Abstract
Simultaneous analyses of peripheral and mucosal immune compartments can yield insight into the pathogenesis of mucosal-associated diseases. Although methods to preserve peripheral immune cells are well established, studies involving mucosal immune cells have been hampered by lack of simple storage techniques. We provide a cryopreservation protocol allowing for storage of gastrointestinal (GI) tissue with preservation of viability and functionality of both immune and epithelial cells. These methods will facilitate translational studies allowing for batch analysis of mucosal tissue to investigate disease pathogenesis, biomarker discovery and treatment responsiveness.
INTRODUCTION
The phenotypic and functional assessment of immune cell populations involved in human disease is critical to understanding the pathogenesis of immune-mediated disorders and the development of personalized treatment approaches. Detailed analysis of mucosal surfaces has proven particularly difficult,1–4 requiring processing of fresh tissue that is often time-consuming and dependent on sophisticated laboratory techniques that are not universally available. Here, we present a simple universal method to immediately cryopreserve mucosal tissue that retains cellular viability, including immune and epithelial components, allowing for successful batched analysis at a later time.
To simultaneously evaluate multiple intestinal immune populations, we have applied mass cytometry (i.e., cytometry by time of flight (CyTOF))5–7 for deep immunophenotyping of lamina propria mononuclear cells (LPMCs) obtained from fresh and frozen tissue. CyTOF combines mass spectrometry and cytometry providing the ability to detect up to 40 antigens, using antibodies tagged with unique heavy metals, in an individual sample at single-cell resolution. It allows for the simultaneous interrogation of all major immune cell lineages as well as the identification of rare subpopulations of cells5–7 in a given tissue or blood sample. CyTOF has been successfully used in a number of settings including detecting extremely rare metastases in peripheral blood,8 immunophenotyping hematopoietic development, as well as characterizing cellular responses to various stimulations.6 A number of unbiased algorithms have been developed that can be applied to CyTOF-generated data to identify unique cell populations as well as perform predictive modeling to characterize particular cellular subtypes with biological parameters.9 With this potential, CyTOF represents a discovery tool that could be harnessed in the setting of clinical trials to help evaluate unique immune cell populations that may predict responsiveness to treatment.
Here, we provide a method to immediately cryopreserve intestinal tissue with retention of viability and functionality of both immune and epithelial cells allowing for subsequent transcriptional and cellular analysis. We show at three independent institutions that cryopreserved tissue can be used to generate single-cell suspensions of live immune cells with maintenance of immune makeup and cytokine expression upon stimulation. Moreover, the cryopreserved tissue allowed for successful generation of enteroids. Additionally, the transcriptional profile of the tissue was preserved with retention of differentially expressed genes (DEGs) between inflamed and uninflamed tissue. Overall, this cryopreservation protocol allows for immediate storage of intestinal tissue for subsequent cellular, functional, and transcriptional analyses facilitating the study of immune and epithelial cell function applicable to a variety of diseases.
Terminology
Throughout the manuscript, we will refer to “fresh” tissue as that obtained from either resected or biopsied (Bx) gastrointestinal (GI) tissue that has been immediately stored in RNAlater for transcriptional analysis or separated into epithelial and immune compartments. Intestinal crypts isolated from the epithelium were used for enteroid cultures while immune analysis came from single-cell suspensions of LPMCs. We will define “frozen cells” as LPMCs obtained after processing fresh tissue and then freezing for future use. Finally, “frozen Bx” will be defined as fresh tissue that is cryopreserved as whole and processed into single cells or isolated for intestinal crypts after thawing.
RESULTS
Gastrointestinal tissue can be cryopreserved with retention of cell viability
Immunophenotyping and functional assessment of GI tissue has largely been performed on either fresh cells or on isolated single cells that were previously frozen. Both of these methods have significant limitations including the requirement of technical expertise to process tissue at the site of collection and the inability to batch-analyze multiple samples. To facilitate multi-site clinical and translational research, a simple preservation protocol is needed that allows for direct and immediate tissue storage at the site of collection and permits for cellular isolation at a later date. We therefore set out to establish a tissue cryopreservation protocol that would allow for preservation of the viability and functionality of intestinal tissue.
We compared fresh intestinal biopsies (2 mm × 2 mm) with biopsies previously frozen slowly in 10% dimethyl sulfoxide (DMSO) and 90% fetal bovine serum (FBS) (Fig. 1a) at several institutions (Boston Children’s Hospital (BCH), Icahn School of Medicine (MSSM), and Perelman School of Medicine (Penn)). No statistically significant differences in total cell counts or CD45+ cell counts were observed between cells obtained from “fresh” or “frozen Bx” at any of the institutions (Fig. 1b), although there was a trend toward a decrease in both total and CD45+ cell count in the “frozen Bx” group. Cell count obtained from matched samples collected at the same time from the same individual similarly did not show any significant difference in total cell count (Fig. S1A) or CD45+ cell count (Fig. S1B) between “fresh” and “frozen bx” samples. At BCH, cell viability was also similar in both methods (Fig. 1d, e, Fig. S1A, and Fig. S1B) with immune cells (CD45+ cells) having greater viability than non-immune cells (CD45− cells) irrespective of the method (Fig. 1d). A slight decrease in cell viability was observed from frozen tissue in CD45+ cells at MSSM, whereas a slight increase in total frozen cell viability was observed at Penn (Fig. 1e). To determine if the duration of freezing has an effect on cellular viability, we evaluated if there was any correlation between the duration of freezing and the viability of the recovered cells (Fig. S1I). Between the three centers, tissue was frozen between 1 and 300 days, and there was no correlation between viability and duration of freezing.
Fig. 1.
