Abstract
Cancer stem cells (CSCs) have recently been identified in leukaemia and solid tumours; however, the role of CSCs in metastasis remains poorly understood. This dearth of knowledge about CSCs and metastasis is due largely to technical challenges associated with the use of primary human cancer cells in pre‐clinical models of metastasis. Therefore, the objective of this study was to develop suitable pre‐clinical model systems for studying stem‐like cells in breast cancer metastasis, and to test the hypothesis that stem‐like cells play a key role in metastatic behaviour. We assessed four different human breast cancer cell lines (MDA‐MB‐435, MDA‐MB‐231, MDA‐MB‐468, MCF‐7) for expression of prospective CSC markers CD44/CD24 and CD133, and for functional activity of aldehyde dehydrogenase (ALDH), an enzyme involved in stem cell self‐protection. We then used fluorescence‐activated cell sorting and functional assays to characterize differences in malignant/metastatic behaviour in vitro (proliferation, colony‐forming ability, adhesion, migration, invasion) and in vivo (tumorigenicity and metastasis). Sub‐populations of cells demonstrating stem‐cell‐like characteristics (high expression of CSC markers and/or high ALDH) were identified in all cell lines except MCF‐7. When isolated and compared to ALDHlowCD44low/– cells, ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells demonstrated increased growth (P < 0.05), colony formation (P < 0.05), adhesion (P < 0.001), migration (P < 0.001) and invasion (P < 0.001). Furthermore, following tail vein or mammary fat pad injection of NOD/SCID/IL2γ receptor null mice, ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ cells showed enhanced tumorigenicity and metastasis relative to ALDHlowCD44low/– cells (P < 0.05). These novel results suggest that stem‐like ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ cells may be important mediators of breast cancer metastasis.
Keywords: breast cancer, metastasis, cancer stem cells, ALDH, CD44
Introduction
Breast cancer is a leading cause of death in women, due primarily to ineffective treatment of metastasis. Metastasis is a multi‐step process that involves tumour cell escape from the primary tumour, migration through the body, adhesion and extravasation at the secondary site, initiation of micrometastatic growth and maintenance of growth into clinically detectable macrometastases [1]. Given the onerous nature of this process, it is not surprising that metastasis is highly inefficient, with the main rate‐limiting steps being initiation and maintenance of growth at secondary sites [1, 2, 3]. Taken together with the heterogeneous nature of solid tumours, this metastatic inefficiency suggests that only a small subset of cells can successfully navigate the metastatic cascade and eventually re‐initiate tumour growth to form metastases. However, the ability to specifically identify and target this deadly subset of cells has remained elusive.
In light of several pivotal studies demonstrating the existence of ‘stem‐like’ cells or cancer stem cells (CSCs) in leukaemia [4, 5] and several types of solid cancer [6, 7, 8, 9, 10, 11], we and others have speculated that CSCs may represent the subset of tumour cells responsible for metastatic disease [12, 13]. However, while several studies have demonstrated the ability of CSCs to initiate and maintain a primary tumour [6, 7, 8, 9, 10, 11], the functional contribution of CSCs to metastatic behaviour remains poorly understood.
In breast cancer, stem‐like cells have been prospectively isolated from primary tumours and pleural effusions based on a CD44+CD24− phenotype [6]. Subsequent experimental studies have also isolated CD44+CD24− breast cancer cells and demonstrated increased in vitro expression of stem cell markers and enhanced capacity for mammosphere formation, invasion and resistance to radiation [14, 15, 16]. Furthermore, clinical studies indicate that CD44+CD24− tumour‐initiating cells express an invasive gene signature and may be associated with distant metastases [17, 18, 19]. Additional putative CSC phenotypes have also been identified in other solid cancers, including CD133+ (brain, colon, pancreatic cancer) [8, 9, 11] and CD44+CD133+ (prostate cancer) [7]. However, because of the heterogeneous nature of solid cancers, the reliability of using cell surface markers as the sole way to isolate CSCs remains controversial [20, 21, 22, 23]. Furthermore, human stem cell sources (including tumours) may contain alternate stem/progenitor cell lineages not efficiently isolated using variably expressed cell surface markers.
