SUMMARY
Microbiota are thought to influence the development and progression of inflammatory bowel disease (IBD), but determining generalizable effects of microbiota on IBD etiology requires larger-scale functional analyses. We colonized germ-free mice with intestinal microbiotas from 30 healthy and IBD donors and determined the homeostatic intestinal T cell response to each microbiota. Compared to microbiotas from healthy donors, transfer of IBD microbiotas into germ-free mice increased numbers of intestinal Th17 cells and Th2 cells and decreased numbers of RORγt+ Treg cells. Colonization with IBD microbiotas exacerbated disease in a model where colitis is induced upon transfer of naive T cells into Rag1−/− mice. The proportions of Th17 and RORγt+ Treg cells induced by each microbiota were predictive of human disease status and accounted for disease severity in the Rag1−/− colitis model. Thus, an impact on intestinal Th17 and RORγt+ Treg cell compartments emerges as a unifying feature of IBD microbiotas, suggesting a general mechanism for microbial contribution to IBD pathogenesis.
In Brief
Britton et al. examine 30 human microbiotas from healthy individuals and individuals afflicted with inflammatory bowel disease (IBD). Their findings define an impact on intestinal Th17 and RORγt+ regulatory T cell compartments as a unifying feature of IBD microbiotas, suggesting a general mechanism for microbial contribution to IBD pathogenesis.
Graphical abstract
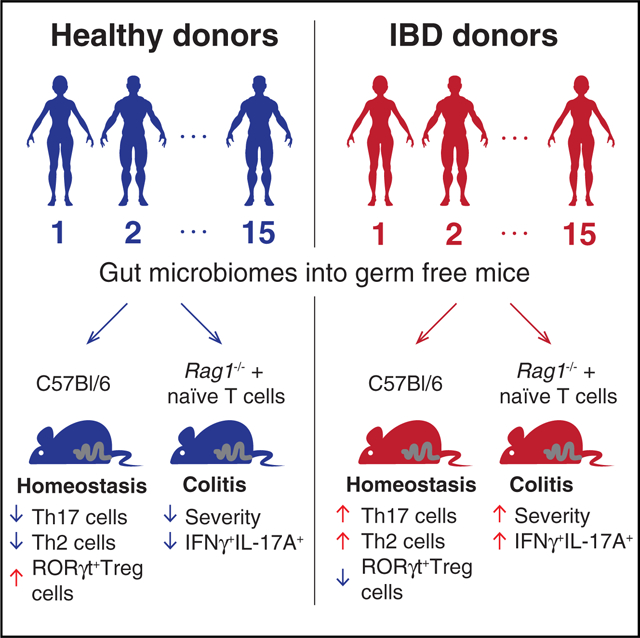
INTRODUCTION
Inflammatory bowel diseases (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), are chronic inflammatory conditions characterized by a dysregulated immune response that results in intestinal inflammation and tissue damage (Sartor, 2008; Khor et al., 2011). Although there is a heritable component to IBD, genome-wide association studies fail to explain the majority of disease risk (Jostins et al., 2012). This and the recent rapid increase in the prevalence of IBD suggest a major role for environmental factors in the etiology of IBD (Sartor, 2008). The composition of the gut microbiota is increasingly appreciated as critical environmental factor with effects on numerous aspects of host physiology. IBD is associated with an altered intestinal microbiota (Frank et al., 2007; Gevers et al., 2014; Jacobs et al., 2016) and genetic defects in microbial handling are risk factors for the disease (Jostins et al., 2012). Therefore, it is widely proposed that IBD occurs as the result of a dysregulated immune response to microbiota and individual susceptibility is determined by both host genetics and the composition of the gut microbiota (Sartor, 2008; Khor et al., 2011).
Culture-independent analyses of the IBD microbiota reveal consistent characteristics that are associated with disease including reduced diversity and an increased ratio of Proteobacteria to Firmicutes compared to healthy individuals (Kostic et al., 2014). However, as changes in the IBD microbiota may be shaped by the disease itself or after exposure to therapies, no definitive causal link has been made between human microbiota composition and IBD (Ni et al., 2017).
Germ-free animals colonized with different microbiotas can be used to test causal relationships between microbiotas and host physiology while maintaining control over host genetics, diet, and environment (Ridaura et al., 2013; Blanton et al., 2016; Sampson et al., 2016; Cekanaviciute et al., 2017; Routy et al., 2018). Such models show that gut microbiota plays a crucial role in shaping the immune system including microbiota-specific pro- and anti-inflammatory effects. Variation in microbiota composition consequently influences host susceptibility to models of autoimmunity, inflammatory disease, and infection both in gut and distant tissue sites (Ivanov et al., 2009; Atarashi et al., 2011, 2017; Palm et al., 2014; Chudnovskiy et al., 2016; De Palma et al., 2017). Germ-free mice have dramatically reduced lamina propria CD4+ T cells and colonization induces rapid expansion and differentiation of effector and regulatory T cell populations (Östman et al., 2006). Colonization with different complex microbiotas or single immunomodulatory strains can induce varied responses and establish diverse gut immune landscapes (Ivanov et al., 2008; Atarashi et al., 2011; Geva-Zatorsky et al., 2017). Among the cells most highly induced upon gut microbiota colonization in ex-germ-free mice are RORγt+FoxP3−Th17 cells (Ivanov et al., 2008) and FoxP3+ regulatory T (Treg) cells (Atarashi et al., 2011; Geuking et al., 2011). Th17 cells are found enriched in human IBD lesions, and microbiotas that strongly induce Th17 cells can exacerbate colitis in mouse models (Fujino et al., 2003; Chudnovskiy et al., 2016; Viladomiu et al., 2017). The majority of gut Th17 cells are specific for microbial antigens (Yang et al., 2014; Tan et al., 2016). Colonization of germ-free mice also increases the frequency of intestinal FoxP3+ Treg cells (Atarashi et al., 2011; Geuking et al., 2011). Specialized subsets of lamina propria Treg cells are distinguished by expression of different transcription factors. GATA3+ Treg cells are particularly responsive to inflammation and have a transcriptional signature associated with tissue repair (Wohlfert et al., 2011; Schiering et al., 2014). Approximately 30%–40% of colon FoxP3+ Treg cells express the transcription factor RORγt (Ohnmacht et al., 2015; Sefik et al., 2015). RORγt+ Treg cells are microbiota dependent, are enriched in gut tissue, and have a strongly suppressive and stable phenotype (Yang et al., 2016). Mice with a selective deficiency of RORγt in Treg cells demonstrate that RORγt+ Treg cells are required to maintain tolerance to microbiota and microbes that favor induction of RORγt+ Treg cells can protect mice from colitis (Sefik et al., 2015). These observations in mouse models support a hypothesis in humans whereby variation in microbiota composition alters the balance between effector and regulatory T cells (Omenetti and Pizarro 2015), particularly Th17 and RORγt+ Treg cells, contributing to the risk of developing intestinal inflammation.
In mouse models of IBD, components of IBD-associated microbiotas can induce Th17-biased effector T cell responses and exacerbate disease severity (Hansen et al., 2010; Eun et al., 2014; Palm et al., 2014; Viladomiu et al., 2017), and complete human fecal microbiotas from both healthy donors and donors with IBD can induce intestinal inflammation in susceptible mice (Moran et al., 2009; Rhee et al., 2009; Eun et al., 2014; Du et al., 2015; Natividad et al., 2015; Nagao-Kitamoto et al., 2016). However, the scale of these studies has precluded identifying specific consequences of colonization with IBD microbiotas compared to healthy microbiotas and defining generalizable functional properties of IBD or healthy donor microbiotas.
Interventions targeting the microbiota are a potential path for IBD treatment. Recent clinical trials have demonstrated the potential of fecal microbiota transplantation (FMT) for treating individuals with ulcerative colitis (Moayyedi et al., 2015; Paramsothy et al., 2017). FMT provides a benefit to approximately 25% of patients in these studies, but efforts to improve efficacy and refine this therapeutic approach are currently hampered by a poor understanding of the differences between gut microbiotas of the population of healthy individuals and individuals with IBD. A better understanding of the functional properties of healthy microbiotas relative to those from individuals with IBD may guide the stratification of patients, the design of more refined microbiota- targeted therapies, and the development of microbiota-focused biomarkers or diagnostics.
A population-scale understanding of the unique immunogenic features of healthy and disease microbiotas is needed to better understand how interpersonal variation in the gut microbiome influences disease risk and the functional biology of complex diseases, including IBD. Here we examined the impact of transferring the gut microbiotas of 32 healthy or IBD donors into ex-germ-free mice. Transfer of microbiotas from IBD donors drove distinct adaptive immune profiles in unchallenged mice, including greater induction of Th2 and RORγt+ Th cells and reduced induction of RORγt+ Treg cells relative to healthy donor microbiotas. IBD donor microbiotas also exacerbated disease in Rag1-deficient mice after transfer of naive CD4+ T cells. Our results demonstrate that IBD-associated microbiotas are consistently more pro-inflammatory than those from healthy donors, suggesting a unifying mechanism for the contribution of gut microbiota to IBD.
RESULTS
IBD and Healthy Donor Fecal Microbiotas Are Compositionally Similar
To gain an insight into unifying properties of IBD and healthy donor microbiotas, we characterized immune responses in mice colonized with more than 30 different human fecal microbiotas. This included both complete fecal microbiotas and cultured collections of microbes isolated from a donor fecal sample (Tables 1 and S1; Goodman et al., 2011; Faith et al., 2014). To determine the microbial composition of these samples, we performed 16S rRNA gene amplicon sequencing of the donor fecal samples. Consistent with previous reports (Gevers et al., 2014), an unweighted UniFrac analysis of the composition of the microbiotas did not distinguish between donors with IBD and healthy donors (Figure 1A). We also compared the composition of the cultured microbiota collections generated from healthy and donor microbiotas. Principle coordinates analysis using a Jac-card distance of species composition failed to discriminate between the cultured collections from healthy and IBD donors (Figure 1B).
Table 1.
