Abstract
Surgery is the main treatment for renal cell carcinoma (RCC); nephron sparing surgery can be performed as a treatment of choice for small peripheral lesions. Epigenetics configures a new entity that regulates gene expression throughout methylation, acetylation and chromatin remodelling. In addition to silencing as a result of mutations, loss of heterozygosity, or classic genetic events, epigenetic modification symbolizes essential events during carcinogenesis and tumour development. We investigated global methylation and histone acetylation expression in a series of small conventional clear cell renal carcinomas (i.e. less than 5 cm) (pT1a) treated with partial nephrectomy, to assess their possible role as diagnostic biomarkers. A total of 54 patients with conventional single RCC were selected and treated with partial nephrectomy; they were followed up to 186 months. Immunohistochemistry was performed on paraffin‐embedded sections, using anti‐5‐methylcytosine (5mc) and anti‐Acetyl‐Histone H3 (Lys 9). Our results confirm that the mean percentage of global cellular methylation in tumoural tissue was significantly higher compared to healthy peritumoural tissue, whereas the mean percentage of histone cellular acetylation in tumoural tissue was significantly lower. The percentage of methylation was significantly higher in grades 3 and 4 (P= 0.033), whereas the percentage of histone acetylation was significantly lower (P= 0.023), suggesting therefore that these markers could correlate with tumour aggressiveness in pT1a RCC. On univariate analysis of patient survival in relation to the different considered factors, Fuhrman grade was the most important survival factor. These epigenetic markers can give us interesting information about chromatin remodelling in RCCs; the percentage of global methylation increases with increasing Fuhrman grade, whereas histone acetylation decreases with increasing grade in small RCC; our results suggest that global hypermethylation and histone hypoacetylation can be assumed to be an early event in RCC and to correlate with tumour aggressiveness.
Keywords: kidney, renal cell carcinoma, global DNA‐methylation, histone acetylation
Introduction
Renal cell carcinoma (RCC) accounts for 3% of all adult malignancies and it is the most lethal of the urological cancers [1]. Approximately 20–30% of patients present with metastatic disease and in 20–40% of patients undergoing nephrectomy for clinically localized RCC metastases develop [2]. During the last 2 decades significant advances in the diagnosis, staging and treatment of patients with RCC have resulted in improved survival of a select group of patients and an overall change in the natural history of the disease. However, despite advances in biological and immune based therapies response rates in patients with metastatic RCC remain at 15–25%[3].
Surgery is the main treatment for RCC, offering a reasonable chance of curing the disease. Radical nephrectomy is usually recommended, whereas nephron sparing surgery can be performed as a treatment of choice for small peripheral lesions [4]; the aim is to preserve maximum renal function while performing a complete cancer surgical ablation. Moreover, some recent studies confirmed the favourable long‐term outcome of nephron sparing surgery in patients with localized RCC and no significant differences in outcome have been observed in patients with T1 RCC and normal contralateral kidney treated with partial or radical nephrectomy [5].
Several anatomical, histological and clinical characteristics, and a lot of emerging tumour markers have been shown to be independent prognostic indicators of disease progression and survival for RCC [4]; however until recently, there were no molecular markers of significant utility, probably in part because of a lack of knowledge regarding the biology of carcinogenesis and progression at the molecular level [6, 7].
Epigenetics, a combination of DNA modifications, chromatin organization and variations in its associated proteins, configures a new entity that regulates gene expression throughout methylation, acetylation and chromatin remodelling. In addition to silencing as a result of mutation, loss of heterozygosity, or classic genetic events, epigenetic modification symbolizes essential events during carcinogenesis and tumour development [8, 9, 10, 11].
There is growing evidence that interplay exists between DNA methylation and histone modifications; it has been shown that DNA hypermethylation, either directly or indirectly, through suppressing transcription, specifies for repressive histone modification at tumour suppressor gene promotion sites and that patterns of DNA and histone methylation are similar in tissue across the spectrum of carcinogenesis; furthermore, there is a significant positive association between these two epigenetic mechanisms [12, 13, 14, 15].
Histone acetylation is another important epigenetic event involved in chromatin remodelling. Histone proteins are the fundamental building blocks of eukaryotic chromatin. Two copies of histones H2A, H2B, H3 and H4, assembled in an octameric core with 146–147 bp of DNA tightly wrapped around it, form a nucleosome. This is the first level of chromatin organization. Core histones are characterized by the presence of N‐terminal tails of variable length that are reversibly modified by acetylation of lysines and methylation of lysines and argynines among others [16, 17, 18, 19, 20].