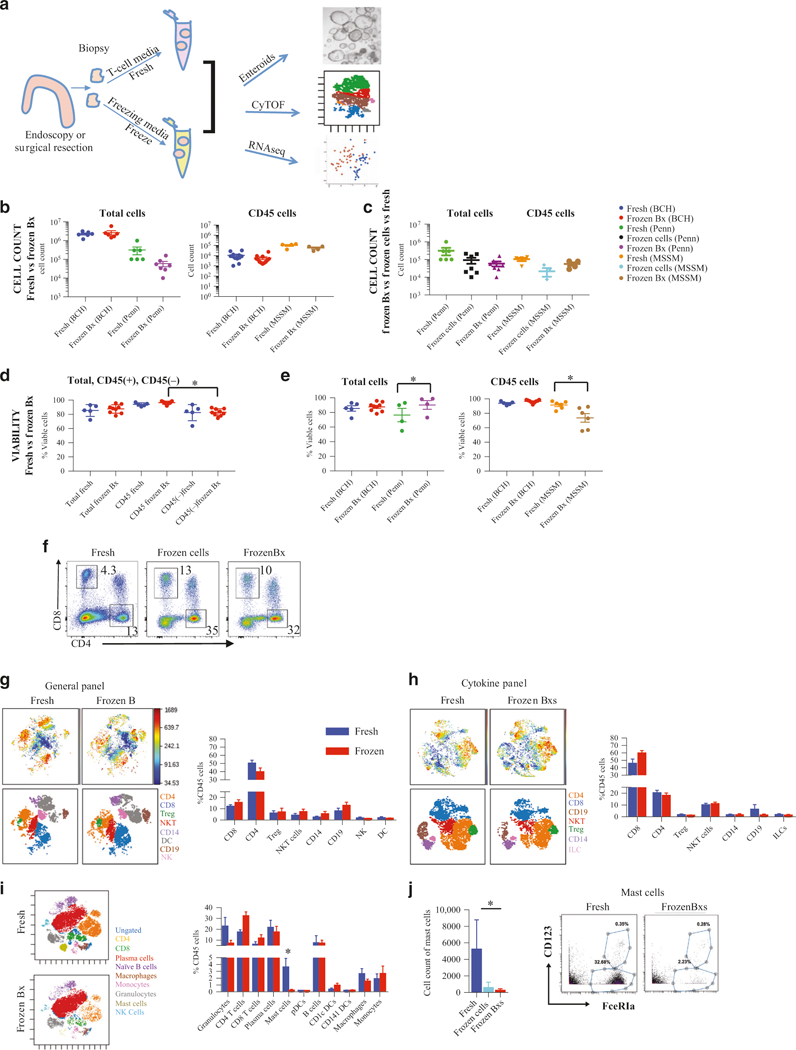
Gastrointestinal tissue can be cryopreserved with retention of cell viability. (a) Schematic obtaining intestinal biopsies by comparing immunophenotyping, establishment of enteroid cultures and transcriptional analysis of fresh or cryopreserved tissue. Intestinal tissue was prepared per the protocol outlined in (a) and single-cell suspensions obtained from either fresh biopsies or tissue frozen as biopsies and then processed were evaluated via CyTOF by staining with the “general” CyTOF panel and Rh for viability. Total and CD45+ cell count (b) from Boston Children’s Hospital (BCH), Mount Sinai School of Medicine (MSSM), and University of Pennsylvania School of Medicine (Penn). Cell count for fresh isolated cells (“fresh”), cells isolated fresh and then frozen (“frozen cells”), and frozen as biopsies and then processed (“frozen Bx”) were compared at MSSM and Penn sites (c). Total cell viability is shown and compared to immune cell viability (CD45+) and non-immune cell viability (CD45−) for fresh versus frozen tissue evaluated at BCH (d) as well as total and CD45+ cell viability at MSSM and Penn (e). FACS analysis (f) of CD8+ and CD4+ T-cells comparing cells obtained “fresh” (cells isolated from fresh tissue), “frozen cells” (cells isolated fresh and then frozen), and “frozen Bx” (cells isolated from previously frozen biopsies). CD45+ viSNE plots from CyTOF analysis, using either the “general” panel (g) or the “cytokine” panel (h) of “fresh” or “frozen Bx.” The top two panels are ungated viSNE analysis for “fresh” (left) and “frozen Bx” (right) samples with a heat map of CD45. The bottom two panels are gated for the various marked immune cells populations. The percent of the various populations are quantified in the graph to the right. (i) CD45+ viSNE analysis of CyTOF using the “MSSM” panel comparing “fresh” (top) to “frozen Bx” (bottom) tissue and quantified to the right. Two-dimensional plots from “MSSM” panel CyTOF analysis of mast cells (j) are shown. Quantifications are to the left
To determine how our freezing protocol compares to previously available means of preserving LPMCs, we compared cell count and viability between all three conditions: “fresh,” “frozen cells” (the more conventional means of storing LPMCs), and “frozen Bx” (our cryopreservation protocol). There were no statistically significant differences in the cell counts (Fig. 1c and Fig. S1C) and viability (Fig. S1C) between any of these methods, however we continued to observe a trend toward lower total and CD45+ cell counts with cryopreservation for both “frozen Bx” and “frozen cell” methods.
To determine whether tissue cryopreservation has a differential effect on various immune cell types, we utilized both standard flourescence activated cell sorting (FACS) analysis and CyTOF to compare LPMCs obtained from either method. FACS analysis of T-cells showed that the ratio of CD4+:CD8+ T-cells is fully preserved with freezing, and the percentage of CD4+ and CD8+ T-cells is very similar whether tissue is frozen whole or cells are isolated from fresh tissue and then frozen as LPMCs (Fig. 1f and Fig. S1D). In fact, there was an increase in CD4+ and CD8+ T-cell viability after freezing the tissue (irrespective of the freezing method) when compared to processing fresh cells (Fig. 1f and Fig. S1D).