Gut Microbiota Donors Included in Gnotobiotic Experiments
| Microbiota Name |
Donor Phenotype |
Active/ Remission |
Cohort | Subject ID | B6 Immune Phenotyping |
RagTCT Model |
BioSample Accession |
|---|---|---|---|---|---|---|---|
| HD2017 | healthy | NA | NYC | 1001099 | C(4) | C(5) | SAMN07747250 |
| HD2018 | healthy | NA | NYC | 1001136 | C(9),S(4) | C(6) | SAMN07747252 |
| HD2019 | healthy | NA | NYC | 1001217 | C(16) | C(5),S(4) | SAMN07747269 |
| HD2020 | healthy | NA | NYC | 1001262 | C(3) | C(5) | SAMN07747287 |
| HD2021 | healthy | NA | NYC | 1001271 | C(6) | C(4),S(5) | SAMN07747295 |
| HD2022 | healthy | NA | NYC | 1001275 | C(3) | C(5) | SAMN07747304 |
| HD2023 | healthy | NA | NYC | 1001283 | C(4) | C(6) | SAMN07747317 |
| CD2028 | CD | remission | NYC | 1100102 | S(5) | S(5) | SAMN07750457 |
| CD2005 | CD | active | NYC | 1101004 | S(3) | S(5) | SAMN07750563 |
| UC2029 | UC | active | NYC | 1101029 | S(3) | S(3) | SAMN07750571 |
| UC2030 | UC | active | NYC | 1101312 | S(6) | C(4),S(5) | SAMN07750587 |
| UC2031 | UC | remission | NYC | 1101437 | S(3) | S(5) | SAMN07750596 |
| CD2006 | CD | active | NYC | 1101653 | S(5) | C(6),S(5) | SAMN07750626 |
| UC2032 | UC | remission | NYC | 1102157 | S(2) | S(5) | SAMN07750657 |
| CD1001 | CD | remission | LA | BSD2780_06_1687 | C(8),S(7) | C(12),S(5) | SAMN08636700 |
| UC1024 | UC | remission | LA | BSD2780_06_1688 | C(5) | C(20),S(6) | SAMN08636698 |
| HD1007 | healthy | NA | LA | BSD2780_06_1689 | C(7) | C(12) | SAMN08636702 |
| UC1025 | UC | remission | LA | BSD2780_12_0874 | C(5) | – | SAMN08636705 |
| CD1002 | CD | remission | LA | BSD2780_12_0875 | C(5) | C(8),S(5) | SAMN08636696 |
| HD1008 | healthy | NA | LA | BSD3178_07_0968 | S(4) | S(4) | SAMN08636690 |
| CD1003 | CD | remission | LA | BSD3178_07_1175 | S(3) | C(7),S(8) | SAMN08636694 |
| UC1026 | UC | remission | LA | BSD3178_07_1176 | S(3) | C(5),S(9) | SAMN08636692 |
| HD1009 | healthy | NA | LA | BSD3178_07_1237 | – | S(4) | SAMN08636695 |
| HD1016 | healthy | NA | LA | BSD3178_12_0969 | S(4) | – | SAMN08636688 |
| HD1010 | healthy | NA | LA | BSD3448_08_0949 | S(3) | C(6),S(6) | SAMN08636693 |
| UC1027 | UC | remission | LA | BSD3448_08_0978 | S(2) | S(10) | SAMN08636689 |
| HD1011 | healthy | NA | LA | BSD3448_12_0968 | S(2) | S(5) | SAMN08636691 |
| HD1012 | healthy | NA | LA | BSD3448_12_1246 | S(2) | S(5) | SAMN08636687 |
| CD1004 | CD | remission | LA | BSD4362_10_1914 | S(4) | S(12) | SAMN08636697 |
| HD1004 | healthy | NA | LA | BSD4362_11_0056 | S(3) | S(5) | SAMN08636704 |
| HD1014 | healthy | NA | LA | BSD4362_11_0057 | S(3) | S(6) | SAMN08636701 |
| HD1015 | healthy | NA | LA | BSD4362_12_1006 | – | S(7) | SAMN08636699 |
Abbreviations: C, gnotobiotic mice colonized with arrayed culture collection derived from donor fecal microbiota; S, gnotobiotic mice colonized with clarified complete stool microbiota from donor. Numbers in parentheses indicate the number of mice per group.
Figure 1. Increased Frequencies of RORγt+ Th Cells in Mice Colonized with IBD-Associated Microbiotas as Compared to Mice Colonized with Healthy Donor Microbiotas.
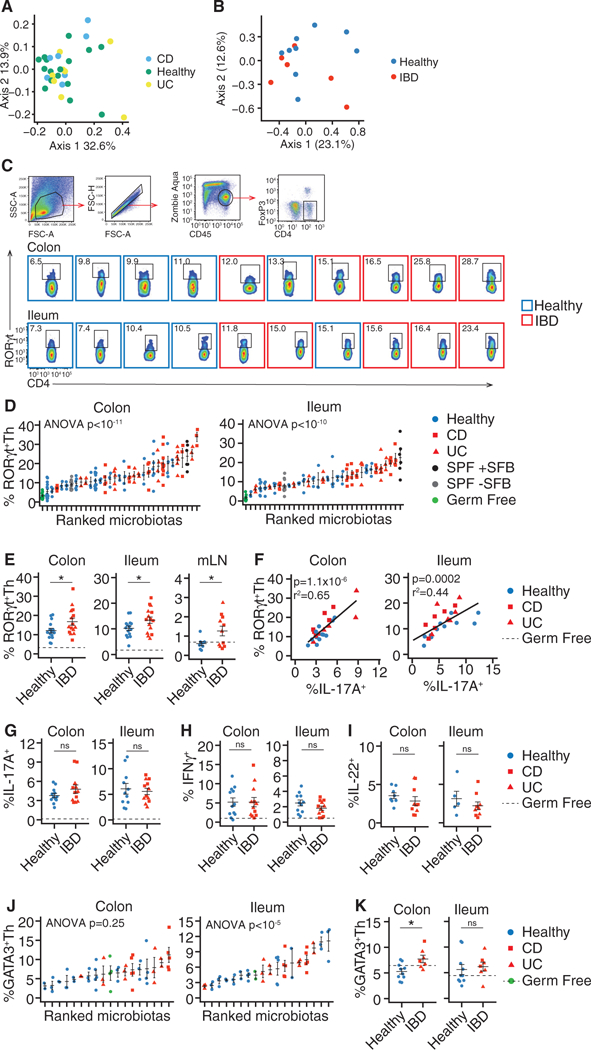
Germ-free B6 mice were colonized with fecal microbiotas from human donors with and without IBDwhose composition was assayed by16S rRNA amplicon sequencing. Effector CD4+ T cells from the colon, ileum, and mLN of these mice were analyzed by flow cytometry.
(A) PCoA based on unweighted UniFrac distances of 16S rRNA amplicon sequencing of the human donor fecal microbiotas used in this study.
(B) PCoA based on Jaccard distances comparing the species-level composition of the arrayed culture collections of microbes used in the study.
(C and D) Colon and ileum RORγt+ Th cells in individual mice.
(E) The mean proportion of RORγt+ Th cells in the colon, ileum, and mLN ofgroupsofmice colonized with the same microbiotas.
(F) Correlation between the proportions of RORγt+ Th cells and IL-17A+CD4+ T cells.
(G–I) The mean proportion of (G) IL-17A+CD4+ Tcells, (H) IFN-γ+CD4+Tcells, and (I) IL-22+Tcells in colon and ileum.
(J) Colon and ileum GATA3+ Th cells in individual mice.
(K) The mean proportion of GATA3+ Th cells in the colon and ileum of groups of mice colonized with the same microbiotas.
The numbers of RORγt+ Th and GATA3+ Th cells are presented as the proportion of live, CD45+, CD4+, FoxP3− cells. The numbers of cytokine+ cells are presented as a proportion of live, CD45+, CD4+ cells. Flow cytometry plots include data acquired at different times, thus gating differs between plots.
(C–K) n = 15 healthy, 8 UC, and 7 CD microbiotas (RORγt), n = 11 healthy, 6 UC, and 7 CD microbiotas (IFN-γ and IL-17A), n = 8 healthy, 4 UC, and 6 CD microbiotas (IL-22), n = 10 healthy, 5 UC, and 2 CD microbiotas (GATA3). (D and J) Each point represents data from one mouse, in all other plots each point representsthe mean value ofagroup of 2–12 mice colonized with a single microbiota. ns, not significant; *p < 0.05, Student’s t test; solid horizontal lines indicate mean ± SEM, dashed horizontal lines represent the mean proportion of the cell type in germ-free mice. Regression p values in (F) calculated by f-test.
See also Figure S1.
Increased Frequencies of RORγt+ Th Cells in Gnotobiotic Mice Colonized with IBD-Associated Microbiotas as Compared to Mice Colonized with Healthy Donor Microbiotas
To determine the functional impact of these gut microbiotas on mucosal T cell populations, we colonized germ-free C57BL/6J (B6) mice with fecal slurries or pools of cultured fecal donor microbes (Goodman et al., 2011; Faith et al., 2014) from two independent cohorts of healthy donors (n = 15) or donors with IBD (n = 15; Table 1) (Jacobs et al., 2016; Contijoch et al., 2018). Since IBD pathophysiology is associated with a dysregulated T cell response (Hegazy et al., 2017), we focused on the gut CD4+ T cell compartment and performed a comprehensive measurement of CD4+ T helper (Th) and T regulatory (Treg) cells in the intestinal lamina propria of each colonized mouse using flow cytometry. Because of the established association of IBD with both Th17 cells and microbiota composition (Fujino et al., 2003; Yang et al., 2014; Omenetti and Pizarro, 2015), we focused first on RORγt+ T helper (Th) cells. The ileum, colon, and mLN of mice colonized with IBD microbiotas contained higher numbers of RORγt+ Th cells (CD4+, FoxP3−, RORγt+) compared to mice colonized with healthy microbiotas (p < 0.05; t test; Figures 1C–1E and S1). RORγt+ Th cells varied over a 6-fold range and included human microbiotas inducing similar proportions to commonly used mouse reference communities (specific-pathogen-free [SPF] microbiotas ± segmented filamentous bacteria [SFB]) (Figure 1D; Ivanov et al., 2009). As expected (Ivanov et al., 2006), the proportion of RORγt+ Th cells correlated with the proportion of IL-17A+CD4+ T cells within each tissue (colon; p = 1.1 × 10–6, R2 = 0.65, ileum; p = 0.0002, R2 = 0.44; Figure 1F). Although the proportion of IFN-γ+ Th1, IL-22+, and IL-17A+CD4+ T cells varied by donor microbiota (Figure S1), these T cell sub-sets were not significantly altered in mice colonized with healthy microbiotas compared with IBD microbiotas (Figures 1G–1I). In contrast, the average proportion of FoxP3−GATA3+ Th2 cells was higher in the colon of mice colonized with IBD microbiotas (p < 0.05, t test; Figures 1J and 1K).