We investigated by immunohistochemistry global methylation and histone acetylation in a series of small conventional clear cell renal carcinomas, treated with partial nephrectomy, to assess their possible role as diagnostic biomarkers. At the best of our knowledge, this is the first immunohistochemical study attempting to demonstrate the overall epigenetic status in RCC.
Material and methods
Patients
A total of 54 patients with conventional single RCC were selected from those who underwent surgery at our Institute of Urology between 1987 and 2000. All patients were treated with partial nephrectomy.
All patients underwent preoperative evaluation, including history and physical examination, urinalysis, blood count and serum chemistry. Preoperative imaging consisted of ultrasound in 100% of cases, excretory urography in 78%, chest and abdominal computerized tomography (CT) in 93%, chest and abdominal magnetic resonance imaging in 17% and renal diaminetetraminepentaacetic acid scan in 92%. In 42 patients contralateral kidney function was normal, whereas in 6 chronic renal failure was present (4) or impending (2). The discriminating factor in favour of conservative surgery was a small renal mass (less than 5 cm, that is pT1a tumours) [21].
The technical principles of partial nephrectomy included resection of the tumour and overlying perinephric fat with at least 0.5 cm margin of normal kidney parenchyma. Intraoperative histological examination was always performed to assess adequate surgical margins. Temporary renal artery and vein occlusion, and surface hypothermia were done.
Follow‐up examination performed at 3‐month intervals in year 1 included urinalysis, serum creatinine determination and abdominal ultrasound. During the following 3 years, abdominal ultrasound was performed every 6 months, and chest and abdominal CT, and bone scan were performed every 12 months for the first 5 years. At year 6 after surgery and thereafter patients underwent yearly follow‐up with serum creatinine determination, abdominal ultrasound and general examination.
Archived materials containing histological sections from our 54 patients were retrieved from the Institute of Pathological Anatomy and used for immunostaining. The features considered when evaluating patients were patient age, tumour size and grade, and methylation and acetylation on the histological specimens. All considered parameters were correlated with patient specific survival.
Tumour grade was based on the Fuhrman scheme [22]. Disease recurrence was defined as evidence of measurable disease on imaging, including CT, magnetic resonance, bone scan and ultrasound, and cytological/histological evaluation of suspected lesions.
Immunohistochemistry
Immunohistochemistry was performed on conventional 6 μm thick histological paraffin‐embedded tissue sections on poly‐L‐lysine‐coated glass slides. After heat‐drying, sections were deparaffinized in xylene and sequentially rehydratated in gradients of ethanol. To better unmask antigenic sites, sections were boiled in 10 nM citrate buffer pH 6.0 for 10 min. and placed in 3.5 N HCl for 30 min. at room temperature for 5‐methylcytosine and boiled in 10 nM citrate buffer pH 6.0 for 20 min. for Acetyl‐Histone H3 (Lys 9). After washes in H2O and in Tris‐HCl buffer solution pH 7.6, they were incubated with normal goat serum for 10 min. and then incubated overnight at 4°C with the following antibodies: monoclonal anti‐5‐methylcytosine (5mc) (dil. 1:200, Aviva System Biology, CA, USA), and polyclonal anti‐Acetyl‐Histone H3 (Lys 9) (AcH3K9) (dil. 1:100, Cell Signaling Technology, MA, USA). The reaction was revealed using the streptoavidin‐biotin‐peroxidase technique (Dako‐Envision Plus/HRP peroxidase kit, Dako SPA, Carpinteria, CA, USA). After incubation with 3.3 diaminobenzidine (0.05 diaminobenzidine in 0.05 M Tris buffer, pH 7.6 and 0.01% hydrogen peroxide) (Sigma‐Aldrich, Milano, Italy), sections were counterstained with Mayer’s Hematoxylin (Bio‐Optica SPA, Milano, Italy), coverslipped with Paramount and observed using a light microscope. Positive controls were represented by paraffin‐embedded sections, previously shown to react with primary antibodies, from human lung squamous cells carcinoma. For negative controls, primary antibodies were replaced with non‐immune sera.
