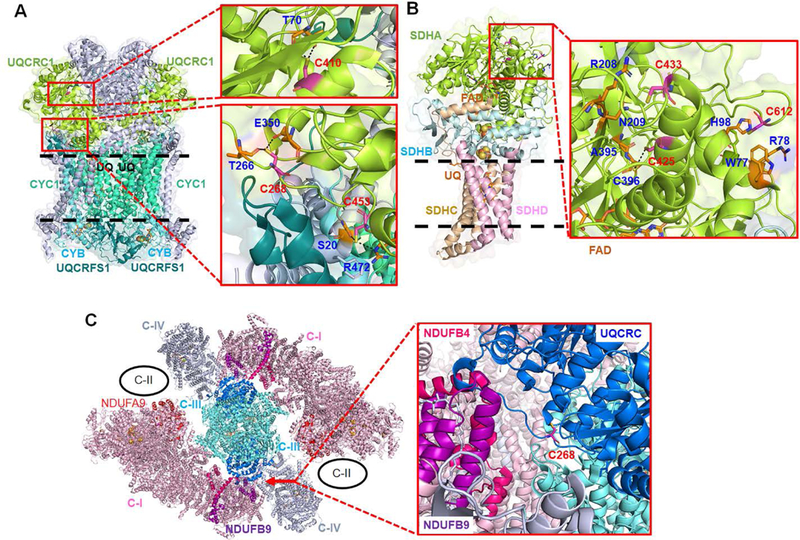
(A) A structure of bovine complex III (PDB: 5XTE), which shows three redox active subunits (CYC1, CYB, UQCRFS1) and UQCRC1 subunit that contains glutathionylated Cys residues. A ubiquinone (UQ) binding site (Qi) is labeled by ‘UQ’. (B) A structure of porcine complex II (PDB: 1ZOY), which shows an SDHA subunit that contains glutathionylated Cys residues. The residues that have potential interaction with the identified Cys (within 4 Å distance) are labeled with a dotted line. (C) A structure of a bovine mega-complex (PDB: 5XTI) that contains complex I (C-I), III (C-III), and IV (C-IV). The position of complex II (C-II) is shown in circle (black).60 Two subunits [NDUFB9 (purple) and NDUFB4 (hot pink)] in complex I are at the interface with UQCRC1 (deep blue) in complex III that contains glutathionylated Cys residues, especially at C268. Complex II is in proximity to NDUFA9 (red) in complex I that contains glutathionylation at C86.
Complex III forms an asymmetric dimer, with each monomer made of three redox-active subunits (CYC1, CYB, UQCRFS1) and 8 other subunits, which catalyzes transfer of two electrons from ubiquinol (UQH2) to two cytochrome C. We identified glutathionylation at UQCRC1 (Table 2 and Figure 6A), which is one of two core subunits (UQCRC1 and UQCRC2) in complex III. Recent structural data show that UQCRC1 of complex III forms significant interactions with NDUFB9 and NDUFB4 of complex I in a mega-complex (Figure 6C).60 Our identified glutathionylated Cys residues (C268, C410, C453) in UQCRC1 (Figure 6A) are located at this interface, especially C268 (about 10 Å away from NDUFB9) (Figure 6C), Thus, glutathionylation of UQCRC1 may disrupt the assembly between complex I and III.
Complex II is made of four subunits, including two hydrophilic subunits (SDHA and SDHB) at the matrix and two membrane-anchor proteins (SDHC and SDHD), which transfer two electrons to UQ resulting from oxidation of succinate to fumarate.63 Our identified glutathionylated Cys residues (C467, C475, C654 in our mass analysis, Table 2, corresponding to C425, C433, C612 after N-terminal cleavage) are in the SDHA subunit that contains a FAD cofactor (Figure 6B). C612 is close to the protein surface, but relatively hidden and surrounded by four aromatic or basic residues (W77, H98, F81, R78). However, C612 is at relatively flexible C-terminus, which may allow for its glutathionylation. C425 and C433 are at α-helix that extends to interact with a FAD cofactor (Figure 6B). Thus, bulky glutathione modifications on C425 and C433 residues may affect the anchored FAD conformation.