A complementary strategy for identifying stem‐like tumour cells involves measurement of aldehyde dehydrogenase (ALDH) activity, an enzyme involved in intracellular retinoic acid production [24]. Retinoic acid signalling is linked to cellular differentiation during development and plays a role in stem cell self‐protection throughout an organism’s lifespan [25]. We and others have previously used this isolation strategy alone or in combination with cell surface markers (i.e. CD133) to successfully isolate normal human haematopoietic progenitor cells [26, 27, 28], and ALDH activity has also been used to identify stem‐like subsets in human haematopoietic cancers [29, 30, 31]. A recent study by Ginestier et al. (2007) elegantly demonstrated that expression of ALDH1 in breast tumours was a predictor of poor clinical outcome, and that high ALDH activity selects for both normal and tumorigenic human mammary epithelial cells with stem/progenitor properties [32]. Therefore, the use of ALDH activity as a purification strategy allows non‐toxic and efficient isolation of human stem‐like cells based on a developmentally conserved stem/progenitor cell function.
Although there is growing evidence supporting the existence of CSCs in solid tumours, what remains less clear is the impact of CSCs on metastatic behaviour. We believe that the dearth of knowledge about CSCs and metastasis is due largely to technical challenges associated with the use of primary human cancer cells in pre‐clinical models of metastasis: even in nonobese diabetic/severe combined immune‐deficient (NOD/SCID) mice, it is very difficult to grow primary cells as xenograft tumours, much less as metastases. Therefore, workable alternative model systems must be developed in order to address this need. It has long been observed by tumour biologists that even with cancer cell lines, large numbers of cells need to be injected to form a tumour and/or metastasize in experimental animals [1, 2, 33]. This suggests that not all cells within a cell line population are equal, and only a small subset of cells is capable of tumour initiation and progression to metastasis. Indeed, studies have shown that commonly used cell lines contain subpopulations of cells with stem‐like properties [14, 15, 16]. Thus, the purification of stem‐like cells from cell lines may provide a valuable model system for starting to investigate the role of such cells in breast cancer metastasis.
In the current study, we assessed four commonly used human breast cancer cell lines (including highly metastatic MDA‐MB‐435 [see note in section ‘Materials and Methods’], moderately metastatic MDA‐MB‐231, weakly metastatic MDA‐MB‐468 and non‐metastatic MCF‐7) for expression of the prospective CSC markers CD44/CD24 and CD133, and for functional activity of ALDH. We then used fluorescence‐activated cell sorting (FACS) and standard functional assays to characterize differences in malignant/metastatic behaviour between cell populations, including cell growth, adhesion, migration, and invasion in vitro, and tumorigenicity and metastasis in vivo. The novel findings presented here suggest that high ALDH activity and CSC marker expression select for stem‐like breast cancer cells with enhanced malignant and metastatic properties, and that ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ stem‐like cells may be important contributors to breast cancer metastasis.
Material and methods
Cell culture
MDA‐MB‐231 and MCF‐7 cells were obtained from ATCC (Manassas, VA, USA). MDA‐MB‐435 and MDA‐MB‐468 cells were obtained from Dr. Janet Price (M.D. Anderson Cancer Center, Houston, TX, USA [34]). MDA‐MB‐435 and MDA‐MB‐468 cells were grown in αMEM + 10% foetal bovine serum (FBS), MCF‐7 cells were grown in DMEM + 10% FBS and MDA‐MB‐231 cells were grown DMEM:F12 + 10% FBS. Media were obtained from Invitrogen (Carlsbad, CA, USA). FBS was obtained from Sigma (St. Louis, MO, USA). It should be noted that the MDA‐MB‐435 cell line was originally isolated from the pleural effusion of a woman with metastatic breast adenocarcinoma [34]. Recently, a debate has arisen over the origins of this cell line, whether it was derived from the M14 melanoma cell or is in fact a true breast cancer cell line [35, 36]. Whether melanoma or breast in origin, the MDA‐MB‐435 cell line does represent a highly metastatic cell line, and thus provides an opportunity to relate functional metastatic ability to expression of CSC‐related markers.
Cell surface marker analysis
MDA‐MB‐435, MDA‐MB‐231, MDA‐MB‐468 and MCF‐7 cells (1 × 105) were incubated with fluorescently conjugated antibodies, including anti‐CD24 (clone ML5; BD Biosciences Canada, Mississauga, ON, Canada) or anti‐CD133 (clone AC133; Miltenyi Biotec, Auburn, CA, USA) conjugated to phycoerytherin (PE); and anti‐CD44 (clone IM7; BD Biosciences) conjugated to fluorescein isothiocyanate (FITC). For supplemental studies, MDA‐MB‐231 and MDA‐MB‐468 cells were also incubated with anti‐CXCR4 (clone 12G5; R&D Systems, Minneapolis, MN, USA) conjugated to PE. Fluorescently conjugated immunoglobulin G (IgG) isotype controls (BD Biosciences) were used as negative controls. Cells were analysed using a Beckman Coulter EPICS XL‐MCL flow cytometer (Beckman Coulter, Fullerton, CA, USA).