Pathological evaluation of immunohistochemical results was performed independently by two observers (A.F., G.L.) for both anti‐5‐methylcytosine and anti‐Acetyl‐Histone H3 antibodies. The immunostaining for anti‐5‐methylcytosine and anti‐Acetyl‐Histone H3 was observed in the nuclei of both neoplastic and non‐neoplastic cells. The percentage of cells at each intensity was estimated and multiplied by the specific intensity score to obtain a weighted average of the intensity score, as previously reported [23]. Each observer graded the intensity of immunostaining on neoplastic cells using a 4‐point scale: 0 = no staining, 1 = mild, 2 = moderate, 3 = high. At least 1000 nuclei were evaluated in each case [1]. The final score reported is the average of the two observers scores.
All the sections of clear cell renal carcinoma had adjacent non‐neoplastic tissue made of proximal and distal tubules for comparative evaluation carried out with the same approach used for cancer tissue.
Statistical analysis
Statistical analysis was performed using the Kolmogorov–Smirnov normality test for all considered parameters. The Mann–Whitney U test, appropriate for nonparametric data, was used to compare the staining intensity and the percentage of the staining cells at each intensity. Correlations between continuous variables were analysed using Spearman’s rank correlation test. The Fischer and chi‐square tests were used to compare nominal data. Kaplan–Meier curves with the log rank test were designed to compare survival parameters. The influence of each parameter on survival was assessed using the Cox proportional hazard models.
Results
Of the patients, 41 were male and 13 were female. Mean age ± S.D. was 58.2 ± 9.5 years (range 33–75). A total of 21 tumours were in the right kidney and 33 in the left kidney. Median diameter was 2.92 ± 0.82 cm (range 1.3–5). Histological grade was 1 to 4 in 17, 31, 3 and 3 patients, respectively.
Median follow‐up was 94.3 months (range 19–186). Four patients with a mean age of 58.7 years died of a disease other than renal cancer after a median follow‐up of 85.5 months (range 20–133). They were cancer‐free at death. Four patients with a mean age of 62.7 years died of metastatic renal cancer after a median follow‐up of 23.5 months (range 11–36). They had multiple metastases, as documented by fine needle aspiration cytology, i.e. in those with lung and liver lesions. None of these patients had local relapse. Of these patients, 1 had grade 2, 1 had grade 3 and 2 had grade 4 RCC.
The mean percentage of global cellular methylation in tumoural tissue (Fig. 1a) was significantly higher compared to healthy peritumoural tissue (Fig. 1b) (mean 77.21 versus 68.8%; IQR: 60–100%versus 50–90%; P= 0.005). The intensity of cellular methylation was not significantly different in tumoural and healthy tissues (P= 0.513). Furthermore, we could observe a positive correlation between methylation in tumoural and healthy tissue in the same patient (r= 0.541, P= 0.000) (Fig. 2a), i.e. to an increase of methylation in tumoural tissue corresponds an increase of methylation in healthy tissue.
Figure 1.
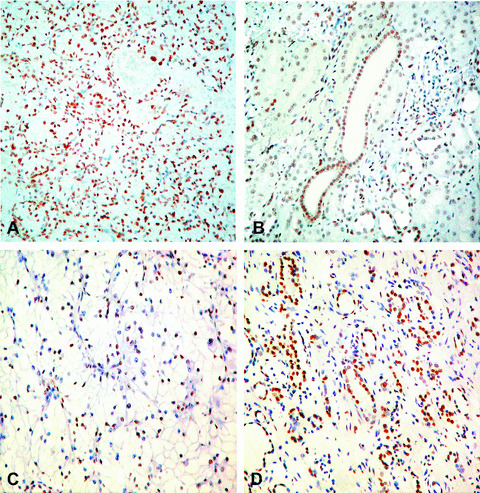
Immunohistochemical expression of 5‐methylcytosine (5mc) and Acetyl‐Histone H3 Lys9 (AcH3K9): 5mc staining in RCC (A) and healthy tissue (B); AcH3K9 staining in RCC (C) and healthy tissue (D) (immunoperoxidase, original magnification ×200).
Figure 2.