Using CyTOF, we performed deep immune phenotyping with two different antibody panels to further characterize the immune populations present in the biopsy tissue. We designed a “general” panel to obtain a global overview of immune cell diversity that contained 32 metal isotope-tagged antibodies (Table 1A) and a “cytokine” panel containing 25 innate and adaptive immune cell surface markers as well as 11 intracellular cytokines and transcription factors (Table 1B). Unbiased viSNE analysis of CD45+ cells using either panel (Fig. 1g, h) showed that fresh and frozen tissue were similar in surface marker expression, as identified by the similarity in location on the viSNE map, as well as in the abundance of the various cell types present. Of note, we observed some differences in the abundance of cellular populations between the panels, with increased CD4+ T-cells, regulatory T-cells (Tregs), and CD14+ cells when using the “general panel” as compared to the “cytokine panel” (Fig. 1g, h). This is likely a combination of downregulation of various cell surface markers, such as CD4, upon stimulation and the differential effect on viability of various cell types upon exposure to PMA/ionomycin and golgiSTOP for 4 h. Additionally, since the biopsies used in the two panels were collected from different individuals, this could also represent the inherent variability between the individuals. Standard two-dimensional (2D) FACS-like plots generated from the same data also showed that the frequency of the cell types studied was not significantly affected by freezing of the tissue (Fig. S1E). Consistent with these data, viSNE analysis of CyTOF data obtained from an immune panel enriched for innate immune cells (MSSM panel containing 33 metal isotype-tagged antibodies, Table 2), also showed no major differences in the cell types that were common between the panels (Fig. 1i). Consistent with previous data, the two cell types affected by freezing, irrespective of the freezing method used (“frozen Bx” versus “frozen cells”), were granulocytes, which showed a trend toward decreased numbers in the frozen groups, and mast cells, which were significantly reduced in the frozen group (Fig. 1j).
Table 1A.
General panel
| Tag | Target | Clone | Source |
|---|---|---|---|
| 89Y | CD45 | HI30 | Fluidigm |
| 113In | |||
| 115In | CD44 | IM7 | Core, self conj |
| 139La | |||
| 141Pr | c-kit | 104D2 | Core, self conj |
| 142Nd | CD19 | HIB19 | Core |
| 143Nd | HLA-DR | L243 | Fluidigm |
| 144Nd | CD64 | 10.1 | Core |
| 145Nd | CD16 | 3G8 | Core |
| 146Nd | CD8a | RPA-T8 | Core |
| 147Sm | CD45RO | UCHL1 | Core |
| 148Nd | CD28 | CD28.2 | Core |
| 149Sm | CD25 | 2A3 | Fluidigm |
| 150Nd | CD38 | 874501 | Core, self conj |
| 151Eu | CD49b | P1E6-C5 | Core |
| 152Sm | CD14 | M5E2 | Core |
| 153Eu | CD45RA | HI100 | Core |
| 154Sm | CD163 | GHI/61 | Fluidigm |
| 155Gd | CD27 | L128 | Fluidigm |
| 156Gd | CD8b | SIDI8BEE | Core |
| 158Gd | CD3 | UCHT1 | Core |
| 159Tb | CD11c | Bu15 | Core |
| 160Gd | |||
| 161Dy | |||
| 162Dy | CD56 | HCD56 | Core |
| 163Dy | CD183 (CXCR) | G025H7 | Fluidigm |
| 164Dy | CD161 | HP-3G10 | Core |
| 165Ho | |||
| 166Er | CD24 | ML5 | Fluidigm |
| 167Er | LAG-3 | HIT2 | Core, self conj |
| 168Er | CCR6 | G034E3 | Core |
| 169Tm | |||
| 170Er | CCR7 | G043H7 | Core |
| 171Yb | CD127 | A019D5 | Core |
| 172Yb | IgM | MHM-88 | Core |
| 173Yb | CD335 | 9E2 | Core, self conj |
| 174Yb | CD4 | SK3 | Fluidigm |
| 175Lu | IgD | IA6–2 | Core |
Table 1B.
Cytokine panel
| Tag | Target | Clone | Source |
|---|---|---|---|
| 89Y | CD45 | HI30 | Fluidigm |
| 113In | |||
| 115In | CD44 | IM7 | Core, self conj |
| 139La | |||
| 141Pr | c-kit | 104D2 | Core, self conj |
| 142Nd | CD19 | HIB19 | Core |
| 143Nd | HLA-DR | L243 | Fluidigm |
| 144Nd | TNFa | A1 | Core |
| 145Nd | IL23 p19 | Core, self conj | |
| 146Nd | CD8a | RPA-T8 | Core |
| 147Sm | CD45RO | UCHL1 | Core |
| 148Nd | CD14 | CD28.2 | Core |
| 149Sm | CD25 | 2A3 | Core |
| 150Nd | IL-22 | 22URTI | Fluidigm |
| 151Eu | CD123 | P1E6-C5 | Core |
| 152Sm | CD152 (CTLA-4) | L3D10 | Fluidigm |
| 153Eu | CD45RA | HI100 | Core |
| 154Sm | CD38 | HIT2 | Core |
| 155Gd | CD27 | C398.4A | Core |
| 156Gd | CCR4 | L291H4 | Core |
| 158Gd | CD3 | UCHT1 | Core |
| 159Tb | CCR7 (cd197) | G043H7 | Fluidigm |
| 160Gd | INFg | 4S.B3 | Core |
| 161Dy | AHR | FF3399 | Core |
| 162Dy | IL-1B | 8D4–8 | Core, self conj |
| 163Dy | CD183 (CXCR3) | G025H7 | Fluidigm |
| 164Dy | CD161 | HP-3G10 | Core |
| 165Ho | FoxP3 | PCH101 | Core |
| 166Er | CD24 | TWAJ | Fluidigm |
| 167Er | LAG3 | 3DS223H | Core, self conj |
| 168Er | CCR6 | G034E3 | Core |
| 169Tm | IL17a | BL168 | Core |
| 170Er | |||
| 171Yb | CD127 | A019D5 | Core |
| 172Yb | IL21 | 3A3-N2 | Fluidigm |
| 173Yb | CD335 (NKp46) | 9E2 | Core, self conj |
| 174Yb | CD4 | SK3 | Fluidigm |
| 175Lu | Tbet | 4B10 | Core |
| 176YB | IL-10 | JES3- 19F1 |
Fluidigm |
Table 2.