Similar Frequencies of FoxP3+ Treg Cells in Mice Colonized with IBD Microbiotas and Those Colonized with Healthy Microbiotas
We hypothesized that the increased numbers of RORγt+ Th and GATA3+ Th2 cells may result from IBD-associated microbiotas failing to promote the differentiation of naive T cells into Foxp3+ Treg cells (Omenetti and Pizarro 2015). As in previous studies (Faith et al., 2014; Geva-Zatorsky etal., 2017), transfer of most microbiotas into gnotobiotic mice led to increased numbers of gut FoxP3+ cells as compared to baseline germ-free levels (Figures 2A and 2B). Although the proportions of FoxP3+Treg cells and IL-10+CD4+ T cells were significantly influenced by donor microbiota (p < 1 × 10–15, p < 0.0001 [FoxP3 colon, ileum], p = 0.006, p = 0.02 [IL-10 colon, ileum]; ANOVA), we observed no difference between the mean proportion of FoxP3+ Treg cells or IL-10+CD4+ T cells between mice colonized with healthy and IBD-associated microbiotas (Figures 2C and 2D). IL-10 can be produced by multiple CD4+ T cell subsets (Ng et al., 2013). In these humanized microbiota mice under homeostatic conditions, the majority of CD4+ T cell-derived IL-10 detected by intracellular cytokine straining was within FoxP3+ T cells (Figure 2E). The proportion of RORγt+ Th and FoxP3+ Treg cells induced by a microbiota were not correlated (Figure 2F). These observations suggested that changes in total FoxP3+ Treg cell number could not explain the expansion of RORγt+ Th and Th2 cells in mice colonized with IBD microbiotas. We therefore sought to better characterize the Treg cells induced by different human donor microbiotas.
Figure 2. Similar Frequencies of FoxP3+ Treg Cells in Mice Colonized with IBD Microbiotas and Those Colonized with Healthy Microbiotas.
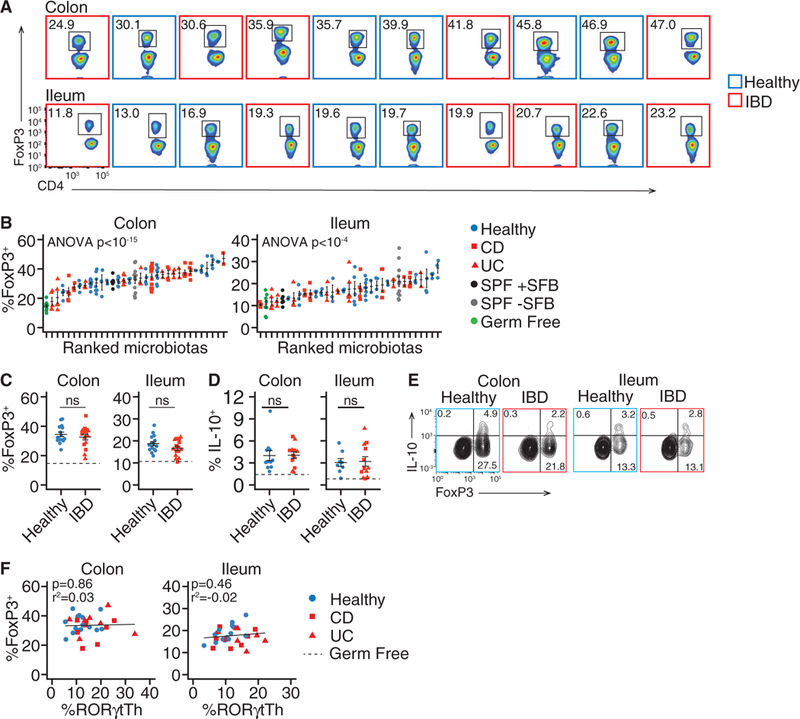
Germ-free B6 mice were colonized with fecal microbiotas from human donors with and without IBD. Regulatory T cells from the colon and ileum of these mice were analyzed by flow cytometry.
(A and B) The proportion of FoxP3+ Treg cells in each mouse colonized with different donor microbiotas.
(C and D) The mean proportion of(C) FoxP3+Treg cells and (D) IL-10+CD4+T cells in the colon and ileum of groups of mice colonized with the same microbiotas.
(E) Co-expression of FoxP3 and IL-10 in CD4+ T cells from colon and ileum.
(F) Correlation between RORγt+ Th cells and FoxP3+ Treg cells from colon and ileum.
The numbers of FoxP3+ and IL-10+ cells are presented as a proportion of live, CD45+, CD4+ cells. Flow cytometry plots include data acquired at different times, thus gating differs between plots. (A–D) n = 11 healthy, 6 UC, and 7 CD microbiotas; (E) representative data from three healthy and three IBD microbiotas. (B) Each point represents data from one mouse, in all other dot plots each point represents the mean value of a group of 3–12 mice colonized with a single microbiota. ns, not significant, Student’s t test; solid horizontal lines indicate mean ± SEM, dashed horizontal lines represent the mean proportion of the cell type in germ-free mice. Regression p values in (F) calculated by f-test.
Transfer of Microbiotas from Healthy Donors Specifically Increases Numbers of RORγt+ Treg Cells
The gut harbors subsets of Treg cells that have non-redundant functions, including those characterized by expression of RORγt and GATA3 (Wohlfert et al., 2011; Ohnmacht et al., 2015; Sefik et al., 2015; Yang et al., 2016; Xu et al., 2018). We therefore used flow cytometry to examine the relative induction of Treg cell subsets by healthy and IBD donor microbiotas. Induction of RORγt+ Treg cells in colon and ileum varied significantly with different microbiotas (p < 1 × 10–15, p < 1 × 10–8 in colon and ileum, ANOVA; Figures 3A and 3B). In contrast to the total FoxP3+ Treg cell population, we observed a significant expansion of RORγt+ Treg cells induced by healthy microbiotas relative to IBD microbiotas in both colon and ileum (p < 0.001, t test; Figure 3C). This difference was significant across the two independent cohorts of microbiota donors (Figure 3D) and in mice colonized with stool or cultured microbiotas (Figure S2). The proportion of total FoxP3+ Treg cells was correlated with RORγt+ Treg cells in ileum (p < 0.001; R2 = 0.39) and weakly correlated in colon (p = 0.04, R2 = 0.1; Figure S2D). Colonization of gnotobiotic mice with healthy or IBD microbiotas increased the numbers of RORγt+ Treg cells in mLN, with no significant difference observed between the two groups (Figure 3E). The proportion of RORγt+ Treg cells in colon and ileum were correlated (p = 4 × 10–5, f-test), but neither correlated with the proportion in mLN (Figure 3F). This confirms previous observations that RORγt+ Treg cells are a gut tissue-specific subset and suggests that the conditions required for RORγt+ Treg cell differentiation are found uniquely in lamina propria (Ohnmacht et al., 2015; Sefik et al., 2015; Yang et al., 2016). Colonic RORγt+ Treg cells from mice colonized with either healthy or IBD microbiotas secreted minimal IL-17A when stimulated ex vivo compared to FoxP3−RORγt+ Th cells (Figure 3G), as reported previously in SPF mice (Sefik et al., 2015).
Figure 3. Transfer of Microbiotas from Healthy Donors Increases Gut RORγt+ Treg Cells.
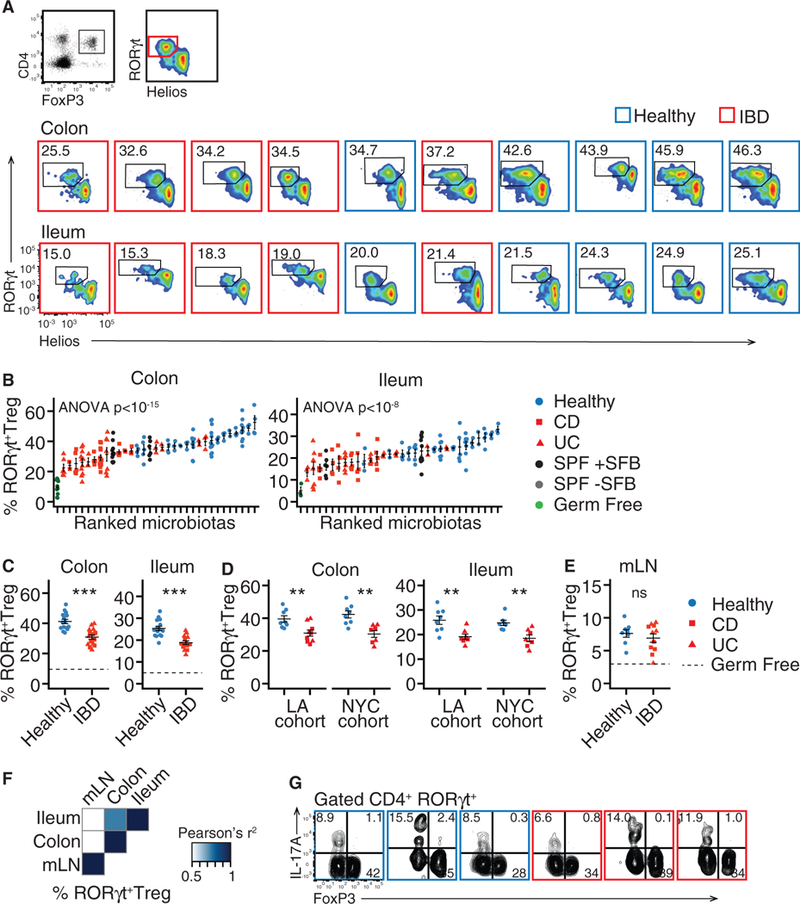
Germ-free B6 mice were colonized with fecal microbiotas from human donors with and without IBD. Regulatory T cell subsets in the colon and ileum of these mice were analyzed by flow cytometry.
(A and B) The proportion of gut RORγt+ Treg cells varies in individual mice.
(C and D) The mean proportion of RORγt+ Treg cells the colon and ileum of groups of mice colonized with the same microbiotas. The data separated according to cohort shown in (D).
(E) The mean proportion of RORγt+ Treg cells the mLN of groups of mice colonized with the same microbiotas.
(F) Correlation between the proportion of RORγt+ Treg cells in different tissues in mice colonized with the same microbiotas.