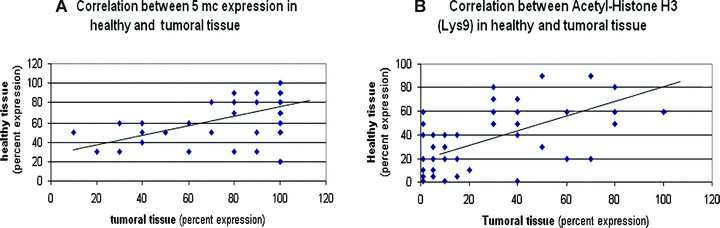
A positive correlation exists for methylation (A) and acetylation (B) between tumoural and healthy tissue in the single patient (Spearman’s rank correlation test; r= 0.541, P= 0.000 and r= 0.634, P= 0.000, respectively).
The mean percentage of histone cellular acetylation in tumoural tissue (Fig. 1c) was significantly lower compared to healthy peritumoural tissue (Fig. 1d) (mean 22.98%versus 30.42%; IQR: 0–40%versus 7.5–50%; P= 0.004). The intensity of mean acetylation was 1 (IQR: 0–2.75) in neoplastic tissue, whereas it was 2 (IQR: 1–3) in healthy tissue (P= 0.007). The percentage of acetylation in tumoural tissue was highly correlated with the intensity of acetylation expression (Rho: 0.856, P < 0.0001). Furthermore, we could observe a positive correlation between acetylation in tumoural and healthy tissue in the same patient (r= 0.634, P= 0.000) (Fig. 2b), i.e. to an increase of acetylation in tumoural tissue corresponds an increase of acetylation in healthy tissue.
We did not find any correlation between variation of global methylation and histone acetylation in tumoural and healthy tissue (r= 0.060, P= 0.0659 and r= 0.039, P= 0.780, respectively).
In Table 1, we divided the patients into three groups, i.e. those with grades 1, 2, and 3 and 4. The latter patients were grouped together because of the small number of those with high grade RCC (4); also grades 1 and 2 were grouped together for comparison purposes. No statistically significant differences were observed for age and tumour size among the three groups (P= 0.992 and 0.120, respectively). No significant correlations were observed between the expression of cellular methylation and acetylation and age, sex and tumour size; the percentage of methylation was significantly higher in grades 3 and 4 (P= 0.033), whereas the percentage of histone acetylation was significantly lower (P= 0.023).
Table 1.
Data and results
| Grade 1 | Grade 2 | (Grades 1–2)* | Grades 3–4 | P‐value | |
|---|---|---|---|---|---|
| Mean age ± S.D. | 59 ± 7.3 | 57.8 ± 10.9 | (58.4 + 8.9) | 57.5 ± 10.8 | 0.992 |
| Median cm size (IQR) | 2.7 (2.2–3.1) | 3 (2.7–3.5) | (2.85) | 2.3 (1.8–3.6) | 0—120 |
| 5‐methylcytosine expression in tumour tissue (%) | 76.25 ± 24.46 | 75.62 ± 27.56 | (75.93 ± 25.94) | 93.33 ± 11.55 (3 casi) | 0.033* |
| Acetyl‐Histone H3 (Lys 9) in tumour tissue (%) | 27.69 ± 26.43 | 18.36 ± 22.39 | (23.02 ± 24.5) | 35.05 ± 49.43 (2 casi) | 0.223* |
All values are expressed as mean ± standard deviation (S.D.). Statistical analysis was performed using the Kolmogorov–Smirnov normality test for clinical parameters and Mann–Whitney U test for immunohistochemical data, respectively.
*Grades 3 and 4 were grouped together because of the small number of high grade RCC; grades 1 and 2 were grouped together because there is recently a tendency to simplify grading in two groups.
On univariate analysis of patient survival in relation to the different considered factors, Fuhrman grade was the most important survival factor. In fact, our data show that patients with grade 1 renal cell cancer showed a 100% survival rate, patients with grade 2 showed a 89.7% survival rate, and patients with grade 3 and 4 showed a 50% survival rate (log rank P= 0.015, Fig. 3).
Figure 3.
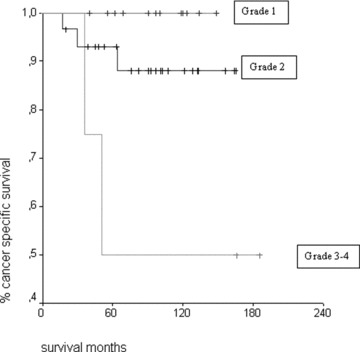
Kaplan–Meier patient survival curves for Fuhrman grades 1, 2 and 3 and 4 (log rank P= 0.015).