MSSM panel
| Tag | Target | Clone | Source |
|---|---|---|---|
| 89Y | CD45 | HI30 | Fluidigm |
| 113In | CD57 | HCD57 | Biolegend |
| 115In | HLA-ABC | W6/32 | Biolegend |
| 141Pr | CD326 | Ep-CAm | Biolegend |
| 142Nd | CD19 | HIB19 | Biolegend |
| 143Nd | CD45RA | HI100 | Biolegend |
| 144Nd | CD141 | M80 | Biolegend |
| 145Nd | CD4 | RPA-T4 | Biolegend |
| 146Nd | CD8a | RPA-T8 | Biolegend |
| 147Sm | IgA | 9H9H11 | Biolegend |
| 148Nd | CD16 | 3G8 | Biolegend |
| 149Sm | CD127 | A019D5 | Biolegend |
| 150Nd | CD1c | L161 | Biolegend |
| 151Eu | CD123 | 6H6 | Biolegend |
| 152Sm | CD66b | G10F5 | Biolegend |
| 153Eu | PD-1 | EH12.2H7 | Biolegend |
| 154Sm | CD86 | IT2.2 | Biolegend |
| 155Gd | CD27 | O323 | Biolegend |
| 158Gd | CD33 | WM53 | Biolegend |
| 159Tb | CD103 | Ber-Act8 | Biolegend |
| 160Gd | CD14 | M5E2 | Biolegend |
| 161Dy | CD56 | B159 | Biolegend |
| 162Dy | CD64 | 10.1 | Biolegend |
| 163Dy | CD172a/b | SIRPa/b | Biolegend |
| 164Dy | CD69 | FN50 | Biolegend |
| 149Sm | FceRIa | AER-37 (CRA-1) | Biolegend |
| 166Er | CD25 | M-A251 | Biolegend |
| 167Er | CD11c | Bu15 | Biolegend |
| 168Er | CD3 | UCHT1 | Biolegend |
| 169Tm | Integrin b7 | FIB504 | Biolegend |
| 170Er | CD38 | HB-7 | Biolegend |
| 164Dy | CD161 | HP-3G10 | Fluidigm |
| 172Yb | CD206 | 15–2 | Biolegend |
| 154Sm | CXCR4 | 12G8 | Biolegend |
| 174Yb | HLADR | L243 | Biolegend |
| 175Lu | CD58 | TS2/9 | Biolegend |
| 176Yb | CD54 | HCD54 | Biolegend |
| 209Bi | CD11b | ICRF44 | Fluidigm |
Preservation of subpopulations of immune cells and their cytokine production
To further characterize the effect of freezing on specific cellular phenotype and function, we compared the subpopulations of various immune cells with CyTOF analysis. Consistent with the data shown above (Fig. 1), viSNE analysis of CD45+ cells did not show any major subpopulation differences between fresh and frozen cells using either the “general” or “cytokine” panels (Fig. 2). Figure 2a shows viSNE analysis of CD45+ cells stained with the “general” panel. The major cell populations are numbered 1–5 (1: CD19+ B-cells, 2: CD11c+ dendritic cells (DCs), 3: CD14+ macrophages, 4: CD4+ T-cells, and 5: CD8+ T-cells). Subgroup analysis of the major cell types is shown below the dotted outline for that cell type. Analysis using the “general” panel showed that cryopreservation did not affect the subpopulations assessed based on surface marker presence as reflected by presence of all characterized subpopulations on viSNE map (Fig. 2a) or by traditional 2D gating (Fig. S1F). These included antibody-producing, naive, transitional, and memory B-cells; naive, central memory, and effector memory T-cells; Th1, Th2 and Th17 CD4+ T-cells; CD163+ macrophages, and CD16+ and HLA-DR+ low (l) and high (h) DCs.
Fig. 2.
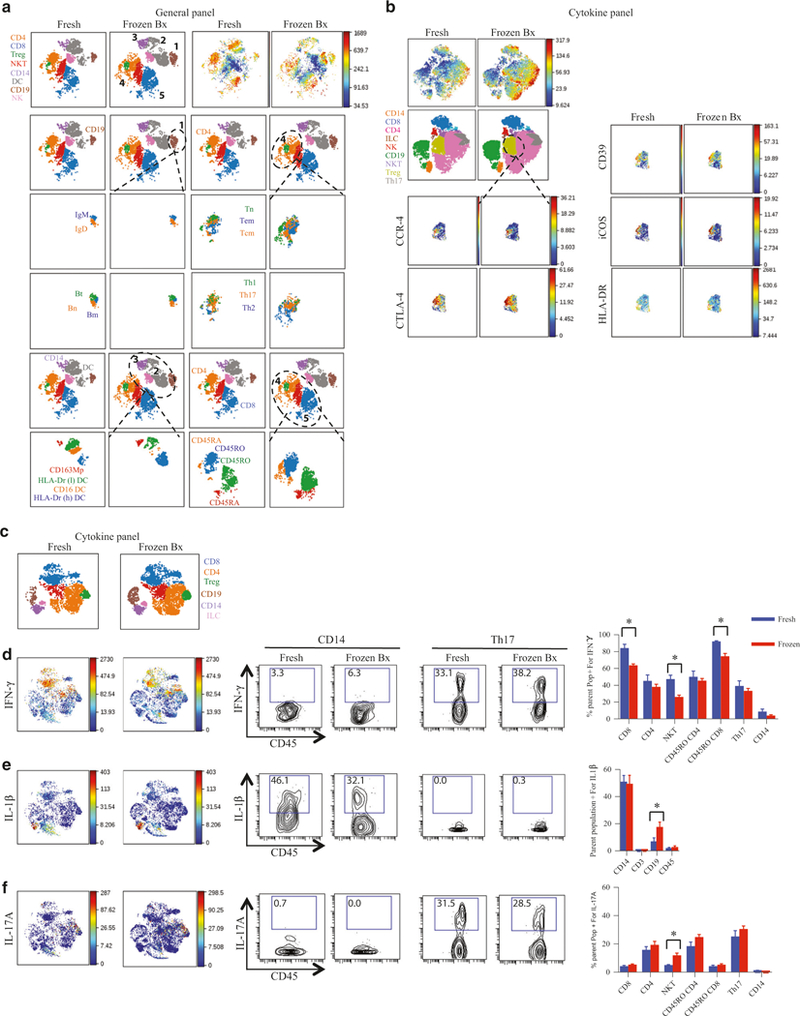
Preservation of subpopulations of immune cells and their cytokine production. Single-cell suspensions obtained from either “fresh,” or “frozen Bx” were stained for CyTOF analysis with either “general panel” (a) or “cytokine panel” (b). All images are representative. CD45 is the marker for the ungated viSNE. a Various major immune populations are numbered 1–5 and then a more detailed analysis of these populations is shown below. The major populations being compared are outlined by the dotted lines. B-cells: CD19+. T-cells CD3+. Bn: CD19+CD27−. Bm: CD19+CD27−. Bt: CD19+CD38+CD24+. Tn: CD4+CCR7+CD45RA+. Tem: CD4+CCR7−CD45RA−. Tcm: CD4+CCR7+CD45RA−. NK cells: CD3−CD56+. Macrophages: CD14+. DC: CD14−CD11c+. b Comparison of the markers expressed on the Treg populations between “fresh” and “frozen Bx” LPMCs. “Cytokine panel” was used for (c–f). c CD45+ viSNE color-coded for various immune populations. d–f Localization of the cells producing the indicated cytokines on the viSNE map on the left-hand side followed by the 2D FACS-like plots of either CD14+ (viable/CD45+/CD3−/CD19−/CD14+) or Th17 cells (viable/CD45+/CD3−/CD4−/CCR6+/CXCR-3−) expressing the cytokines indicated (in the middle panels; number represent percentages of cells in gate) and then the quantification of the production of the cytokines indicated by various cell types (panel on the right). *p value <0.05
To evaluate the effect of freezing on the specific antigens of a more focused cell population, we concentrated on evaluating the expression of surface and intracellular markers on regulatory T-cells (Tregs) as this cell type plays an important role in mucosal homeostasis. We used the “cytokine” panel to assess surface antigens and intracellular markers present on Tregs. Employing viSNE analysis, we show that CCR4, HLA-DR, CD39, iCOS, and CTLA-4 expression levels did not differ between fresh and frozen samples (Fig. 2b). Standard 2D FACS plots similarly showed no difference in expression of key surface markers associated with Tregs when “frozen Bx” and “fresh” tissue was compared (Fig. S1G and Fig. S1H).