DISCUSSION
The significance of ROS or oxidative stress in myocytes is documented extensively in various muscle-relevant diseases, including cardiomyopathy, heart failure, and muscle dystrophy,1, 4 which are in part attributed to oxidative modifications, including glutathionylation, of many muscle proteins.8 In this report, we have used a clickable glutathione approach that allows for direct identification of glutathionylated peptides in proteomic mass analysis, without a need to reduce and re-alkylate glutathionylated Cys residues in a common biotin-switch method.20 With this approach, we identified over 1,000 proteins that are susceptible to glutathionylation in HL-1 cells in response to H2O2 with over 1,700 glutathionylated peptides, which include muscle-specific proteins. In our approach, we have adopted a cleavable linker, which leaves a small fragment of modification on glutathionylated peptides after cleavage or elution, thus increasing possibility of direct identification by mass spectrometry analysis. Interestingly, our procedure of enrichment with a biotin-DDE-linker resulted in a small number of identifications, as opposed to its several successful uses in previous studies,37–38 suggesting relative challenge for identification of glutathionylated proteins under oxidizing conditions. Overall, we found a silyl ether-based DADPS linker is suitable for our clickable glutathione approach that involves copper-mediated click chemistry and disulfide-bonds in glutathionylation.
DAVID GO analysis indicated a large number of glutathionylated mitochondrial proteins, together with cytosolic proteins, which may be expected due to high dependency of mitochondria in myocytes while suggesting significance of glutathionylation on mitochondrial proteins in myocytes. Our STRING and cluster analyses suggest 125 glutathionylated proteins that are functionally relevant to cardiomyopathy. It is notable to find clustered proteins in the biological process of sarcomere maintenance, chaperone, and metabolism, many of which are known to be largely redox-sensitive. In our analysis, we have focused on sarcomere-associated proteins that are essential for contractile functions in muscle. It is interesting to find that many of our identified sarcomeric proteins are located at the Z-disk, which is thought to have limited accessibility of oxidants due to a high level of organization,64 as opposed to I-band that is more accessible to oxidants due to many stretchable domains during relaxation.65 Due to the importance of Z-disk for sarcomere stability and mechano-transduction signaling, our analysis suggests that glutathionylation at the Z-disk may impact sarcomeric contractile functions.
Identification of glutathionylated proteins serves as an initial step toward understanding the functional significance of glutathionylated proteins.9 Importantly, identification of specific glutathionylated Cys residues, when combined with available structural information, could allow for predicting functional alteration of identified proteins at a molecular level. We have highlighted such examples with a sarcomere-associated protein, CSRP/MLP, and three mitochondrial complexes I, II, and III. To our knowledge, functional implication of CSRP3/MLP oxidation has not been investigated. Our mass analyses identified glutathionylation of MLP, but also other LIM-containing proteins, including LBD3 and PDLI5, that are important for sarcomere stability at the Z-disk.42 LIM domains contain many Cys residues that bind with Zn atoms. Oxidation of such Cys residues would impair protein-protein interactions mediated by LIM domains in these proteins. Recently, complex I, II, III and their mega-complex structures were analyzed.59–60, 62–63 Our analysis of available structures suggests a potential impact of individual glutathionylation on the mega-complex formation in addition to catalytic functions. Previously, glutathionylation of complex I was investigated, finding glutathionylation at two major Cys residues (C704 and C531) in NDUFS1 (75-kD) and others (e.g. C367) in NDUFV1 (51-kD)13, 66, which decreases complex I catalytic activity.13–14 Two Cys residues (C704 and C531) in NDUFS1 were also found in our mass analysis (corresponding to C727 and C554 in our study), but we did not find any Cys residue in NDUFV1 (51-kD). C704 and C531 were found to be close to the protein surface, but relatively far away from catalytic cofactors (~40 Å). In addition, glutathionylation of complex II was previously reported at C90 (C47 after N-terminal cleavage), which was present at a basal level, but de-glutathionylated upon ischemic-reperfusion in the heart.57 C47 is located at a beta-strand relatively buried in the subunit and may interact with G26 (3.9 Å) that makes a binding surface for FAD (Figure S9). The basal level of C47 glutathionylation is likely to disrupt a FAD binding site. However, our analysis did not identify C47 glutathionylation at the basal or stressed conditions. Lastly, complex III glutathionylation was shown in previous mass analyses,24 but its functional implication is yet unknown. Our structural analysis suggests that complex III glutathionylation may interfere with mega-complex assembly. In addition to these highlighted examples, our proteomic mass identification would contribute to understanding the functional impact of glutathionylation at a molecular level with other available structures.