Discussion
The last decade has witnessed the gradual transition from the use of solitary clinical factors as prognostic markers for patients with RCC to the introduction of systems that integrate multiple factors to the beginning of the use of molecular and genetic markers [24]. Diagnostic markers can be identified by comparing gene and protein expression profiles from normal and malignant tissue [7].
The organization of nuclear chromatin changes according to the metabolic state of the cells, which might correlate with the prognosis of the individual patient [12]; therefore, studying the nuclear chromatin phenotype may be helpful in understanding the genetic, molecular and biological changes involved in cancer development and progression, because it is a sensitive indicator of cell development phenotype.
A link between DNA methylation and cancer was first demonstrated in 1983, when it was shown that the genomes of cancer cells are hypomethylated relative to their normal counterparts [13]. Loss of genomic methylation is a frequent event in cancer, and correlates with disease severity and metastatic potential in many tumour types [25, 26].
Methylation of DNA, which occurs at cytosine residues of CpG dinucleotides by an enzymatic reaction that produces 5‐methylcytosine (5‐mc), is an extensively characterized mechanism for epigenetic regulation. Neoplastic cells may simultaneously harbour widespread (global) genomic hypomethylation, regional areas of hypermethylation and increased DNA‐methyltransferase activity [23, 27]. The immunohistochemical evaluation of global DNA methylation by using an antibody specific to 5‐mc has advantages over radio‐labelled methyl incorporation assay, especially when methylation related to cancer cannot be normalized by methylation of normal tissue, or when the number of samples available for evaluation is small [28].
Histone modifications in the basic N‐terminal tails result in changes in the cell transcriptional state. The best studied of these modifications is histone acetylation. These dynamic post‐translation modifications are required for active chromatin configuration. In particular, acetylation of specific residues such as lysine 9 (K9) of histone 3 (H3K9) has been associated with an open chromatin configuration and a permissive transcription state. Histone H3 is primarily acetylated at lysine 9, 14, 18 and 23. Modification at lysine 9 is thought to play a dominant role in histone mobilization and transcriptional regulation. Genes that are methylated are usually related to deacetylated and inactive chromatin, whereas unmethylated promoters and active genes are associated with an open hyperacetylated euchromatin [8]. Methylation of a specific lysine residue (at positions 4 and 9 in the amino acid chain) in histone H3 has been shown to be involved in the regulation of chromatin structure [29]. Moreover, Fahrner et al.[30] showed that DNA hypermethylation, either directly or indirectly, through suppressing transcription, specifies for repressive histone modification at tumour suppressor gene promotion sites. More recently, Piyathilake et al.[13] found that patterns of DNA and histone methylation are similar in tissue across the spectrum of oral carcinogenesis, and there is a significant positive association between the two epigenetic mechanisms of acetylation and methylation.
Several authors have investigated the epigenetic mechanisms in clear cell RCCs: Christoph et al.[31] identified methylated APAF1 and DAPK1 genes as independent prognostic markers; Yamada et al.[32] evaluated methylation of the gene encoding DAL‐1/4.1B in 55 clear cell RCC and found that it correlated with metastatic recurrence and provided independent prognostic information, suggesting that methylation of the gene encoding DAL‐1/4.1B predicts recurrence in clear cell RCC.
By using gene array analysis in 177 primary clear cell RCCs, Zhao et al. [33] defined expression profiles that correlated with survival in long‐term follow‐up, independent of grade, stage and performance status; in this analysis, 259 genes predicted survival after surgery, independent of clinical prognostic factors.
Furge et al. [34], evaluating DNA copy numbers and signalling abnormalities, found that an inactivated Myc signalling pathway was associated with aggressive type 2 papillary RCC; regulation of the Myc signalling pathway may represent a therapeutic option in type 2 papillary RCC. Met oncogene is known to be involved in hereditary papillary RCC; it was strongly expressed in papillary RCC and urothelial carcinoma of the renal pelvis, whereas in clear cell RCC and chromophobe RCC Met expression was negative or only focally positive; marked expression of Met correlated with aggressive tumour behaviour in papillary RCCs [35].
In recent studies [12, 36], it has been observed that in PIN (prostatic intraepithelial neoplasia) lesions, there was a high chromatin content with DNA‐hypermethylation, whereas in prostatic adenocarcinoma there was a lower chromatin content with DNA‐hypomethylation; in another study [37] new histone deacetylase inhibitors have been suggested to have potential therapeutic role for advanced prostatic carcinoma.