Analysis of ALDH activity
To assess ALDH activity of the different cell lines, the ALDEFLUOR® assay kit (StemCell Technologies, Vancouver, BC, Canada) was used. The basis for this assay is that uncharged ALDH substrate (BODIPY‐aminoacetaldehyde [BAAA]) is taken up by living cells via passive diffusion. Once inside the cell, BAAA is converted into negatively charged BODIPY‐aminoacetate (BAA) by intracellular ALDH. BAA− is then retained inside the cell, causing the cell to become highly fluorescent. Only cells with an intact cell membrane can retain BAA−, so only viable cells can be identified [24]. The ALDEFLUOR® assay was performed essentially as described previously [26, 27, 28]. Briefly, MDA‐MB‐435, MDA‐MB‐231, MDA‐MB‐468 and MCF‐7 cells were harvested, placed in ALDEFLUOR® assay buffer (2 × 106/ml) and incubated with the ALDEFLUOR® substrate for 45 min. at 37°C to allow substrate conversion. As a negative control for all experiments, an aliquot of ALDEFLUOR®‐stained cells was immediately quenched with 1.5‐mM diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor. Cells were analysed using the green fluorescence channel (FL1) on a Beckman Coulter EPICS XL‐MCL flow cytometer. Human umbilical cord blood (discard product) was labelled and assayed for ALDH activity as described previously [27] in order to optimize and validate the flow cytometry protocol.
FACS isolation of cells
Based on cell surface marker phenotype and ALDH activity, MDA‐MB‐468 and MDA‐MB‐231 cell lines were chosen for further cell isolation by FACS and functional analysis. Cells were concurrently labelled with 7‐Amino‐actinomycin (7‐AAD), fluorescent antibodies (CD44‐APC + CD24‐PE [MDA‐MB‐231]or CD44‐APC + CD133‐PE [MDA‐MB‐468]) and the ALDEFLUOR™ assay kit as described above. Cell subsets were isolated using a four‐colour analysis protocol on a FACS Vantage/Diva cell sorter (BD Biosciences), including ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ subsets (from MDA‐MB‐231 cells) and ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− subsets (from MDA‐MB‐468 cells). For both cell lines, ALDH activity was used as the primary sort criteria (top ∼20%= ALDHhi; bottom ∼20%= ALDHlow) and CD44+CD24− (MDA‐MB‐231 cells) or CD44+CD133+ (MDA‐MB‐468 cells) phenotype as the secondary sort criteria (top ∼10% gated on ALDHhi; bottom ∼10% gated on ALDHlow). Cell viability was assessed by 7‐AAD staining during cell sorting, and confirmed by trypan blue exclusion after sorting. Following FACS isolation, cells were used immediately for functional in vitro and in vivo assays.
Cell proliferation assays
Sorted cell populations (ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ from MDA‐MB‐231 cells; ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− from MDA‐MB‐468 cells) were plated at a density of 5.0 × 104 cells/60 mm plate (n= 3 for each time‐point) and maintained in regular growth media. Every 48 hrs for 14 days, triplicate cultures were trypsinized and counted by hemocytometer. Doubling time of each cell population was estimated during the exponential growth phase according to T d= 0.693t/ln (Nt/N 0), where t is time (in hours), N t is the cell number at time t and N 0 is the cell number at initial time. Lag time was taken as the time for each population to reach the exponential growth phase.
Colony forming assays
In preparation for the assay, 60 mm dishes were coated with 1% agarose (Bioshop; Burlington, ON, Canada) in normal growth media and allowed to set for 1 hr. Cell suspensions (1.0 × 104 cells/60 mm plate) for each sorted population (ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ from MDA‐MB‐231 cell line; ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− from MDA‐MB‐468 cell line) were prepared using 0.6% agarose in normal growth media and plated on top of the 1% agarose base layer (n= 4 for each time‐point). Normal growth media were added on top of the cell layer and changed every 3–4 days for 4 weeks, at which time media were removed and plates were fixed in 10% neutral‐buffered formalin (EM Sciences, Gladstone, NJ, USA). Five fields of view (100×) were counted for each dish, and mean number of colonies/field and mean colony diameter was calculated.