(G) Co-expression of FoxP3 and IL-17A in CD4+ T cells. Each plot shows data from a different microbiota.
The numbers of RORγt+ Treg cells are presented as a proportion of live, CD45+, CD4+, FoxP3+ cells. Flow cytometry plots include data acquired at different times, thus gating differs between plots.
(A–F) n = 15healthy, 8 UC, and 7 CD microbiotas; (B) each point represents data from one mouse, in all other dot plots each point represents the mean value of a group of 2–12 mice colonized with a single microbiota. Data in (G) is representative of three mice colonized per microbiota. ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, Student’s t est; solid horizontal lines indicate mean ± SEM, dashed horizontal lines represent the mean proportion of the cell type in germ free mice.
Regression p values calculated by f-test. See also Figure S2.
We considered that reduced induction of RORγt+ Treg cells in IBD donor colonized mice could be compensated for by expansion of other Treg cell subsets. We therefore further characterized the fraction of RORγt−FoxP3+ Treg cells that were more abundant in IBD donor colonized mice. Whereas gut RORγt+ Treg cells are assumed to be induced in response to peripheral stimulus from microbiota, many lamina propria RORγt− Treg cells express the transcription factor Helios, indicating a possible thymic origin (Ohnmacht et al., 2015). In the ileum of mice colonized with IBD microbiotas, there was a greater proportion of Helios+ Treg cells relative to mice colonized with healthy donor microbiotas and, as expected (Sefik et al., 2015), RORγt+ Treg and Helios+ Treg cells were inversely correlated (Figure S2D). A lower proportion of FoxP3+CD4+ T cells expressed neither RORγt nor Helios (Figure S2). This “double negative” population was enriched in the colon mice colonized with IBD microbiotas relative to those colonized with healthy microbiotas (Figure S2B). As previous described (Ohnmacht et al., 2015), the proportion of FoxP3+GATA3+ Treg cells was not significantly increased in colonized mice relative to germ-free mice, and they were not differentially modulated by healthy and IBD microbiotas (Figure S2C). These data, and the inverse correlation of RORγt+ Treg with GATA3+ and Helios+ Treg cells (Figure S2D), support the hypothesis that there is a specific reduction in the expansion of RORγt+ Treg in IBD-donor colonized mice, while changes in the relative proportion of other Treg cell subsets are a consequence of the microbiota-altered RORγt+ Treg cell frequency. Cytokines secreted by type 3 innate lymphoid cells (ILC3s) play roles in the maintenance of mucosal homeostasis, including Treg cell induction (Morthaetal., 2014). We found no significant difference in the proportion of IL-17A+, IL-22+, or Csf2+ (GM-CSF+) ILC3s in colon lamina propria of mice colonized with healthy or IBD microbiotas (Figure S2E).
It has been suggested that RORγt+ Treg cells are uniquely positioned to regulate Th2 cell responses (Ohnmacht et al., 2015). Although we observed a significant expansion of Th2 (GATA3+FoxP3−CD4+) cells in the colon of gnotobiotic mice colonized with IBD microbiotas relative to healthy microbiotas (p < 0.05, t test; Figures 1J and 1K), the proportion of Th2 cells was uncorrelated with RORγt+ Treg cells (p = 0.09, p = 0.9 in colon and ileum; Figure S3A). We also found no correlation between the proportion of RORγt+ Treg cells and RORγt+ Th or IFN-γ+CD4+ T cells (Figures S3A). FoxP3-cre × RORγt-flox mice, deficient in RORγt+ Treg cells, show increased lamina propria dendritic cell (DC) activation (Ohnmacht et al., 2015). We sought to determine whether the deficit in RORγt+Treg cells observed in mice colonized with IBD microbiotas was sufficient to influence DC phenotype. In B6 mice colonized with healthy and IBD microbiotas representing the extremes of RORγt+ Treg cell induction, we found that a low proportion of RORγt+ Treg cells correlated with increased expression of CD80 and CD86 on CD11c+CD64− DCs and CD64+ macrophages/monocytes (Figure S3B).
IBD-Associated Microbiotas Transmit Enhanced Colitis Severity to Susceptible Mice
To assess whether IBD-associated microbiotas influence colito-genesis, we tested healthy-and IBD-donor microbiotas in a gnotobiotic mouse model of colitis. Given the known importance of T cells in IBD pathophysiology, we chose a model of colitis that is dependent on both T cells and microbiota. Transfer of CD45RBhi (naive) CD4+ T cells to Rag1-deficient mice induces colitis-like pathology, but only in the presence of an immunogenic microbiota (hereafter the Rag T cell transfer [RagTCT] model) (Powrie et al., 1993; Stepankova et al., 2007). At 4–8 weeks prior to T cell transfer, we colonized germ-free Rag1–/– mice with fecal microbiotas from both healthy (n = 16) or IBD (n = 14) human donors (see Table 1). The alpha diversity (Shannon) of microbiota from B6 and Rag–/– colonized with the same human donor microbiota were significantly correlated (r2 = 0.6, p = 0.002, f-test), an indication of similar engraftment between the mouse models. A control microbiota included in every iteration of the colitis model demonstrated low inter-experiment variation (Figure S4A). As measured by loss in body mass, histology, and elevation of fecal lipocalin2 (LCN2), colitis was more severe in mice colonized with fecal microbiotas from individuals with IBD than those colonized with microbiotas from healthy donors (p = 4.2 × 10–5, p = 0.0058 6 weeks after T cell transfer for body mass and LCN2, respectively, t test; Figures 4A and 4B). Loss in body mass was correlated with elevated fecal LCN2 (R2 = 0.33, p = 1.4 × 10–7; Figure S4B). Remarkably, a significant difference in weight loss between healthy and IBD microbiotas was already detectable 7 days after T cell transfer and became more prominent overtime (Figure S4C). There was no significant difference in colitis severity between mice colonized with microbiotas from donors with UC compared to CD (p = 0.59, t test; Figure 4D), and CD and UC microbiotas each independently induced colitis that was more severe than in mice colonized with healthy donor microbiotas (p < 0.01, p < 0.001 for UC and CD, respectively, ANOVA; Figure 4D). We replicated these findings in two independent cohorts of donors (Figures 4E and S4D). We also found both stool microbiotas and cultured collections of microbes from donors with IBD were similarly able to increase colitis susceptibility in mice, relative to healthy donor microbiotas (Figures 4F and S4E). Colitis was not significantly different in mice colonized with IBD microbiotas from donors with active disease or in remission (Figure S4F). For ten donors, we assayed the colitogenicity of both the stool and the cultured microbiota collection derived from the stool. Eight of the ten cultured microbiotas transferred colitis of equivalent severity as the total stool microbiota derived from the same donor (Figure S4G).
Figure 4. IBD-Associated Microbiotas Transmit Enhanced Colitis Severity to Susceptible Mice.
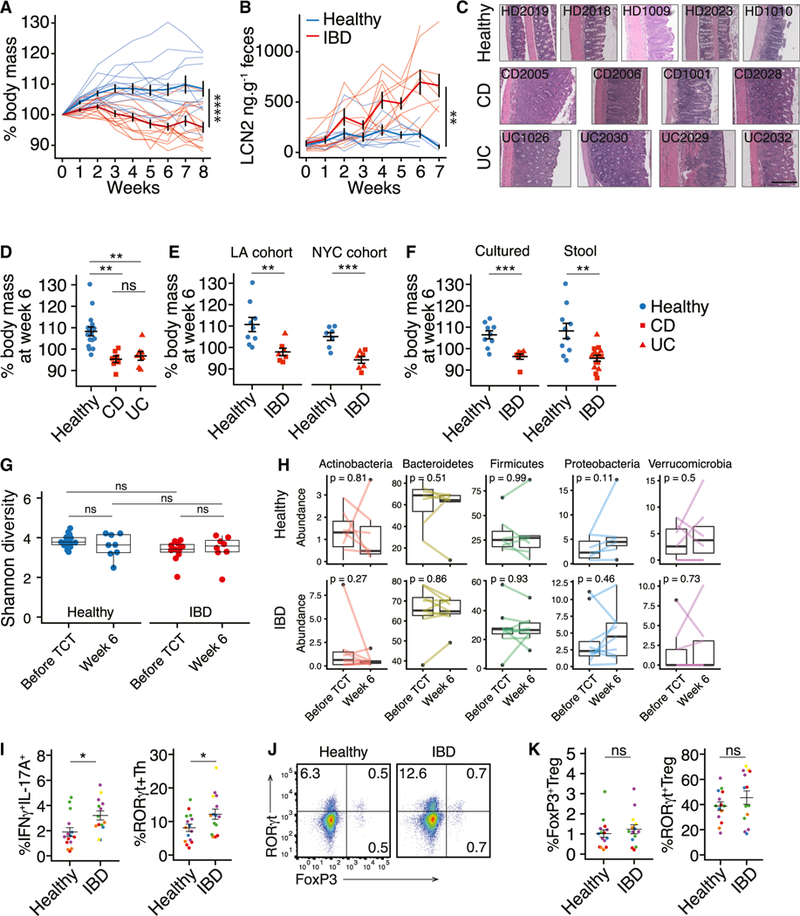
Colitis was induced by transferring naive CD4+ T cells into Ragl-deficient mice colonized with healthy or IBD donor microbiotas.
(A and B) Loss of body mass and fecal lipocalin2 (LCN2) in RagTCT mice colonized with IBD and healthy microbiotas. Thin lines represent the mean data from a group of5–15 mice colonized with a single microbiota and bold lines represent the mean ± SEM of all groups of mice colonized with either healthy donor or IBD donor microbiotas.
(C) Representative H&E-stained colon sections from RagTCT mice colonized with different human donor microbiotas 5–7 weeks after T cell transfer. Scale bar = 200 μm.
(D) Change in body mass at week 6 in RagTCT mice colonized with healthy, UC, or CD microbiotas. (E and F) Colitis severity in RagTCT mice colonized with microbiotas from (E) two cohorts and (F) with stool and cultured IBD microbiotas.
(G) Shannon diversity of RagTCT mouse fecal microbiotas before and after colitis induction, based on 16S rRNA gene amplicon sequencing.
(D-G) Each point shows the mean weight change of a group of5–15 mice 6 weeks after T cell transfer (n=16 healthy donors, n = 6 CD donors, n = 6 UC donors [of which 2 UC and 2 CD had active disease]).