However, it is important to indicate limitations of current studies. We have used a relatively high amount of glucose oxidase to identify a large number of glutathionylated proteins. Such a high level of H2O2 may not be achievable in a biological environment, which suggests that a portion of our identified proteins may be only susceptible to glutathionylation at high levels of oxidative stress. It is also important to point out that we have not detected some of the previously reported glutathionylated proteins, including titin and troponin I,15, 17 which may be due to different experimental conditions or suggest that further optimization of our approach would be necessary. In a similar context, quantification of the extent of glutathionylation on individual Cys residues will be necessary to identify functionally-relevant proteins for glutathionylation in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Health Grant R01 HL131740 (Y.-H.A). The Wayne State University Proteomics Core was supported through the NIH Center Grant P30 ES 020957, the NIH Cancer Center Support Grant P30 CA 02253 and the NIH Shared Instrumentation Grant S10 OD 010700. We thank Dr. Joseph Caruso and Dr. Paul Stemmer in WSU proteomics center for analysis. We thank Dr. Eranthie Weerapana and Tyler Bechtel for help and discussion.
Footnotes
NOTES
The authors declare no competing financial interest.
The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE [1] partner repository with the dataset identifier PXD012171 (https://www.ebi.ac.uk/pride/archive/projects/PXD012171).
REFERENCES
- 1.Burgoyne JR; Mongue-Din H; Eaton P; Shah AM, Redox Signaling in Cardiac Physiology and Pathology. Circ. Res. 2012, 111 (8), 1091–1106. [DOI] [PubMed] [Google Scholar]
- 2.Chen YR; Zweier JL, Cardiac Mitochondria and Reactive Oxygen Species Generation. Circ. Res. 2014, 114 (3), 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Granger DN; Kvietys PR, Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol. 2015, 6, 524–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano FJ, Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Invest. 2005, 115 (3), 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandasamy AD; Chow AK; Ali MAM; Schulz R, Matrix metalloproteinase-2 and myocardial oxidative stress injury: beyond the matrix. Cardiovasc. Res. 2010, 85 (3), 413–423. [DOI] [PubMed] [Google Scholar]
- 6.Letavernier E; Zafrani L; Perez J; Letavernier B; Haymann JP; Baud L, The role of calpains in myocardial remodelling and heart failure. Cardiovasc. Res. 2012, 96 (1), 38–45. [DOI] [PubMed] [Google Scholar]
- 7.Willis MS; Schisler JC; Portbury AL; Patterson C, Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc. Res. 2009, 81 (3), 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckendorf L; Linke WA, Emerging importance of oxidative stress in regulating striated muscle elasticity. J. Muscle Res. Cell. Motil. 2015, 36 (1), 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen CE; Carroll KS, Cysteine-Mediated Redox Signaling: Chemistry, Biology, and Tools for Discovery. Chem. Rev. 2013, 113 (7), 4633–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mailloux RJ; Willmore WG, S-glutathionylation reactions in mitochondrial function and disease. Front. Cell Dev. Biol. 2014, 2, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adachi T; Weisbrod RM; Pimentel DR; Ying J; Sharov VS; Schoneich C; Cohen RA, S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat. Med. 2004, 10 (11), 1200–7. [DOI] [PubMed] [Google Scholar]
- 12.Aracena P; Sanchez G; Donoso P; Hamilton SL; Hidalgo C, S-glutathionylation decreases Mg2+ inhibition and S-nitrosylation enhances Ca2+ activation of RyR1 channels. J. Biol. Chem. 2003, 278 (44), 42927–42935. [DOI] [PubMed] [Google Scholar]