Finally, cytokine expression from fresh and frozen samples was measured following stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin to induce cytokine production. The percentage of cells expressing cytokines and their specificity of cytokine production were similar for the majority of cell types and cytokines in both groups, i.e., T-cells produced interferon (IFN)γ and interleukin (IL)-17α and macrophages produced IL1β(Fig. 2c–f). IFNγ production was slightly decreased in CD8+ and natural killer T-cells (NKT) upon cryopreservation (Fig. 2d), IL1β production slightly increased in cryopreserved B-cells and IL17α production was mildly increased in cryopreserved NKT-cells. However, cellular trends of cytokine production (cells that produce the most of a certain cytokine and those that produce the least) were fully preserved by cryopreservation. Taken together, analysis of “fresh” and “frozen Bx” samples show similar viability, cell count, immunophenotyping (with the exception of reduced mast cells and a trend toward a reduction of granulocytes), and cytokine production.
Successful generation of intestinal enteroids from cryopreserved tissue
Given that the viability of immune cells was similar between fresh and frozen tissues, we next sought to determine whether epithelial stem cells also remain viable during cryopreservation. Despite a decrease in overall viability of CD45–, cells as compared to CD45+ with cryopreservation (Fig. 1d), enteroid cultures could be readily established from both fresh and frozen samples, confirming the viability of intestinal stem cells in frozen samples (Fig. 3a–d). When samples were obtained under standard clinical conditions, including from inflamed regions of intestine affected by IBD, enteroids could still be generated and successfully passaged from ~72% of samples, whether fresh or frozen (Fig. 3a–d). The success rate for generating and passaging enteroids from both fresh and frozen samples rose to 100% when 2 mm × 2 mm mucosal fragments were dissected from surgically resected duodenum under more controlled conditions (Fig. 3d). The growth and proliferation rates of enteroids obtained from fresh or cryopreserved tissue were also similar (data not shown). Taken together, these data establish that intestinal stem cells remain viable following cryopreservation of intestinal tissue biopsies.
Fig. 3.
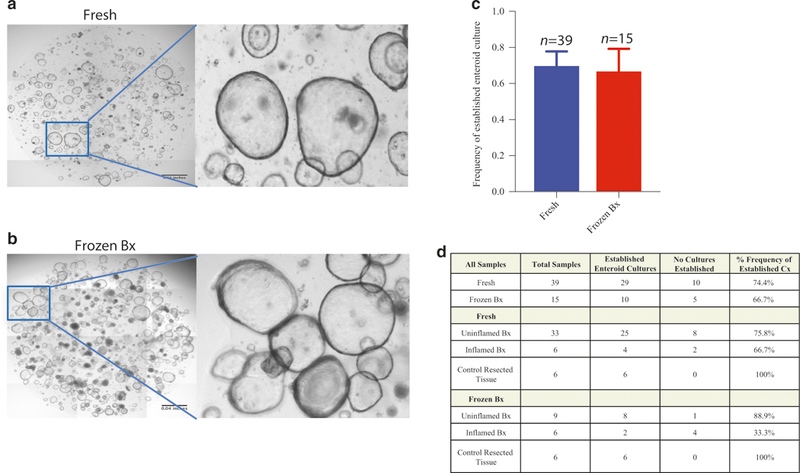
Successful intestinal enteroid generation from cryopreserved tissue. Organoids were either generated from fresh GI tissue obtained from IBD subjects, “fresh” (a) or from “Bx” (b). a, b Representative enteroid cultures. c Quantification of success rate of establishment of enteroid cultures. d Quantification of success rate of establishment of enteroid cultures segregated by patient conditions
Maintenance of unique gene expression patterns between inflamed and uninflamed tissue with cryopreservation
To determine if gene expression patterns were analogous in tissue frozen in DMSO freezing medium, corresponding to “frozen Bx,” compared to tissue stored in RNAlater, corresponding to “fresh”, we performed RNAseq analysis on RNA obtained from matched samples. RNA isolated from 36 ulcerative colitis (UC) subjects (with matched inflamed and uninflamed samples) were compared. All RNAlater samples had high-quality RNA as revealed by RNA concentration, RNA quality number (RQN), percentage of RNA fragments >200 bp, and intact 28S/18S RNA peaks (Fig. S2A, B). The RNA obtained from DMSO samples was similar in concentration to RNAlater preserved samples, but showed a decreased RQN, decreased percentage of RNA fragments >200 bp in length, and contained a mixture of intact 28S/18S RNA peaks and varying levels of degraded RNA fragments (Fig. S2A, B). Although there was an overall reduction in RQN in the RNA obtained from DMSO frozen biopsies as compared to that isolated from RNAlater, there was no correlation between the duration of freezing of the biopsies and the RNA RQN (Fig. S2C). Given that RNA from the DMSO samples contained degraded material (DV200 values >50) (Fig. S2B), to optimize the number of detected transcripts, library preparation was performed with total RNAseq. We were able to generate libraries meeting Illumina standards from all samples without clear outliers from either group. No differences in the percentages of unique, multiple, or unmapped reads were observed between inflamed and uninflamed tissue using either preservation method (Fig. S2D, E). However, because the RNAlater preservation method resulted in a greater percentage of intact RNA species (Fig. S2B), these samples had a higher rate of unique gene mapping with subsequently lower rates of multiple and unmapped reads than the DMSO-preserved samples (Fig. S2E). The gene mapping rate was nearly 40% across both sample types (typical for total RNAseq analysis) with no obvious difference between the two conditions (data not shown) and ~17,000 transcripts showed expression at >1 reads per kilobase of transcript per million mapped reads (RPKM) across all samples (Fig. S2F, G). There were no differences in the number of transcripts detected between the two storage methods except for the most stringent cutoff of 10 RPKM where RNAlater-stored samples had a slightly higher number of transcripts detected than the DMSO-stored samples (Fig. S2G).