(H) Relative abundance of major phyla in RagTCT mouse fecal microbiotas before and after colitis induction. Lines connect the mean abundances from groups of mice colonized with the same microbiota, before and after colitis induction.
(I) The proportion of RORγt+ Th cells and IFNγ+IL-17A+CD4+ T cells in the colon of RagTCT mice 4 weeks after TCT. Each point represents data from one mouse, each color represents mice colonized with different microbiotas.
(J) RORγt+ and Foxp3+ cells in the colon lamina propria 4 weeks after TCT.
(K) The proportion of FoxP3+ Treg cells and RORγt+ Treg cells in the colon of RagTCT mice4weeks after TCT. Each point represents data from one mouse; each color represents a different microbiota.
The numbers of FoxP3+ and cytokine+ cells are presented as a proportion of live, CD45+, CD4+ cells. The numbers of RORγt+ Treg cells are presented as a proportion of live, CD45+, CD4+, FoxP3+ cells.
Boxplots show the median and interquartile range. P values are calculated using ANOVA with Tukey’s correction for multiple comparisons(D and G), paired t test (H), or unpaired Student’s t test (all other panels). ns, not significant, **p < 0.01, ***p < 0.001, ****p < 0.0001; Student’s t test. See also Figures S3, S4G, and S4I.
Each point represents data from one mouse, each color represents a different microbiota.
We considered that induction of intestinal inflammation could alter the composition of the microbiota and that outgrowth of pathogenic strains could contribute to colitis progression. We performed 16S rRNA amplicon sequencing on feces from the RagTCT mice both before colitis induction and 6 weeks after T cell transfer, around the peak of disease for susceptible mice. We observed no difference in the alpha diversity of the engrafted healthy or IBD microbiotas, either before or after colitis induction (Figure 4G). There were also no consistent changes in the broad taxonomic composition of the fecal microbiotas of the RagTCT mice after colitis induction (Figure 4H).
Within groups of mice colonized with one of five healthy or six IBD microbiotas, we characterized the activation and differentiation of the progeny of the transferred CD45RBHIT cells 4 weeks after transfer. Interpretation of immune population variation between these groups is complicated by the microbiota-induced variation in disease severity 4 weeks post-transfer. It was previously demonstrated that exacerbation of colitis in mice is associated with an increased proportion of IFN-γ+IL-17A+CD4+ T cells (Ahern et al., 2010). In line with these observations, we find the same population expanded in T cell transfer mice colonized with the IBD microbiotas (Figure 4I). We also found an increased proportion of RORγt+ Th (FoxP3−) cells in the colon of these mice (Figure 4J). As previously reported (Uhlig et al., 2006), between 1% and 2% of the expanded cells in lamina propria expressed FoxP3 (Figures 4J and 4K). This proportion was not significantly different between mice colonized with healthy or IBD microbiotas (Figures 4J and 4K). It was notable that an average of 40%–50% of the FoxP3+ cells co-expressed RORγt, indicating that the splenic origin of the naive T cells did not hamper RORγt+ Treg cell development (Figures 4J and 4K). However, the proportion of RORγt+ Treg cells was highly variable between animals and there was no significant difference in the proportion of RORγt+ Treg cells between mice colonized with healthy or IBD microbiotas (Figure 4K).
Homeostatic Induction of RORγt+ Treg and RORγt+ Th Cells Predicts Colitis Severity in Susceptible Mice Colonized with the Same Microbiota
Finally, we sought to understand how the variation in CD4+ T cell responses we observed in unchallenged gnotobiotic B6 mice correlated with colitis severity in RagTCT mice colonized with the same donor microbiotas. A total of 15 healthy and 14 IBD microbiotas were tested in both models (Table 1). While colitis severity was not correlated with GATA3+ Th or FoxP3+ Treg cells (Figures 5A), the proportion of colon and ileum RORγt+ Th cells induced by a microbiota in B6 mice was positively correlated with colitis severity in RagTCT mice colonized with the same microbiota (R2 = 0.32, p = 0.001; R2 = 0.26, p = 0.004 for colon and ileum, respectively; Figure 5A). Induction of RORγt+ Treg cells in B6 mice was inversely correlated with colitis severity in RagTCT mice (R2 = 0.29, p = 0.002; R2 = 024, p = 0.006 for colon and ileum, respectively; Figure 5A). The proportion of IL-17A+CD4+ T cells induced in colon was also weakly associated with colitis severity (R2 = 0.25, p = 0.013; Figure S5). A linear model explained 53% of the variation in colitis severity (weight loss at week 6) as a function of the proportion of both RORγt+ Th and RORγt+ Treg cells in both tissues (R2 = 0.53, p = 0.002; F-test). Colitis severity was not associated with Helios+ Treg, IL-10+, or IFN-γ+CD4+T cells (Figure S5). We used receiver operating characteristic (ROC) curves to assess the value of the humanized-microbiota mouse data in predicting the health status of the human microbiota donor using a logistic model (Figure 5B). The proportion of colon RORγt+ Th cells had reasonable predictive value (AUC = 0.71), but the proportion of colon RORγt+ Treg cells was more informative (AUC = 0.92) (Figure 5B). Colitis severity as measured by weight loss 6 weeks after T cell transfer was also highly predictive of donor health (AUC = 0.93) (Figure 5B). We found the best predictive power when the proportion of colon RORγt+ Treg cells induced under homeostatic conditions and colitis severity at week 6 in RagTCT mice were combined in the logistic model (AUC = 0.95) (Figure 5B).
Figure 5. Homeostatic Induction of RORγt+ Treg and RORγt+ Th Cells Predicts Experimental Colitis Severity and Human Microbiota Donor Health.
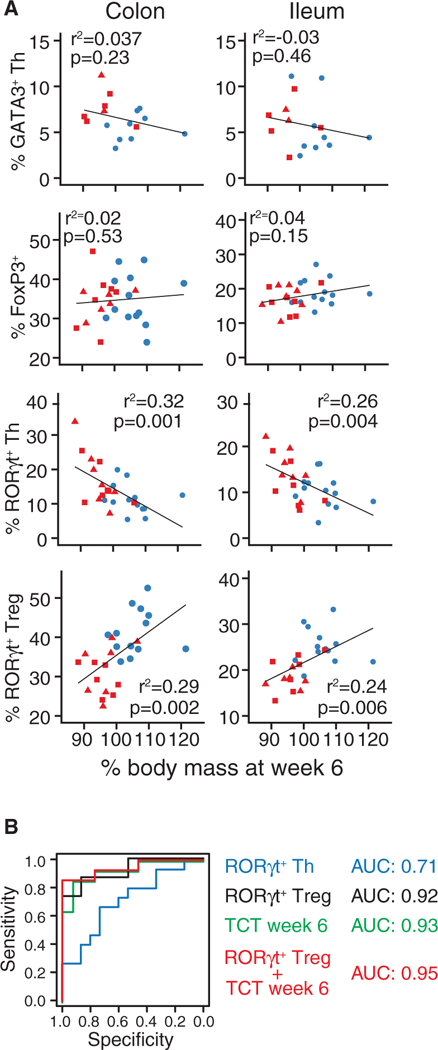
Data on the effect of the donor microbiotas on T cell populations in B6 mouse gut and on colitis severity in RagTCT mice were combined and used to generate a logistic model that accurately predicted the health of the microbiota donor.
(A) Correlations between colitis severity in RagTCT mice and the proportion of gut T cell subsets in unchallenged B6 mice colonized with the same microbiota.(B) Receiver operating characteristic (ROC) curves assessing the value of logistic models based on measurements made in humanized microbiota mice as binary classifiers to predict the health of the microbiota donor. “TCT week 6” refers to body mass data from RagTCT mice 6 weeks after T cell transfer. Other data used in the models are the proportions of RORγt+ Treg and RORγt+ Th cells measured in the colon of B6 mice.
The body weight data represent the mean measurements of groups of 5–15 RagTCT mice colonized with a single human donor microbiota and the phenotyping data is the mean value of a group of 2–12 B6 mice colonized with the same single microbiota. p values are calculated by f-test. See also Figure S5.
DISCUSSION
By colonizing germ-free mice with fecal microbiotas from more than 30 human donors, we demonstrated that there are consistently altered immune responses induced by human gut microbiotas from donors with IBD relative to healthy donors. Specifically, mice colonized with IBD microbiotas had a greater number of gut Th17 cells and fewer gut RORγt+ Treg cells than mice colonized with healthy donor microbiotas. In a T cell transfer model of colitis, mice colonized with IBD microbiotas experienced more severe disease than those colonized with healthy donor microbiotas. Culture-independent analyses of the composition of the gut microbiota in individuals with IBD, together with animal models and genetic risk factors, have led to a widely held belief that the gut microbiota plays a role in the development and progression of the disease in genetically susceptible individuals. Through the data presented here, we provide further evidence that supports this hypothesis.
Genetic association studies implicate Th17 cells in IBD, and these cells are enriched in tissue of individuals with IBD (Fujino et al., 2003; Jostinset al., 2012). In small clinical studies, RORγt+ Treg cells are described in several human tissues (Voo et al., 2009), but both RORγt+FoxP3+ and IL-17+FoxP3+ cells are particularly abundant in gut (Hovhannisyan et al., 2011; Han et al., 2014; Sefik et al., 2015). No specific correlation between RORγt+ Treg cells and IBD has been made, but our observations in this manuscript suggest that a well-powered clinical study is warranted to assess the role microbiota-induced RORγt+ Treg cells may play in the pathobiology of IBD. The mechanisms that drive divergent induction of Th17 cells or RORγt+ Treg cells in mice colonized with different microbiotas are unclear. These cell populations share many common features, including a requirement for IL-6, IL-23, and Stat3 signaling (Ohnmacht et al., 2015). It is therefore intriguing to consider what might drive induction of one subset over the other. Dietary retinoic acid (RA) can boost RORγt+ Treg cell induction (Ohnmacht etal., 2015) but can also modulate microbiota composition, making it hard to determine direct verses indirect effects (Cha et al., 2010). Th17 cells can be induced in gut tissue after adhesion of microbes to the epithelium (Atarashi et al., 2015; Sano et al., 2015). This is particularly notable as adhesive/invasive strains of E. coli (AIEC) have been found enriched in the mucosa of individuals with IBD (Darfeuille-Michaud et al., 2004) and at least some AIEC are able to induce Th17 cells (Viladomiu et al., 2017). The Th17 cell-inducing microbiotas identified in this study may harbor strains that induce Th17 cells through adhesion to the epithelium. Further studies are clearly warranted to determine the mechanisms by which healthy and IBD microbiotas alter the immune landscape in the gut. In mice colonized with IBD microbiotas, we also observed significantly more GATA3+ Th2 cells as compared to mice colonized with healthy donor microbiotas. Although Th2 cell-biased immune responses are typically associated with intestinal homeostasis and tissue repair (Allen and Sutherland 2014), elevated Th2 cytokines have been observed in mucosal tissue from individuals with ulcerative colitis, and certain haptan-induced mouse models of colitis are Th2 cell dependent (Heller et al., 2002). Th2 cells may be specifically sensitive to regulation by RORγt+ Treg cells (Ohnmacht et al., 2015). It is possible that dysregulation of RORγt+ Treg cells in IBD microbiota-colonized mice contributes to the greater proportion of Th2 cells observed in the gut tissue of these animals.