- 13.Hurd TR; Requejo R; Filipovska A; Brown S; Prime TA; Robinson AJ; Fearnley IM; Murphy MP, Complex I within oxidatively stressed bovine heart mitochondria is glutathionylated on Cys-531 and Cys-704 of the 75-kDa subunit - Potential role of Cys residues in decreasing oxidative damage. J. Biol. Chem. 2008, 283 (36), 24801–24815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mailloux RJ; Xuan JY; McBride S; Maharsy W; Thorn S; Holterman CE; Kennedy CR; Rippstein P; deKemp R; da Silva J; Nemer M; Lou M; Harper ME, Glutaredoxin-2 is required to control oxidative phosphorylation in cardiac muscle by mediating deglutathionylation reactions. J. Biol. Chem. 2014, 289 (21), 14812–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alegre-Cebollada J; Kosuri P; Giganti D; Eckels E; Rivas-Pardo JA; Hamdani N; Warren CM; Solaro RJ; Linke WA; Fernandez JM, S-Glutathionylation of Cryptic Cysteines Enhances Titin Elasticity by Blocking Protein Folding. Cell 2014, 156 (6), 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen FC; Ogut O, Decline of contractility during ischemia-reperfusion injury: actin glutathionylation and its effect on allosteric interaction with tropomyosin. Am. J. Physiol. Cell Physiol. 2006, 290 (3), C719–C727. [DOI] [PubMed] [Google Scholar]
- 17.Mollica JP; Dutka TL; Merry TL; Lamboley CR; McConell GK; McKenna MJ; Murphy RM; Lamb GD, S-Glutathionylation of troponin I (fast) increases contractile apparatus Ca2+ sensitivity in fast-twitch muscle fibres of rats and humans. J. Physiol. 2012, 590 (6), 1443–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munkanatta Godage DNP; VanHecke GC; Samarasinghe KTG; Feng HZ; Hiske M; Holcomb J; Yang Z; Jin JP; Chung CS; Ahn YH, SMYD2 glutathionylation contributes to degradation of sarcomeric proteins. Nat. Commun. 2018, 9 (1), 4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong EM; Monasky MM; Gu L; Taglieri DM; Patel BG; Liu H; Wang Q; Greener I; Dudley SC Jr.; Solaro RJ, Tetrahydrobiopterin improves diastolic dysfunction by reversing changes in myofilament properties. J. Mol. Cell. Cardiol. 2013, 56, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samarasinghe KTG; Ahn Y-H, Synthesizing Clickable Glutathione by Glutathione Synthetase Mutant for Detecting Protein Glutathionylation. Synlett 2015, 26 (03), 285–293. [Google Scholar]
- 21.Brennan JP; Miller JIA; Fuller W; Wait R; Begum S; Dunn MJ; Eaton P, The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Mol. Cell. Proteomics 2006, 5 (2), 215–225. [DOI] [PubMed] [Google Scholar]
- 22.Brennan JP; Wait R; Begum S; Bell JR; Dunn MJ; Eaton P, Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. J. Biol. Chem. 2004, 279 (40), 41352–41360. [DOI] [PubMed] [Google Scholar]
- 23.Pan KT; Chen YY; Pu TH; Chao YS; Yang CY; Bomgarden RD; Rogers JC; Meng TC; Khoo KH, Mass Spectrometry-Based Quantitative Proteomics for Dissecting Multiplexed Redox Cysteine Modifications in Nitric Oxide-Protected Cardiomyocyte Under Hypoxia. Antioxid. Redox Signal. 2014, 20 (9), 1365–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer PA; Duan J; Gaffrey MJ; Shukla AK; Wang L; Bammler TK; Qian WJ; Marcinek DJ, Fatiguing contractions increase protein S-glutathionylation occupancy in mouse skeletal muscle. Redox Biol. 2018, 17, 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samarasinghe KTG; Godage DNPM; VanHecke GC; Ahn YH, Metabolic Synthesis of Clickable Glutathione for Chemoselective Detection of Glutathionylation. J. Am. Chem. Soc. 2014, 136 (33), 11566–11569. [DOI] [PubMed] [Google Scholar]
- 26.Kekulandara DN; Samarasinghe KTG; Godage DNPM; Ahn YH, Clickable glutathione using tetrazine-alkene bioorthogonal chemistry for detecting protein glutathionylation. Org. Biomol. Chem. 2016, 14 (46), 10886–10893. [DOI] [PubMed] [Google Scholar]
- 27.Samarasinghe KTG; Godage DNPM; Zhou YN; Ndombera FT; Weerapana E; Ahn YH, A clickable glutathione approach for identification of protein glutathionylation in response to glucose metabolism. Mol. Biosyst. 2016, 12 (8), 2471–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szychowski J; Mahdavi A; Hodas JJ; Bagert JD; Ngo JT; Landgraf P; Dieterich DC; Schuman EM; Tirrell DA, Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J. Am. Chem. Soc. 2010, 132 (51), 18351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAllister TE; Nix MG; Webb ME, Fmoc-chemistry of a stable phosphohistidine analogue. Chem. Commun. 2011, 47 (4), 1297–9. [DOI] [PubMed] [Google Scholar]