Using principal component analysis (PCA), the primary determinant of the difference between the samples (PCA-1 29.3%) was their inflammation status. Analysis of all samples showed a clear separation of inflamed UC tissue from uninflamed UC (Fig. 4a). Although the DMSO preservation method contained a lower percentage of uniquely mapped reads (Fig. S2D), the differential analysis of inflamed versus uninflamed tissue showed 70% concordance between collection methods (Fig. 4c). Additionally, majority of differentially expressed pathways between inflamed and uninflamed tissue identified by fold change were similar between the “fresh” and “frozen Bx” samples (Fig. 4d). Moreover, there were many more pathways that were differentially regulated between inflamed and uninflamed samples than between “fresh” and “frozen Bx” samples (Fig. 4d). Both preservation types identified differentially regulated genes commonly found when comparing actively inflamed to inactive inflamatory bowel disease (IBD) tissue10 including DUOX2, DUOXA2, and LCN-2 (Fig. 4e).
Fig. 4.
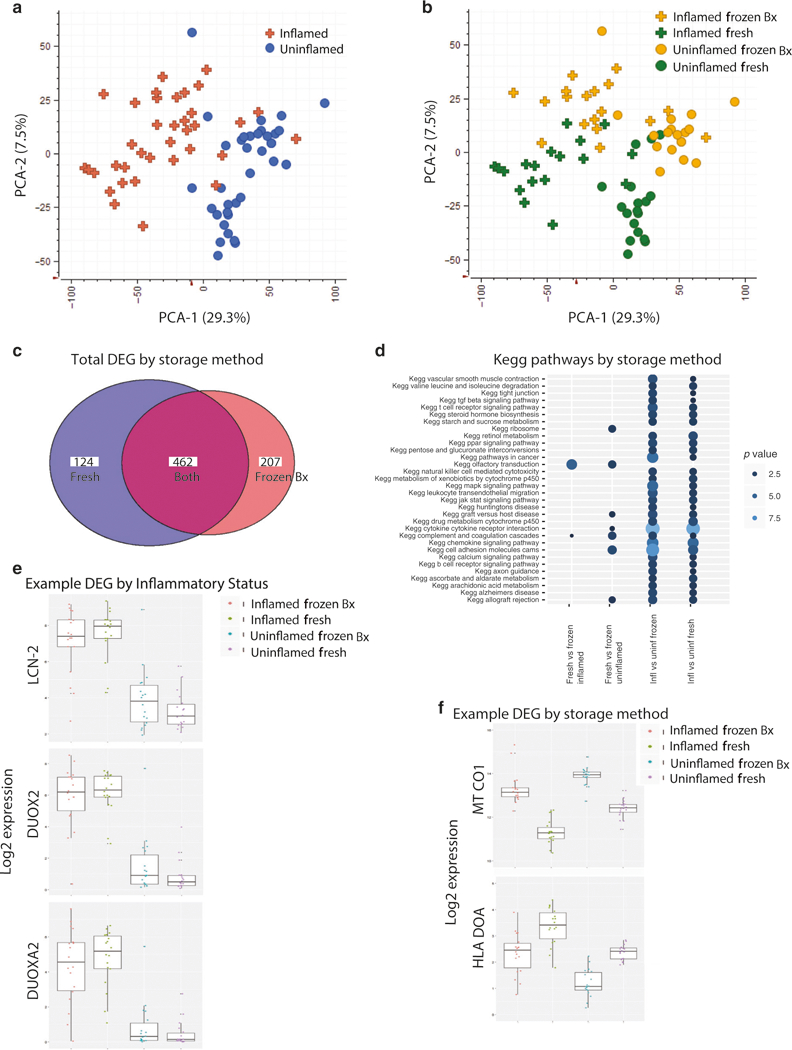
Maintenance of unique gene expression patterns between inflamed and uninflamed tissue with cryopreservation. Matched inflamed and uninflamed tissue from UC subjects were stored in RNAlater (“fresh”) or in DMSO (“frozen Bx”) and then analyzed together by total RNAseq. PCA plots of the DEG showing segregation by inflammatory status (a) and method of storage (b) are shown. (c) Total number of DEG by the two methods. (d) Top affected pathways by the preservation method or by inflammatory status. (e) Representative genes that are differentially expressed depending on the inflammation status of the tissue. (f) Representative genes that are differentially affected by freezing method
The factor driving PCA-2 (7.5%) for the comparison between all samples was the method of preservation (i.e., RNAlater versus DMSO), independent of inflammatory status (Fig. 4b). Consistent with previous work showing increased mitochondrial damage from thawing cells,11,12 differential pathway analysis revealed that the two major classes of genes most affected by the preservation method were: (1) mitochondrial and oxidative phosphorylation genes (commonly observed in cells under significant stress), which had increased expression in the DMSO samples, and (2) genes related to multiple immunological processes, including HLA and IG as well as various chemokines and their receptors, which had increased expression in the DMSO samples (Fig. 4f).