At presentation, incidental renal tumours are of significantly lower stage and grade than tumours producing symptoms. These clinically and histologically less aggressive lesions lead to better patient survival and decreased recurrence.
In our immunohistochemical study, 54 patients with small RCCs were selected and closely followed for a maximum of more than 15 years. Our study shows that the percentage of global methylation is higher in tumoural tissue than in healthy tissue, whereas the percentage of histone acetylation is lower in tumoural tissue. We have also observed that a positive correlation exists, respectively, for methylation and acetylation between tumoural and normal tissue in the single patient, suggesting that these epigenetic mechanisms could happen early in healthy tissue and could be involved in RCCs tumourigenesis.
Interestingly, we have observed that the percentage of global methylation increases with increasing Fuhrman grade, whereas histone acetylation decreases with increasing grade, suggesting therefore that these markers could correlate with tumour aggressiveness.
In spite of these observations, we can conclude that at present Fuhrman grade is the most important factor for patient survival. We can also say that these epigenetic markers can give us interesting information about chromatin remodelling in RCCs and that global hypermethylation and histone hypoacetylation is an early event in RCCs. A further step will be the analysis of global methylation and histone acetylation in pT2–4 RCCs [21], to observe whether these data will be confirmed also in higher diameter renal tumours.
References
- 1. Jemal A, Tiwari RC, Murray T, et al . Cancer statistics 2004. CA Cancer J Clin. 2004; 54: 8–29. [DOI] [PubMed] [Google Scholar]
- 2. Janzen NK, Kim HL, Figlin RA, et al . Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol Clin North Am. 2003; 30: 843–52. [DOI] [PubMed] [Google Scholar]
- 3. Gitlitz BJ, Figlin RA. Cytokine based therapy for metastatic renal cell cancer. Urol Clin North Am. 2003; 30: 589–600. [DOI] [PubMed] [Google Scholar]
- 4. Minardi D, Lucarini G, Mazzucchelli R, et al . Prognostic role of Fuhrman grade and vascular endothelial growth factor in pT1a clear cell renal carcinoma in partial nephrectomy specimens. J Urol. 2005; 174: 1208–12. [DOI] [PubMed] [Google Scholar]
- 5. Leibovich BC, Blute ML, Cheville JC, et al . Nephron sparing surgery for appropriately selected renal cell carcinoma between 4 and 7 cm results in outcome similar to radical nephrectomy. J Urol. 2004; 171: 1066–70. [DOI] [PubMed] [Google Scholar]
- 6. Lam JS, Shvarts O, Leppert JT, et al . Renal cell carcinoma 2005: new frontiers in staging, prognostication and targeted molecular therapy. J Urol. 2005; 173: 1853–62. [DOI] [PubMed] [Google Scholar]
- 7. Ljungberg B. Prognostic markers in renal cell carcinoma. Curr Op Urol. 2007; 17: 303–8. [DOI] [PubMed] [Google Scholar]
- 8. Herranz M, Esteller M. DNA methylation and histone modifications in patients with cancer: potential prognostic and therapeutic targets. Methods Mol Biol. 2007; 361: 25–62. [DOI] [PubMed] [Google Scholar]
- 9. Collas P, Noer A, Timoskainen S. Programming the genome in embryonic and somatic stem cells. J Cell Mol Med. 2007; 11: 602–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Recillas‐Targa F, De La Rosa‐Velázquez IA, Soto‐Reyes E, et al . Epigenetic boundaries of tumour suppressor gene promoters: the CTCF connection and its role in carcinogenesis. J Cell Mol Med. 2006; 10: 554–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cassiman JJ. Epigenetics: a major cause of variations on the same theme. J Cell Mol Med. 2006; 10: 552–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mohamed MA, Greif PA, Diamond J, et al . Epigenetic events, remodelling enzymes and their relationship to chromatin organization in prostatic intraepithelial neoplasia and prostatic adenocarcinoma. BJU Int. 2007; 99: 908–15. [DOI] [PubMed] [Google Scholar]