- 30.Bhatti HN; Madeeha M; Asgher M; Batool N, Purification and thermodynamic characterization of glucose oxidase from a newly isolated strain of Aspergillus niger. Can. J. Microbiol. 2006, 52 (6), 519–24. [DOI] [PubMed] [Google Scholar]
- 31.Keller A; Nesvizhskii AI; Kolker E; Aebersold R, Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002, 74 (20), 5383–92. [DOI] [PubMed] [Google Scholar]
- 32.Nesvizhskii AI; Keller A; Kolker E; Aebersold R, A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75 (17), 4646–4658. [DOI] [PubMed] [Google Scholar]
- 33.Ashburner M; Ball CA; Blake JA; Botstein D; Butler H; Cherry JM; Davis AP; Dolinski K; Dwight SS; Eppig JT; Harris MA; Hill DP; Issel-Tarver L; Kasarskis A; Lewis S; Matese JC; Richardson JE; Ringwald M; Rubin GM; Sherlock G, Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25 (1), 25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D; Morris JH; Cook H; Kuhn M; Wyder S; Simonovic M; Santos A; Doncheva NT; Roth A; Bork P; Jensen LJ; von Mering C, The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45 (D1), D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claycomb WC; Lanson NA Jr.; Stallworth BS; Egeland DB; Delcarpio JB; Bahinski A; Izzo NJ Jr., HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 1998, 95 (6), 2979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller S; Millonig G; Waite GN, The GOX/CAT system: a novel enzymatic method to independently control hydrogen peroxide and hypoxia in cell culture. Adv. Med. Sci. 2009, 54 (2), 121–35. [DOI] [PubMed] [Google Scholar]
- 37.Yang YL; Verhelst SHL, Cleavable trifunctional biotin reagents for protein labelling, capture and release. Chem. Commun. 2013, 49 (47), 5366–5368. [DOI] [PubMed] [Google Scholar]
- 38.Xiao H; Wu R, Global and Site-Specific Analysis Revealing Unexpected and Extensive Protein S-GlcNAcylation in Human Cells. Anal. Chem. 2017, 89 (6), 3656–3663. [DOI] [PubMed] [Google Scholar]
- 39.Chen YJ; Lu CT; Huang KY; Wu HY; Chen YJ; Lee TY, GSHSite: Exploiting an Iteratively Statistical Method to Identify S-Glutathionylation Sites with Substrate Specificity. PloS One 2015, 10 (4), e0118752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun CL; Shi ZZ; Zhou XB; Chen LN; Zhao XM, Prediction of S-Glutathionylation Sites Based on Protein Sequences. PloS One 2013, 8 (2), e55512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang DW; Sherman BT; Tan Q; Collins JR; Alvord WG; Roayaei J; Stephens R; Baseler MW; Lane HC; Lempicki RA, The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8 (9), R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank D; Frey N, Cardiac Z-disc Signaling Network. J. Biol. Chem. 2011, 286 (12), 9897–9904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knoll R; Buyandelger B; Lab M, The sarcomeric Z-disc and Z-discopathies. J. Biomed. Biotechnol. 2011, 2011, 569628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arber S; Hunter JJ; Ross J Jr.; Hongo M; Sansig G; Borg J; Perriard JC; Chien KR; Caroni P, MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 1997, 88 (3), 393–403. [DOI] [PubMed] [Google Scholar]
- 45.Lange S; Gehmlich K; Lun AS; Blondelle J; Hooper C; Dalton ND; Alvarez EA; Zhang XY; Bang ML; Abassi YA; dos Remedios CG; Peterson KL; Chen J; Ehler E, MLP and CARP are linked to chronic PKC alpha signalling in dilated cardiomyopathy. Nat. Commun. 2016, 7, 12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLendon PM; Robbins J, Desmin-related cardiomyopathy: an unfolding story. Am. J. Physiol. Heart Circ. Physiol. 2011, 301 (4), H1220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shah SB; Davis J; Weisleder N; Kostavassili I; McCulloch AD; Ralston E; Capetanaki Y; Lieber RL, Structural and functional roles of desmin in mouse skeletal muscle during passive deformation. Biophys. J. 2004, 86 (5), 2993–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vatta M; Mohapatra B; Jimenez S; Sanchez X; Faulkner G; Perles Z; Sinagra G; Lin JH; Vu TM; Zhou Q; Bowles KR; Di Lenarda A; Schimmenti L; Fox M; Chrisco MA; Murphy RT; McKenna W; Elliott P; Bowles NE; Chen J; Valle G; Towbin JA, Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 2003, 42 (11), 2014–27. [DOI] [PubMed] [Google Scholar]