In B6 mice colonized with IBD microbiotas, the reduced proportion of RORγt+ Treg cells was balanced by an increased proportion of Helios+ Treg cells and a smaller population RORγt−Helios− Treg cells. Although evidence points to the RORγt+ Treg cell population as the major regulator of tolerance to microbiota (Ohnmacht et al., 2015; Sefik et al., 2015), under conditions where this population is deficient, it is possible that the RORγt− Treg cell populations compensate to maintain homeostasis. There may be a specific role for gut GATA3+ Treg cells during inflammation (Wohlfert et al., 2011), and an analysis of the induction and function of these cells in colitic animals is warranted. Although we found that an increased proportion of RORγt+ Treg cells correlated with reduced DC activation, we did not find a correlation of RORγt+ Treg cells with any specific T helper cell subset that is generalizable across all microbiotas that could explain how RORγt+ Treg cells may protect from colitis. In SPF conditions, mice lacking RORγt+ Treg cells show a specific expansion of Th2 cells in one study (Ohnmacht et al., 2015) and expanded Th1 and Th17 cells in another (Sefik et al., 2015), demonstrating the varied roles RORγt+ Treg cells may play in regulating T cell responses.
Tracking genetic and environmental differences between populations with and without disease has been an essential scientific paradigm to attribute individual traits or exposures to a disease. Numerous population-scale host genetics initiatives have led to the identification of genetic variants that explain 18%–26% of IBD susceptibility risk (Jostins et al., 2012; Liu et al., 2015)—a critical advance in our knowledge of IBD that still leaves the majority of risk unexplained. Across two cohorts of individuals, we describe immunomodulatory capabilities that are found extensively and specifically in the gut microbiota of individuals with IBD compared to healthy control subjects. Thus, we can ascribe a critical role for microbiome in the etiology of IBD and provide an immunological basis underlying this inference. Furthermore, it offers a mechanistic hypothesis explaining gut microbiota as a risk factor for IBD, whereby individuals harboring communities that enrich tolerogenic RORγt+ Treg cells are at reduced risk, while those harboring communities that enrich Th17 cells are at increased risk.
The lack of differences between healthy and IBD donor gut microbiome composition and diversity suggest that the functional impact of the microbiota lies in the strain-level composition of each unique community or in the unique combination of strains in each individual. The consistency in data from mice colonized with cultured and complete microbiotas from donors with IBD provide the prospect of determining the relative contribution of individual isolated strains to specific host phenotypes (Faith et al., 2014). Finally, these data further support the microbiota as a viable target for therapeutic intervention in IBD and provide a hypothesis for rational design of microbiota-directed preventative strategies and treatment in IBD.
STAR✰METHODS
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antbodies | ||
| Ant-mouse CD4 (GK1.5) APC | Biolegend | Cat#: 100412; RRID: AB_312697 |
| Ant-mouse CD4 (GK1.5) PerCP-Cy5.5 | Biolegend | Cat#: 100432; RRID: AB_893323 |
| Ant-mouse IL-17A (TC11–18H10.1) PE | Biolegend | Cat#: 506904; RRID: AB_315464 |
| Ant-mouse IFNγ (XMG1.2) PE-Cy7 | Biolegend | Cat#: 505826; RRID: AB_2295770 |
| Ant-mouse IL-10 (JESE-16E3) BrilliantViolet421 | Biolegend | Cat#: 505022; RRID: AB_3563240 |
| Ant-mouse RORγt (Q31–378) PerCP-Cy5.5 | BD Biosciences | Cat#: 562683; RRID: AB_2737720 |
| Ant-mouse FoxP3 (FJK-16 s) PE | eBioscience | Cat#: 12–5773-80; RRID: AB_465935 |
| Ant-mouse GATA3 (16E10A23) BrilliantViolet421 | BD Biosciences | Cat#: 560405; RRID: AB_1645544 |
| Ant-mouse CD45RB (C363–16A) FITC | Biolegend | Cat#: 103306; RRID: AB_313013 |
| Ant-mouse CD25(PC61) PE | Biolegend | Cat#: 102008; RRID: AB_312857 |
| Ant-mouse Helios (22F6) FITC | Biolegend | Cat#: 137214; RRID: AB_10662745 |
| Ant-mouse I-A/I-E (M5/114.15.2) Pacific Blue | Biolegend | Cat#: 107620; RRID: AB_493527 |
| Ant-mouse CD11c (N418) PE-Cy7 | Biolegend | Cat#: 117318; RRID: AB_493568 |
| Ant-mouse CD11b (M1/70) PerCP-Cy5.5 | Biolegend | Cat#: 101230; RRID: AB_2129374 |
| Ant-mouse CD64 (X54–5/7.1) PE | Biolegend | Cat#: 139304; RRID: AB_10612740 |
| Ant-mouse CD86(GL-1) FITC | Biolegend | Cat#: 105006; RRID: AB_313149 |
| Ant-mouse CD80 (16–10A1) APC | Biolegend | Cat#: 104714; RRID: AB_313135 |
| Ant-mouse CD3 (17A2) BrilliantViolet421 | Biolegend | Cat#: 100228; RRID: AB_2562553 |
| Ant-mouse CD19 (6D5) BrilliantViolet421 | Biolegend | Cat#: 115538; RRID: AB_11203527 |
| Ant-mouse IL-22 (Poly5164) PE | Biolegend | Cat#: 366704; RRID: AB_2565568 |
| Ant-mouse IL-17A (TC11–18H10.1) PE-Cy7 | Biolegend | Cat#: 506922; RRID: AB_2125010 |
| Ant-mouse NKp46 (29A1.4) APC | Biolegend | Cat#: 137608; RRID: AB_10612758 |
| Ant-mouse Csf2 (MP1–22E9) FITC | Biolegend | Cat#: 505404; RRID: AB_315380 |
| Biological Samples | ||
| 32 human stool samples | Jacobs et.al. 2016 and this study | N/A |
| 17 arrayed culture collections of human microbiotas | This study | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| DNase 1 | Sigma Aldrich | Cat#: DN25 |
| Collagenase | Sigma Aldrich | Cat#: C5138 |
| EDTA | Ambion | Cat# AM9260G |
| HEPES | Caisson | Cat#: HOL06 |
| Fetal bovine serum | Thermo Fisher Scientific | Cat#: 10438026 |
| ACK Lysis Buffer | GIBCO | Cat#: A1049201 |
| Zombie Aqua Fixable Viability Dye | Biolegend | Cat#: 423102 |
| Phorbal 12-myristate 13-acetate | Sigma Aldrich | Cat#: P8139 |
| lonomycin | Sigma Aldrich | Cat#: I3909 |
| Monensin | Biolegend | Cat#: 420701 |
| Brefeldin A | Biolegend | Cat#: 420601 |
| Critical Commercial Assays | ||
| Mouse Lipocalin-2/NGAL Duoset ELISA | R&D Systems | Cat#: DY1857 |
| FoxP3/Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat#: 00–5523-00 |
| Intracellular Fixation and Permeabilization Buffer Set | Thermo Fisher Scientific | Cat#: 88–8824-00 |
| MagniSort Mouse CD4 T cell Enrichment Kit | Thermo Fisher Scientific | Cat#: 8804–6821-74 |
| QIAquick 96 PCR Purification Kit | QIAGEN | Cat# 28181 |
| Quant-IT dsDNA Assay Kit - High Sensitivity | Thermo Fisher Scientific | Cat# Q33130 |
| Quant-IT dsDNA Assay Kit - Broad Range | Thermo Fisher Scientific | Cat# Q32853 |
| Deposited Data | ||
| 16S rDNA amplicon sequencing | Contijoch et al. (2018) and this manuscript | NCBI: PRJNA436992 |
| 16S rDNA amplicon sequencing | Contijoch et al. (2018) and this manuscript | NCBI: PRJNA413199 |
| Greengenes reference database version 13_8 | DeSantis et al., 2006 | http://greengenes.lbl.gov |
| Experimental Models: Organisms/Strains | ||
| Germ free C57BL/6J | Mount Sinai Gnotobiotic Facility | N/A |
| Germ free B6.129S7-Rag1tm1Mom/J | Mount Sinai Gnotobiotic Facility | N/A |
| Specific Pathogen Free C57BL/6J | Jackson Labs | MGI:5657312 |
| Specific Pathogen Free C57BL/6NTac | Taconic Farms | MGI:5658006 |
| Oligonucleotides | ||
| 16S V4 (515–806) F 5’-GTGCCAGCAGCCGCGGTAA-3’ | IDT (Relman et al., 1992) | N/A |
| 16S V4 (515–806) R 5’-GGACTACCAGGGTATCTAAT-3’ | IDT (Relman et al., 1992) | N/A |
| Software and Algorithms | ||
| R Studio | R Studio | N/A |
| FlowJo X | Tree Star | N/A |
| MacQIIME | Caporaso et al., 2010 | N/A |
| FACSDiva | BD Biosciences | N/A |
| Phyloseq | McMurdie and Holmes, 2013 | N/A |
| Prism 6 | GraphPad | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeremiah Faith (jeremiah.faith@mssm.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Detail of the human microbiota donors can be found in Table 1. All fecal samples were obtained from a stool biobank from two prior IRB approved clinical studies (Jacobs et al., 2016, Contijoch et al., 2018). All subjects were given a study identification number that all their study samples were labeled with. All study samples were processed with no identifiers linked to them other than their study ID. Germ free C57BL/6J and Rag1-deficient C57BL/6J (B6.129S7-Rag1tm1Mom/J) mice were bred in-house at the Mount Sinai Immunology Institute Gnotobiotic Facility. Mice were colonized at 4–6 weeks old and flow cytometry analysis or T cell transfer was performed 8–12 weeks old. In total, 74 female and 79 male gnotobiotic C57BL/6 mice and 120 female and 147 male Rag1–/– mice were used in this study. Specific pathogen free C57BL/6J mice from Jackson Labs were used as T cell donors at 8–10 weeks old. All phenotyping experiments in B6 mice included both male and female mice. Of the 30 microbiotas screened in the Rag1-deficient colitis model, 26 were tested in both male and female mice. All mouse experiments were approved by the Mount Sinai Institutional Animal Care and Use Committee.