Cell adhesion assays
Cells were plated onto sterile 96‐well non‐tissue culture plates (Titertek, Flow Laboratories, Inc.; McLean, VA, USA) treated with either 10 μg/ml of human fibronectin (Sigma) (MDA‐MB‐231 cells), 5 μg/ml of human vitronectin (Sigma) (MDA‐MB‐468 cells), or phosphate‐buffered saline (PBS) (negative control), using 1 × 104 cells/well (n= 3) for each sorted cell population (ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ from MDA‐MB‐231 cell line; ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− from MDA‐MB‐468 cell line). Vitronectin and fibronectin were chosen based on previous experiments in our laboratory that have demonstrated that MDA‐MB‐231 and MDA‐MB‐468 cells differentially express integrin receptors for fibronectin and vitronectin, respectively ([37] and our unpublished data). Cells were allowed to adhere for 5 hrs, after which the media were removed and non‐adhered cells were rinsed away. Adhered cells were fixed with 2% gluteraldehyde and stained using Harris’ haematoxylin. Five high‐powered fields (HPF) (400×) were counted for each well, and mean numbers of adhered cells/field were calculated.
Cell migration and invasion assays
Transwell plates (6.5 mm, 8 μm pore size; Becton Dickinson; Franklin Lakes, NJ, USA) were coated with 6 μg/well of gelatin (Sigma) (migration assay) or 10 μg/well of Matrigel (Becton Dickinson) (invasion assay) as described previously [38, 39]. Control [0.01% bovine serum albumin (BSA)] or chemoattractant (5% FBS) media were placed in the bottom portion of each well. A total of 3.5 × 104 cells of each sorted population (ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ from MDA‐MB‐231 cell line; ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− from MDA‐MB‐468 cell line) were plated on top of the transwells. Cells were allowed to migrate or invade for 24 hrs, after which the upper transwell was removed, inverted, fixed with 1% gluteraldehyde and stained with Harris’s haematoxylin. Non‐migrated or non‐invaded cells on the inner surface of the transwell were carefully removed using a cotton swab. Five HPF were counted for each well, and mean numbers of migrated or invaded cells/field were calculated.
In vivo animal experiments
All in vivo work was carried out using NOD/SCID‐IL2γ receptor null mice, which permit increased tissue engraftment of human cells without rejection due to reduced innate immunity (NOD mutation), complete T‐ and B‐cell deficiency (SCID mutation) and reduced natural killer (NK)‐cell function (IL‐2Rγ mutation) [40]. Animal procedures were conducted in accordance with the recommendations of the Canadian Council on Animal Care, under a protocol approved by the University of Western Ontario Council on Animal Care.
Following cell sorting, sorted populations (ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ from MDA‐MB‐231 cell line; ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− from MDA‐MB‐468 cell line) were resuspended in sterile Hank’s buffered salt solution (MDA‐MB‐231) or PBS (MDA‐MB‐468) at 5 × 106 cells/ml. Using established models of experimental metastasis (tail vein injection) or spontaneous metastasis (mammary fat pad injection) [33], 100 μl (5 × 105 cells) of each sorted cell population were injected into the lateral tail vein or right thoracic mammary fat pad of 7–10‐week‐old female NOD/SCID‐IL2Rγ null mice (n= 4 mice/group). Primary tumour growth was evaluated weekly by calliper measurement in two perpendicular dimensions, and tumour volume was estimated using the following formula: (volume = 0.52 ×[width]2×[length]) for approximating the volume (mm3) of an ellipsoid. Metastatic growth in the lung and extrapulmonary organs was allowed to develop for 5–7 weeks (MDA‐MB‐231) or 12 weeks (MDA‐MB‐468), at which point the mice were killed and assessed for metastatic burden. End‐points and cell numbers for injection were chosen based on preliminary experiments using unsorted cell populations and NOD/SCID‐IL2Rγ null mice (data not shown).
Tissues and organs were examined superficially for evidence of gross macroscopic metastases at necropsy. Tissues collected at necropsy were then fixed in 10% neutral‐buffered formalin, embedded in paraffin, sectioned randomly (4‐μm sections; at least 100 μm apart; five sections/tissue) and subjected to standard haematoxylin and eosin staining. Stained slides were evaluated by light microscopy in a blinded fashion in order to identify and characterize incidence and regions of micrometastatic involvement. Metastatic tumour burden in the lung (tumour area/total organ area) was determined quantitatively by manual outlining and analysis by ImageJ software (NIH, Bethesda, MD, USA) as described previously [38].