- 49.Zheng M; Cheng H; Li X; Zhang J; Cui L; Ouyang K; Han L; Zhao T; Gu Y; Dalton ND; Bang ML; Peterson KL; Chen J, Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 2009, 18 (4), 701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassel D; Dahme T; Erdmann J; Meder B; Huge A; Stoll M; Just S; Hess A; Ehlermann P; Weichenhan D; Grimmler M; Liptau H; Hetzer R; Regitz-Zagrosek V; Fischer C; Nurnberg P; Schunkert H; Katus HA; Rottbauer W, Nexilin mutations destabilize cardiac Z-disks and lead to dilated cardiomyopathy. Nat. Med. 2009, 15 (11), 1281–8. [DOI] [PubMed] [Google Scholar]
- 51.Arber S; Halder G; Caroni P, Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell 1994, 79 (2), 221–31. [DOI] [PubMed] [Google Scholar]
- 52.Geier C; Perrot A; Ozcelik C; Binner P; Counsell D; Hoffmann K; Pilz B; Martiniak Y; Gehmlich K; van der Ven PF; Furst DO; Vornwald A; von Hodenberg E; Nurnberg P; Scheffold T; Dietz R; Osterziel KJ, Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation 2003, 107 (10), 1390–5. [DOI] [PubMed] [Google Scholar]
- 53.Geier C; Gehmlich K; Ehler E; Hassfeld S; Perrot A; Hayess K; Cardim N; Wenzel K; Erdmann B; Krackhardt F; Posch MG; Osterziel KJ; Bublak A; Nagele H; Scheffold T; Dietz R; Chien KR; Spuler S; Furst DO; Nurnberg P; Ozcelik C, Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum. Mol. Genet. 2008, 17 (18), 2753–65. [DOI] [PubMed] [Google Scholar]
- 54.Knoll R; Hoshijima M; Hoffman HM; Person V; Lorenzen-Schmidt I; Bang ML; Hayashi T; Shiga N; Yasukawa H; Schaper W; McKenna W; Yokoyama M; Schork NJ; Omens JH; McCulloch AD; Kimura A; Gregorio CC; Poller W; Schaper J; Schultheiss HP; Chien KR, The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 2002, 111 (7), 943–55. [DOI] [PubMed] [Google Scholar]
- 55.Hoffmann C; Moreau F; Moes M; Luthold C; Dieterle M; Goretti E; Neumann K; Steinmetz A; Thomas C, Human Muscle LIM Protein Dimerizes along the Actin Cytoskeleton and Cross-Links Actin Filaments. Mol. Cell. Biol. 2014, 34 (16), 3053–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papalouka V; Arvanitis DA; Vafiadaki E; Mavroidis M; Papadodima SA; Spiliopoulou CA; Kremastinos DT; Kranias EG; Sanoudou D, Muscle Lim Protein Interacts with Cofilin 2 and Regulates F-Actin Dynamics in Cardiac and Skeletal Muscle. Mol. Cell. Biol. 2009, 29 (22), 6046–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen YR; Chen CL; Pfeiffer DR; Zweier JL, Mitochondrial complex II in the post-ischemic heart - Oxidative injury and the role of protein S-glutathionylation. J. Biol. Chem. 2007, 282 (45), 32640–32654. [DOI] [PubMed] [Google Scholar]
- 58.Hirst J, Mitochondrial complex I Annu. Rev. Biochem. 2013, 82, 551–75. [DOI] [PubMed] [Google Scholar]
- 59.Zhu J; Vinothkumar KR; Hirst J, Structure of mammalian respiratory complex I. Nature 2016, 536 (7616), 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo R; Zong S; Wu M; Gu J; Yang M, Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170 (6), 1247–1257. [DOI] [PubMed] [Google Scholar]
- 61.Elurbe DM; Huynen MA, The origin of the supernumerary subunits and assembly factors of complex I: A treasure trove of pathway evolution. Biochim. Biophys. Acta 2016, 1857 (7), 971–9. [DOI] [PubMed] [Google Scholar]
- 62.Wu M; Gu J; Guo R; Huang Y; Yang M, Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell 2016, 167 (6), 1598–1609. [DOI] [PubMed] [Google Scholar]
- 63.Sun F; Huo X; Zhai Y; Wang A; Xu J; Su D; Bartlam M; Rao Z, Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 2005, 121 (7), 1043–57. [DOI] [PubMed] [Google Scholar]
- 64.Sumandea MP; Steinberg SF, Redox Signaling and Cardiac Sarcomeres. J. Biol. Chem. 2011, 286 (12), 9921–9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinberg SF, Oxidative Stress and Sarcomeric Proteins. Circ. Res. 2013, 112 (2), 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen CL; Zhang LW; Yeh A; Chen CA; Green-Church KB; Zweier JL; Chen YR, Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry 2007, 46 (19), 5754–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