METHOD DETAILS
Human samples and bacterial culture
Human stool samples from were frozen at –80°C before processing. Samples were pulverized under liquid nitrogen. Under strict anaerobic conditions ~500mg of pulverized stool from each donor was blended into a slurry (40–50mg/mL) in pre-reduced bacterial culture media (LYBHIv4 media (Sokol et al., 2008); 37 g/l Brain Heart Infusion [BD], 5g/l yeast extract [BD], 1 g/l each of D-xylose, D-fructose, D-glactose, cellubiose, maltose, sucrose, 0.5 g/l N-acetylglucosamine, 0.5 g/l L-arabinose, 0.5 g/l L-cysteine, 1g/l malic acid, 2 g/l sodium sulfate, 0.05% Tween 80, 20 μg/mL menadione, 5 mg/l hemin (as histidine-hemitin), 0.1 M MOPS, pH 7.2). The slurries were passed through sterile 100 μm strainers to remove large debris. To store for later administration to mice, slurries were diluted 1:20 in LYBHIv4 media containing 15% glycerol (final concentration) and stored at –80°C.
Arrayed culture collections were generated for selected donors as previously described. Briefly, clarified and diluted donor stool was plated onto a variety of solid selective and non-selective media under anaerobic, micro-aerophilic and aerobic conditions. Plates were incubated for 48–72 hours at 37°C. 384 single colonies from each donor microbiota were individually picked and regrown in liquid LYBHIv4 media for 48 hours under anaerobic conditions. Regrown isolates were identified at the species level using a combination of MALDI-TOF mass spectrometry (Bruker Biotyper) and 16S rDNA amplicon sequencing. An average of 16 unique species were isolated from each fecal sample (range 10–29; Supplemental Data Table 1). There was no significant difference in the number of unique isolates obtained from healthy donor and IBD donors (p = 0.19, Mann-Whitney). All regrown isolates were stored in LYBHIv4 with 15% glycerol at –80°C as a pooled cocktail for administration to mice.
Gnotobiotic mice
Germ free C57BL/6J and C57BL/6J Rag1-deficient (B6.129S7-Rag1tm1Mom/J) mice were bred in-house at the Mount Sinai Immunology Institute Gnotobiotic Facility in flexible vinyl isolators. To facilitate high-throughput studies in gnotobiotic mice we utilized “out-of-the-isolator” gnotobiotic techniques (Faith et al., 2014). Shortly after weaning (28–42 days old) and under strict aseptic conditions, germ-free mice were transferred to autoclaved filter-top cages outside the of the breeding isolator and colonized with human microbiotas; Mice were colonized with 200–300 μL of a fecal slurry or pooled cocktail of cultured strains by oral gavage, given only once. Alternatively, mice were colonized with a mouse specific pathogen free microbiota, with or without segmented filamentous bacteria, from the cecal contents of C57BL/6J or C57BL/6NTac mice from Jackson Labs orTaconic farms respectively. All experiments were performed at least 28 days after colonization.
16S rDNA sequencing and analysis
The composition of human fecal samples was analyzed by 16S rRNA gene amplicon sequencing as previously described (Faith et al., 2013, Reyes et al., 2013) and in Contijoch et al. BioRxiv, 2018. DNA was extracted by bead-beating followed by QiaQuick columns (QIAGEN) and quantified by Qubit assay (Life Technologies). The V4 region of the 16S gene was amplified by PCR and paired-end 250bp reads sequenced on an Ilumina MiSeq. Analysis was performed with MacQIIME 1.9.1.8 (Caporaso et al., 2010) and using open source R packages. OTUs were picked with 97% sequence similarity. OTUs were aligned to the Greengenes reference set, requiring 150bp minimum sequence length and 75% ID.
Lymphocyte isolation
Spleen and mesenteric lymph nodes were collected into RPMI containing 5% fetal bovine serum (FBS). Single cell suspensions were obtained by pressing though 40 μm strainers. Red blood cells were removed with ACK Lysing Buffer (GIBCO). Gut tissues were separated, opened longitudinally and washed in Hanks Buffered salt solution (HBSS) to remove intestinal contents. Ileum was defined as the distal 1/3 of the small intestine. Peyers patches were removed. Epithelial cells were removed by gentle shaking in HBSS (Ca/Mg free) with 5 mM EDTA, 15 mM HEPEs and 5% FBS for 30 min. The remaining tissue was washed in HBSS before mincing with scissors into digestion buffer (HBSS, 2% FBS, 0.4mg/mLCollagenaseType IV [Sigma Aldrich C5138], 0.1–0.25 mg/mL DNase1 [Sigma Aldrich DN25]) and incubated at 37°C with gentle shaking for 30–40 min. The resulting suspensions were passed sequentially though 100 μm and 40 μm strainers. No gradient centrifugation enrichment of lymphocytes was performed, except for in preparation of the data in Figures S2E and S2G where mononuclear lymphocytes were enriched in a discontinuous gradient of 40%–80% Percoll.
Flow cytometry
For analysis of intracellular T cell cytokines (IL-10, IFNγ, IL-17A, IL-22), lamina propria lymphocytes were restimulated in complete RPMI with 5 ng/mL phorbal 12-myristate 13-acetate (PMA) and 500 ng/mL ionomycin in the presence of monensin (Biolegend) for 3.5 hours at 37°C. For analysis of intracellular ILC cytokines, lamina propria lymphocytes were incubated at 37°C for 4 hours with 5 mg/mL Brefeldin A (Biolegend) without additional restimulation. Dead cells were excluded from all analyses using Zombie Aqua Fixable Viability dye (Biolegend). For intracellular cytokine staining, cells were fixed with IC Fixation Buffer (eBioscience) and transcription factors were detected in unstimulated cells fixed with FoxP3 Fixation/Permeabilization buffers (eBioscience). Simultaneous detection of cytokines and transcription factors was achieved by sequential staining; first of cytokines using IC Fixation buffer followed by transcription factors in Fixation/Permeabilization buffers. All data was acquired on the same LSRII instrument (BD Biosciences), with the exception of the data in Figure S3B that was acquired on a FACSAriaII (BD Biosciences), and analyzed using FlowJoX (TreeStar).
T cell transfer colitis
T cell transfer colitis experiments were performed as previously described (Llewellyn et al., 2018). Briefly, naive (CD45RBHI, CD25-) CD4 T cells were isolated from the spleen and subcutaneous lymphnodes of 7–9 week old specific pathogen free C57BL/6J mice (The Jackson Laboratory). Following tissue dissociation and red blood cell lysis CD4+ T cells were enriched using negative magnetic selection (Magnisort, eBioscience). The resulting cells were stained for expression of CD4, CD25 and CD45RB. A fraction representing ~50% of the total CD4+ population, selected on the basis of absent CD25 staining and high CD45RB staining was sorted using a FACSAria (BD Biosciences). Purity of the sorted fraction was checked and routinely exceeded 98%. Sorted cells were washed multiple times with sterile PBS. Rag1–/– mice received 1×106CD45RBHIT cells in 200 μL of sterile PBS by intraperitoneal injection. Donor cells were sex-matched to recipients. Mice colonized with healthy and IBD donor microbiotas had no significant difference in initial body mass before T cell transfer (22.36 ± 3.8 versus 22.95 ± 3.8 g, p = 0.2; t test).
Mice were weighed and fecal pellets were collected at the time of T cell transfer and weekly thereafter. Any mouse experiencing > 80% loss in body weight or which was deemed otherwise moribund was euthanized. In these cases, the last measurements of body mass or LCN2 taken for that mouse were carried forward and included in the data for subsequent time points. Inter-experimental variation was assessed across the screen by including one cultured donor microbiota in each experiment. This donor (UC1024) induced highly reproducible colitis in many repeats over ~2 years (Figure S4A). We set pre-determined exclusion criteria for any experiment where mice colonized with UC1024 did not develop colitis in this reproducible manner.
Histology
Histology was performed by HistoWiz Inc (https://home.histowiz.com). Tissue was fixed in 10% buffered formalin, embedded in paraffin and 4 mm sections were cut before staining with hematoxylin and eosin. Slides were scanned with an Aperio AT2 (Leica).
LCN2 measurements
Lipocalin2 concentrations were measured in feces as a biomarker of intestinal inflammation (Chassaing et al., 2012). Fecal pellets were collected into sterile pre-weighed and barcoded tubes and frozen at −20°C until the time of analysis. Pellets were weighed and suspended in 500 μL of sterile PBS by shaking in a BeadBeater (with no beads in the tube) for 2 min. Tubes were centrifuged at 4000rpm for 20 min. The resulting supernatant was assayed for LCN2 by sandwich ELISA (R&D systems). The concentration of LCN2 was normalized to the weight of the input feces.
DATA AND SOFTWARE AVAILABILITY
16S rDNA datasets analyzed in the manuscript are available through NCBI under accession numbers PRJNA436992 and PRJNA413199. Specific BioSample accession numbers are listed in Table 1.
Supplementary Material
Highlights.
Fecal microbiotas from humans with IBD alter gut CD4+ T cell homeostasis in mice
Microbiotas from individuals with IBD induce more Th2 and Th17 cells
Microbiotas from healthy individuals induce more RORgt+ Treg cells
In a model of colitis, mice colonized with IBD microbiotas get more severe disease
ACKNOWLEDGMENTS
We thank C. Fermin, E. Vazquez, and G.N. Escano of the Mount Sinai Immunology Institute Gnotobiotic Facility for technical support. This work was supported by grants from the NIH (NIGMS GM108505 and NCCIH AT008661), Janssen Human Microbiome Institute, CCFA Microbiome Innovation Award (362048), and the New York Crohn’s Foundation to J.J.F., NIH DK108487 to S.R.L., NIH DK112679 to E.J.C., and NIH DK085691, CA016042, and UL1TR000124 to J.B. G.J.B. is supported by a Research Fellowship Award from the Crohn’s and Colitis Foundation of America. Next-generation sequencing was performed at NYU School of Medicine bythe Genome Technology Center partially supported by the Cancer Center Support Grant, P30CA016087. This work was supported in part by the staff and resources of Scientific Computing and of the Flow Cytometry Core at the Icahn School of Medicine at Mount Sinai.