- 13. Piyathilake CJ, Bell WC, Jones J, et al . Patterns of global DNA and histone methylation appear to be similar in normal, dysplastic and neoplastic oral epithelium of humans. Dis Markers. 2005; 21: 147–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y, Peters CJ, Fitzgerald RC, et al . Progressive silencing of p14ARF in oesophageal adenocarcinoma. J Cell Mol Med. 2008; 13: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piperi C, Vlastos F, Farmaki E, et al . Epigenetic effects of lung cancer predisposing factors: impact on clinical diagnosis and prognosis. J Cell Mol Med. 2008; 12: 1495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997; 389: 349–52. [DOI] [PubMed] [Google Scholar]
- 17. Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004; 117: 721–33. [DOI] [PubMed] [Google Scholar]
- 18. Marino‐Ramirez L, Kann MG, Shoemaker BA, et al . Histone structure and nucleosome stability. Expert Rev Proteomics 2005; 2: 719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orr JA, Hamilton PW. Histone acetylation and chromatin pattern in cancer. A review. Anal Quant Cytol Histol. 2007; 29: 17–31. [PubMed] [Google Scholar]
- 20. Turner BM. Histone acetylation as an epigenetic determinant of long‐term transcriptional competence. Cell Mol. Life Sci. 1998; 54: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sobin LH, Wittheking CH, editors. International Union Against Cancer (UICC). TNM classification of malignant tumours. 6th ed. New York : Wiley‐Liss; 2002. pp. 193–5. [Google Scholar]
- 22. Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982; 6: 655–63. [DOI] [PubMed] [Google Scholar]
- 23. Piyathilake CJ, Henao O, Frost AR, et al . Race‐ and age‐dependent alterations in global methylation of DNA in squamous cell carcinoma of the lung (United States). Cancer Causes Control. 2003; 14: 37–42. [DOI] [PubMed] [Google Scholar]
- 24. Lam JS, Belldegrun AS, Figlin RA. Tissue array‐based predictions of pathobiology, prognosis and response to treatment for renal cell carcinoma therapy. Clin Cancer Res. 2004; 10: 6304–9. [DOI] [PubMed] [Google Scholar]
- 25. Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000; 1: 11–9. [DOI] [PubMed] [Google Scholar]
- 26. Widschwendter M, Jiang G, Woods C, et al . DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004; 64: 4472–80. [DOI] [PubMed] [Google Scholar]
- 27. Brothman AR, Swanson G, Maxwell TM, et al . Global hypomethylation is common in prostate cancer cells. A quantitative predictor for clinical outcome Cancer Genet Cytogenet 2005; 156: 31–6. [DOI] [PubMed] [Google Scholar]
- 28. Piyathilake CJ, Johanning GL, Frost AR, et al . Immunohistochemical evaluation of global DNA methylation: comparison with in vitro radiolabeled methyl incorporation assay. Biotech Histochem. 2000; 75: 251–8. [DOI] [PubMed] [Google Scholar]
- 29. Rea S, Eisenhaber F, O’Carroll D, et al . Regulation of chromatin structure by site‐specific histone H3 methyltransferases. Nature. 2000; 406: 593–9. [DOI] [PubMed] [Google Scholar]
- 30. Fahrner JA, Eguchi S, Herman JG, et al . Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002; 62: 7213–8. [PubMed] [Google Scholar]
- 31. Christoph F, Weikert S, Kempkensteffen C, et al . Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications. Clin Cancer Res. 2006; 12: 5040–6. [DOI] [PubMed] [Google Scholar]
- 32. Yamada D, Kikuchi S, Williams YN, et al . Promoter hypermethylation of the potential tumor suppressor DAL‐1/4.1B gene in renal clear cell carcinoma. Int J Cancer. 2006; 118: 916–23. [DOI] [PubMed] [Google Scholar]
- 33. Zhao H, Ljungberg B, Grankvist K, et al . Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006; 3: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Furge KA, Chen J, Koeman J, et al . Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high‐grade papillary renal cell carcinoma. Cancer Res. 2007; 67: 3171–6. [DOI] [PubMed] [Google Scholar]
- 35. Choi JS, Kim MK, Seo JW, et al . MET expression in sporadic renal cell carcinomas. J Korean Med Sci. 2006; 21: 672–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schulz WA, Hatina J. Epigenetics of prostate cancer: beyond DNA methylation. J Cell Mol Med. 2006; 10: 100–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wedel SA, Sparatore A, Del Soldato P, et al . New histone deacetylase inhibitors as potential therapeutic tools for advanced prostate carcinoma. J Cell Mol Med. 2008; 12: 2457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]