DISCUSSION
A goal of human mucosal translational research is to obtain insight into the pathophysiology of disease and to provide biomarkers that correlate with remission and relapse to help guide therapy. However, one of the limiting factors to performing such work is the ability to procure and store fresh human intestinal tissue samples. This is especially true for orphan disorders where patients with the particular diseases are rare, longitudinal studies where repeated samples are obtained from the same individual, and large clinical trials involving numerous recruiting sites. The two most widely used ways of processing these samples have been to either isolate the cell of interest from procured intestinal tissue “fresh” followed by immediately analysis, or to cryopreserve the isolated “frozen cells” for later analysis. Both methods are labor-intensive and require the technical expertise that is not universally available, typically limiting such work to large centers with significant resources. Therefore, a major unmet need of translational research focusing on gastrointestinal disorders has been the development of a freezing method where tissue can be frozen directly at the sites of procurement and analyzed later. A key advance of this study is a simple cryopreservation protocol that increases accessibility of tissue storage and allows for successful subsequent tissue analysis. We showed that intestinal tissue can be frozen directly after procurement and prior to any processing, stored, and then processed at centers that have the technical expertise to evaluate these samples. Moreover, the viability of cells was not affected by independent of the duration of cryopreservation. We were able to perform these experiments at four separate sites on both pediatric and adult samples ensuring the reproducibility of the protocol.
This manuscript significantly expands in a number of ways upon recently published data13 demonstrating preservation of CD3+, CD14+, and CD66b+ leukocytes using a similar methodology. First, our evaluation of multiple cellular populations via deep immunophenotyping with up to 40 simultaneous antigen markers5 (via CyTOF) enabled us to not only profile the major immune cell populations in fresh diseased tissue, but to demonstrate the retention of this profile in tissue that was cryopreserved. The two cellular populations that exhibited decreased survival after freezing were granulocytes and mast cells. The decrease in these cell types was secondary to cryopreservation itself and not specific to our method, as similar decreases were observed when cryopreservation occurred at the LPMC stage. This is in accordance with published data suggesting limited survival of granulocytes and mast cells after cryopreservation.14–17 Additionally, we expanded on the published data previously limited to CD8+ T-cells, by evaluating cytokine production upon stimulating immune cell populations that were either obtained from “fresh” or “frozen Bx” tissue. We demonstrated that both innate and adaptive immune cells obtained from “frozen bx” tissue maintained their specificity of cytokine production upon stimulation and secreted similar amounts of cytokines as compared to “fresh” cells.
Secondly, we demonstrated preservation of non-lymphoid cells using this protocol. One advantage to cryopreservation of whole tissue, as opposed to solely the LPMCs alone, is the ability to preserve a variety of cell types. Importantly, we showed that the protocol preserves epithelial cells. Epithelial stem cells cryopre- served in tissue retain their ability to form enteroids with similar growth and proliferation rates when compared to enteroids generated from fresh tissue.
Finally, we demonstrated that the transcriptional analysis of cells obtained from “fresh” or “frozen Bx” is comparable. PCA analysis of RNAseq data of inflamed and uninflamed tissue showed separation of samples by inflammatory status irrespective of the storage methodology. Moreover, the majority of DEG between the inflamed and uninflamed tissue were similar between the two preservation methods and identified differentially regulated genes previously reported to be differentially expressed between active and inactive IBD tissue.10 Nevertheless, the method of storage remained a component in the PCA analysis. The DEG that were most affected by direct cryopreservation were transcripts related to mitochondria and oxidative stress, a well-described consequence of cryopreservation.11,12,18 An additional benefit of storing tissue in DMSO cryopreservation medium as opposed to RNAlater is that this method preserves the ability to subsequently isolate individual cell allowing for downstream analysis at a single-cell level such as with single-cell RNAseq or single-cell ATACseq.
In summary, this manuscript provides a cryopreservation method for storing intestinal tissue that preserves viability and functionality of epithelial and immune cells and allows for transcriptional analysis of the tissue (SOP provided with the manuscript). Our cryopreservation protocol provides a simple single-storage preservation method for tissue allowing for the convenience of subsequent multifunctional analysis. As a result, this protocol has the potential to change how we conduct translational studies, allowing for longitudinal preservation of tissue at multiple sites followed by batch analysis of mucosal tissue to investigate disease pathogenesis and discovery of disease-specific biomarkers as well as a patient response to treatment.
METHODS
Please see supplementary methods and attached SOP for detailed information.
Cryopreservation
Fresh biopsies, surgical tissue (four to five pieces), and isolated LPMCs were slow-frozen in 1 mL of freeze medium (10% DMSO) (Sigma) and 90% FBS (Gibco).
LPMC isolation of fresh and frozen biopsies and tissue samples
LPMC isolation (Boston).
Samples were digested overnight in digestion media at 37 °C and made into single-cell suspensions.
LPMC isolation (MSSM).
Samples were digested for 45 min at 37 ° C in digestion medium (HBSS with Ca+ + Mg+ containing 2% FBS, 0.5 mg/ml DNaseI, and 0.5 mg/ml collagenase IV).
LPMC isolation (Penn).
Samples were digested for 20 min at 37 °C in digestion media.
Primary human enteroid culture
Biopsies were digested and single-cell suspensions were added to Matrigel (Corning). The cultures were maintained in complete human small intestinal medium at 37 °C with 5% CO2 as previously described.19
CyTOF analysis (Boston and MSSM)
Boston.
LPMC suspensions of 1.5–2 million cells were stained with either only cell surface antibodies or with cell surface and intracellular antibodies per previously described CyTOF protocol. Rh103 was used as a viability marker. The samples were run on Helios or CyTOF2 (Fluidigm) equipped with a SuperSampler fluidics system (Victorian Airships) at an event rate of <500 events per s.
MSSM.
LPMC suspensions were first labeled with Rh103 as a viability marker, and then labeled with a panel of metal-labeled antibodies. The samples were then fixed and permeabilized with BD Cytofix/Cytoperm buffer and incubated in Ir intercalator and stored until acquisition. The samples were acquired on a CyTOF2 equipped with a SuperSampler fluidics system (Victorian Airships) at an event rate of <500 events per s.