Footnotes
DECLARATION OF INTERESTS
D.G. and A.D. are employees of Janssen Research & Development LLC. J.B. is on the scientific advisory boards of Prolacta Bioscience, Inc. and Janssen Research & Development LLC. J.J.F. is on the scientific advisory board of Vedanta and is a consultant for Janssen Research & Development LLC.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figuresand onetableand can befound with this article online at https://doi.org/10.10167j.immuni.2018.12.015.
REFERENCES
- Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, and Powrie F (2010). Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JE, and Sutherland TE (2014). Host protective roles of type 2 immunity: parasite killing and tissue repair, flip sides of the same coin. Semin. Immunol. 26, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. (2017). Ectopic colonization oforal bacteria inthe intestine drivesTH 1 cell induction and inflammation. Science 358, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, et al. (2016). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK, Hauser SL, et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. USA 114, 10713–10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha HR, Chang SY, Chang JH, Kim JO, Yang JY, Kim CH, and Kweon MN (2010). Downregulation of Th17 cells in the small intestine by disruption of gut flora in the absence of retinoic acid. J. Immunol. 184, 6799–6806. [DOI] [PubMed] [Google Scholar]
- Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, and Vijay-Kumar M (2012). Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS ONE 7, e44328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudnovskiy A, Mortha A, Kana V, Kennard A, Ramirez JD, Rahman A, Remark R, Mogno I, Ng R, Gnjatic S, et al. (2016). Host-protozoan interactions protect from mucosal infections through activation of the inflam- masome. Cell 167, 444–456.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contijoch E, Britton GJ, Yang C, Mogno I, Li Z, Ng R, Llewellyn SR, Hira S, Johnson C, Rabinowitz KM, et al. (2018). Gut microbiota density influences host physiology and is shaped by host and microbial factors. bioRxiv. https://doi.org/10.110½77095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, and Colombel JF (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421. [DOI] [PubMed] [Google Scholar]
- De Palma G, Lynch MD, Lu J, Dang VT, Deng Y, Jury J, Umeh G, Miranda PM, Pigrau Pastor M., Sidani S, et al. (2017). Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci. Transl. Med. 9, eaaf6397. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, and Andersen GL (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Hudcovic T, Mrazek J, Kozakova H, Srutkova D, Schwarzer M, Tlaskalova-Hogenova H, Kostovcik M, and Kverka M (2015). Development of gut inflammation in mice colonized with mucosa-associated bacteria from patients with ulcerative colitis. Gut Pathog. 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun CS, Mishima Y, Wohlgemuth S, Liu B, Bower M, Carroll IM, and Sartor RB (2014). Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10−/− mice. Infect. Immun. 82, 2239–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. (2013). The long-term stability of the human gut microbiota. Science 341, 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, and Gordon JI (2014). Identifying gut microbehost phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6, 220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Amand St A.L., Feldman RA, Boedeker EC, Harpaz N, and Pace NR (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104, 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, and Fujiyama Y (2003). Increased expression of interleukin 17 in inflammatory bowel disease. Gut 52, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, and Macpherson AJ (2011). Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34, 794–806. [DOI] [PubMed] [Google Scholar]
- Geva-Zatorsky N, Sefik E, Kua L, Pasman L, Tan TG, Ortiz-Lopez A, Yanortsang TB, Yang L, Jupp R, Mathis D, et al. (2017). Mining the human gut microbiota for immunomodulatory organisms. Cell 168,928–943.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, and Gordon JI (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 108, 6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Glanville J, Hansmann L, and Davis MM (2014). Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 32, 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Gulati A, and Sartor RB (2010). The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr. Opin. Gastroenterol. 26, 564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy AN, West NR, Stubbington MJT, Wendt E, Suijker KIM, Datsi A, This S, Danne C, Campion S, Duncan SH, et al. ; Oxford IBD Cohort Investigators (2017). Circulating and tissue-resident CD4(+) T cells with reactivity to intestinal microbiota are abundant in healthy individuals and function is altered during inflammation. Gastroenterology 153, 1320–1337.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, and Strober W (2002). Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity 17, 629–638. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan Z, Treatman J, Littman DR, and Mayer L (2011). Characterization of interleukin-17-producing regulatory T cells in inflamed intestinal mucosa from patients with inflammatory bowel diseases. Gastroenterology 140, 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, and Littman DR (2006). The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde.L., Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, and Littman DR. (2008). Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4, 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JP, Goudarzi M, Singh N, Tong M, McHardy IH, Ruegger P, Asadourian M, Moon BH, Ayson A, Borneman J, et al. (2016). A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell Mol Gastroenterol Hepatol 2, 750–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. ; International IBD Genetics Consortium (IIBDGC) (2012). Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor B, Gardet A, and Xavier RJ (2011). Genetics and pathogenesis of inflammatory bowel disease. Nature 474, 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, and Gevers D (2014). The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium (2015). Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 47, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn SR, Britton GJ, Contijoch EJ, Vennaro OH, Mortha A, Colombel JF, Grinspan A, Clemente JC, Merad M, and Faith JJ (2018). Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology 154,1037–1046.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, and Holmes S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, Onischi C, Armstrong D, Marshall JK, Kassam Z, Reinisch W, and Lee CH (2015). Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6. [DOI] [PubMed] [Google Scholar]
- Moran JP, Walter J, Tannock GW, Tonkonogy SL, and Sartor RB (2009). Bifidobacterium animal is causes extensive duodenitis and mild colonic inflammation in monoassociated interleukin-10-deficient mice. Inflamm. Bowel Dis. 15, 1022–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, and Merad M (2014). Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 343, 1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao-Kitamoto H, Shreiner AB, Gillilland MG 3rd, Kitamoto S, Ishii C, Hirayama A, Kuffa P, El-Zaatari M, Grasberger H, Seekatz AM, et al. (2016). Functional characterization of inflammatory bowel disease-associated gut dysbiosis in gnotobiotic mice. Cell Mol Gastroenterol Hepatol 2, 468–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad JM, Pinto-Sanchez MI, Galipeau HJ, Jury J, Jordana M, Reinisch W, Collins SM, Bercik P, Surette MG, Allen-Vercoe E, and Verdu EF (2015). Ecobiotherapy rich in firmicutes decreases susceptibility to colitis in a humanized gnotobiotic mouse model. Inflamm. Bowel Dis. 21, 1883–1893. [DOI] [PubMed] [Google Scholar]
- Ng THS, Britton GJ, Hill EV, Verhagen J, Burton BR, and Wraith DC (2013). Regulation of adaptive immunity; the role of interleukin-10. Front. Immunol. 4, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Wu GD, Albenberg L, and Tomov VT (2017). Gut microbiota and IBD: causation orcorrelation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, et al. (2015). MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORγt+T cells. Science 349, 989–993. [DOI] [PubMed] [Google Scholar]
- Omenetti S, and Pizarro TT (2015). The Treg/Th17 axis: A dynamic balance regulated by the gut microbiome. Front. Immunol. 6, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östman S, Rask C, Wold AE, Hultkrantz S, and Telemo E (2006). Impaired regulatory T cell function in germ-free mice. Eur. J. Immunol. 36, 2336–2346. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramsothy S, Kamm MA, Kaakoush NO, Walsh AJ, van den Bogaerde J, Samuel D, Leong RWL, Connor S, Ng W, Paramsothy R, et al. (2017). Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet 389, 1218–1228. [DOI] [PubMed] [Google Scholar]
- Powrie F, Leach MW, Mauze S, Caddle LB, and Coffman RL (1993). Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 5, 1461–1471. [DOI] [PubMed] [Google Scholar]
- Relman DA, Schmidt TM, MacDermott RP, and Falkow S (1992). Identification of the uncultured bacillus of Whipple’s disease. N. Engl. J. Med. 327, 293–301. [DOI] [PubMed] [Google Scholar]
- Reyes A, Wu M, McNulty NP, Rohwer FL, and Gordon JI (2013). Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. USA 110, 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KJ, Wu S, Wu X, Huso DL, Karim B, Franco AA, Rabizadeh S, Golub JE, Mathews LE, Shin J, et al. (2009). Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect. Immun. 77, 1708–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, et al. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, Lee JY, Ziel JW, Miraldi ER, Domingos AI, et al. (2015). An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 163, 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor RB (2008). Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594. [DOI] [PubMed] [Google Scholar]
- Schiering C, Krausgruber T, Chomka A, Fröhlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, et al. (2014). The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. (2015). MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiotaanalysisofCrohn disease patients. Proc. Natl. Acad. Sci. USA 105, 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada O, et al. (2007). Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm. Bowel Dis. 13, 1202–1211. [DOI] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D,Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc. Natl. Acad. Sci. USA 113, E8141–E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlig HH, Coombes J, Mottet C, Izcue A, Thompson C, Fanger A, Tannapfel A, Fontenot JD, Ramsdell F, and Powrie F (2006). Characterization of Foxp3+CD4+CD25+ and IL-10-secreting CD4+CD25+ T cells during cure of colitis. J. Immunol. 177, 5852–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, et al. (2017). IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci. Transl. Med. 9, eaaf9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voo KS, Wang YH, Santori FR, Boggiano C, Wang YH, Arima K, Bover L, Hanabuchi S, Khalili J, Marinova E, et al. (2009). Identification of IL-17-producing FOXP3+ regulatory T cells in humans. Proc. Natl. Acad. Sci. USA 106, 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, et al. (2011). GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest. 121, 4503–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, and Littman DR (2018). c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 554, 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, et al. (2014). Focused specificity of intestinal TH17 cellstowards commensal bacterial antigens. Nature 510,152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BH, Hagemann S, Mamareli P, Lauer U, Hoffmann U, Beckstette M, Föhse L, Prinz I, Pezoldt J, Suerbaum S, et al. (2016). Foxp3(+) Tcells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 9, 444–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
16S rDNA datasets analyzed in the manuscript are available through NCBI under accession numbers PRJNA436992 and PRJNA413199. Specific BioSample accession numbers are listed in Table 1.