Flow cytometry (Penn)
LPMCs that had been rested overnight were stained according to standard FACS protocols. Data analysis was performed using FlowJo (version 10.3) software (LLC, Ashland, OR). Dead cells were removed by gating on a LIVE/DEAD Aqua kit (Invitrogen, Carlsbad, CA) versus forward scatter (FSC-H).
RNAseq (Pfizer)
RNA was extracted from individual biopsies utilizing the Qiagen (Valencia, CA) miRNA kit and homogenized on a Bertin Precellys Homogenizer (Rockville, MD) system using ceramic beads. RNAseq was performed using the Truseq Total RNAseq kit with RiboZero (Illumina, San Diego CA). Libraries were pooled to equimolar concentrations and sequenced on a Nextseq 500 system (Illumina, San Diego CA). Processing of the fastq files was performed using the QuickRNAseq20 (pipeline utilizing Hg38 for the genome and Gencode v24 for annotation).
QUANTIFICATION AND STATISTICAL ANALYSIS
CyTOF data analysis
CyTOF files were analyzed by premium Cytobank. Statistical analysis was performed using GraphPad Prism 7.
Enteroid culture analysis
Cultures were imaged using the EVOS FL Auto 2 (ThermoFisher).
RNAseq analysis
RNAseq counts were normalized using the EdgeR algorithm and the voom function of limma.21,22 Differential analysis was performed using limma22 to compare samples by inflammatory status and collection method. Genes with expression <10 count per million (c.p.m.) in all samples were removed from analysis, and genes with a p value <0.001 and FC >±2 are reported as significant. Output of the limma analysis was used for functional enrichment analysis for KEGG pathways using the tmod R package v 0.31 (1, see below) and the output was visualized using ggplot2 v 2.2.1.
Supplementary Material
ACKNOWLEDGEMENTS
L.K. is a recipient of a Career Development Award Grant from the Crohn’s and Colitis Foundation of America (CDA 422348). S.B.S. is supported by NIH Grants: R01 DK115217, R56 AI125766; P30DK034854, the Helmsley Charitable Trust, and the Wolpow Family Chair in IBD Treatment and Research. C.R. is supported by NIH K08 DK106562–01A1. D.T.B. is supported by R01DK084056, the Timothy Murphy Fund, the IDDRC P30HD18655 and the HDDC P30DK034854. V.M. is supported by 5 T32 DK7533
Footnotes
These authors jointly supervised this work: Scott B. Snapper, Miriam Merad.
Liza Konnikova and Scott B. Snapper are members of the VEOIBD Consortium.
ADDITIONAL INFORMATION
The online version of this article (https://doi.org/10.1038/s41385-018-0047-y) contains supplementary material, which is available to authorized users.
Competing interests: Conflicts of Interest: S.B.S. is supported by grants or in-kind contributions from Pfizer, Janssen, Merck, and Regeneron. He is on the scientific advisory boards of Pfizer, Janssen, IFM Therapeutics, Lycera, Inc., Celgene, Pandion Therapeutics, and Applied Molecular Transport. He has consulted for Amgen and Hoffman La-Roche. Except for support from Pfizer that performed the RNA seq experiments in this work and assisted in the analytics, there are no conflicts of interest that are related to this work.
REFERENCES
- 1.Neurath MF Current and emerging therapeutic targets for IBD. Nat. Rev. Gastroenterol. Hepatol. 14, 269–78 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Starr AE et al. Proteomic analysis of ascending colon biopsies from a paediatric inflammatory bowel disease inception cohort identifies protein biomarkers that differentiate Crohn’s disease from UC. Gut 66, 1573–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnes EL, Liew CC, Chao S & Burakoff R Use of blood based biomarkers in the evaluation of Crohn’s disease and ulcerative colitis. World J. Gastrointest. Endosc. 7, 1233–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burakoff R et al. Blood-based biomarkers used to predict disease activity in Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 21, 1132–40 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Bendall SC, Nolan GP, Roederer M & Chattopadhyay PK A deep profiler’s guide to cytometry. Trends Immunol. 33, 323–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–96 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ & Nolan GP Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry A 81, 552–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine JH et al. Data-driven phenotypic dissection of aml reveals progenitor like cells that correlate with prognosis. Cell 162, 184–97 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amir el AD et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31, 545–52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirza AH et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. 7, 39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reardon AJ, Elliott JA & McGann LE Investigating membrane and mitochondrial cryobiological responses of HUVEC using interrupted cooling protocols. Cryobiology 71, 306–17 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Keane KN, Calton EK, Cruzat VF, Soares MJ & Newsholme P The impact of cryopreservation on human peripheral blood leucocyte bioenergetics. Clin. Sci. 128, 723–33 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Hughes SM et al. Cryopreservation of human mucosal leukocytes. PLoS ONE 11, e0156293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishimura M, Mitsunaga S & Juji T Frozen-stored granulocytes can be used for an immunofluorescence test to detect granulocyte antibodies. Transfusion 41, 1268–72 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Lionetti FJ, Hunt SM, Lin PS, Kurtz SR & Valeri CR Preservation of human granulocytes. II. Characteristics of granulocytes obtained by counterflow centrifugation. Transfusion 17,465–72 (1977). [DOI] [PubMed] [Google Scholar]
- 16.Frim J & Mazur P Approaches to the preservation of human granulocytes by freezing. Cryobiology 17, 282–6 (1980). [DOI] [PubMed] [Google Scholar]
- 17.Rowe AW & Lenny LL Cryopreservation of granulocytes for transfusion: studies on human granulocyte isolation, the effect of glycerol on lysosomes, kinetics of glycerol uptake and cryopreservation with dimethyl sulfoxide and glycerol. Cryobiology 17, 198–12 (1980). [DOI] [PubMed] [Google Scholar]
- 18.Baust JM, Corwin W, Snyder KK, Van Buskirk R & Baust JG Cryopreservation: evolution of molecular based strategies. Adv. Exp. Med. Biol. 951, 13–29 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Sato T & Clevers H Primary mouse small intestinal epithelial cell cultures. Methods Mol. Biol. 945, 319–28 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Zhao S et al. QuickRNASeq lifts large-scale RNA-seq data analyses to the next level of automation and interactive visualization. BMC Genomics 17, 39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law CW, Chen Y, Shi W & Smyth GK voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