Data analysis
Experiments analysing CSC marker phenotype and ALDH activity were performed a minimum of three times. In vitro experiments with FACS‐isolated cells were performed twice (after four separate cell sorts) with at least three biological replicates included within each experiment. In vivo studies were carried out using multiple animals (n= 4 per cell population) and using cells obtained from five separate cell sorts. In all cases, quantitative data were compiled from all experiments. Statistical analysis was performed with GraphPad Prism 4.0 software© (San Diego, CA, USA) using either t‐test (for comparison between two groups) or anova with Tukey post‐test (for comparison between more than two groups). Differences between means were determined using the Student’s t‐test when groups passed both a normality test and an equal variance test. When this was not the case, the Mann‐Whitney rank sum test was used. Linear regression was used to assess the relationship between primary tumour size and metastatic burden in the lung. Unless otherwise noted, data are presented as the mean ± S.E.M. P‐values of ≤0.05 were regarded as being statistically significant.
Results
Human breast cancer cell lines contain sub‐populations of cells expressing prospective CSC markers
Flow cytometry was used to assess the expression of prospective CSC markers, including CD44/CD24 and CD133. Interestingly, the most aggressive cell line (MDA‐MB‐435) had the highest proportion of CD44+CD24− cells (96.6 ± 1.2%) (Fig. 1A), whereas the least aggressive cell line (MCF‐7) had the lowest proportion of CD44+CD24− cells (0.05 ± 0.01%) (Fig. 1D). The moderately metastatic MDA‐MB‐231 cell line contained 79.5 ± 2.7% CD44+CD24− cells (Fig. 1B). In contrast, the weakly metastatic MDA‐MB‐468 cell line contained very few cells (0.09 ± 0.05%) with a CD44+CD24− phenotype; however, the majority of cells (92.9 ± 4.4%) did express both CD44 and CD24 (CD44+CD24+) (Fig. 1C). Quantitative analysis demonstrated that the MDA‐MB‐435 cell line contained significantly more cells with a CD44+CD24− phenotype than any of the other cell lines (P < 0.01), and that the MDA‐MB‐231 cell line had significantly more cells with a CD44+CD24− phenotype than either the MDA‐MB‐468 or MCF‐7 cell lines (P < 0.01) (Fig. S1).
Figure 1.
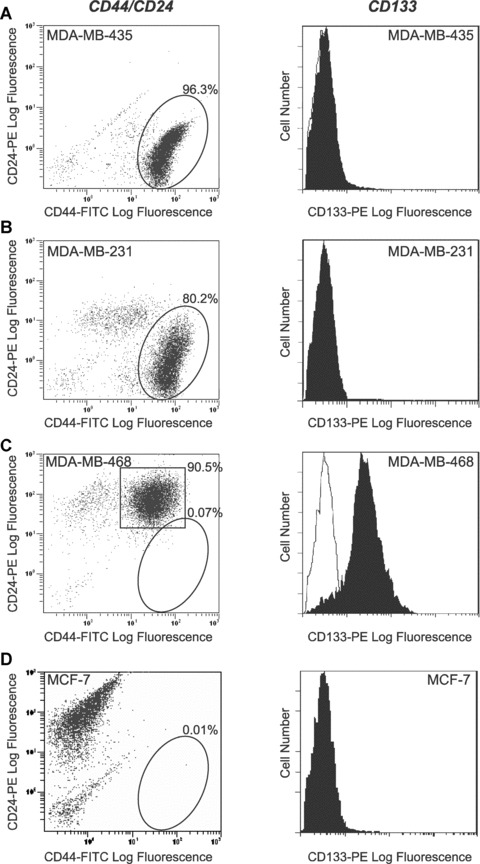
Human breast cancer cell lines contain sub‐populations of cells expressing prospective CSC markers. (A–D) Flow cytometry analysis of CD44/CD24 and CD133. Antibodies used included an anti‐CD44 antibody (clone IM7) conjugated to fluorescein isothiocyanate (FITC), an anti‐CD24 antibody (clone ML5) conjugated to phycoerytherin (PE), an anti‐CD133 antibody (clone AC133) conjugated to PE, or appropriate FITC and PE‐conjugated IgG isotype controls. (A) MDA‐MB‐435 cells; (B) MDA‐MB‐231 cells; (C) MDA‐MB‐468 cells and (D) MCF‐7 cells. Left‐hand panels show representative dot plots of CD44‐FITC versus CD24‐PE expression. Cells which fell within the circled regions were considered to express the phenotype of interest (CD44+CD24−). Compiled quantitative analysis of CD44+CD24− phenotype expression from three separate experiments is shown in Fig. S1. In (C), cells which fell within the square region were considered to express the phenotype CD44+CD24+. Right‐hand panels show representative histograms of CD133 expression (black profiles) relative to the IgG isotype control (white profiles). A minimum of 10,000 events were collected per sample.
MDA‐MB‐435, MDA‐MB‐231 and MCF‐7 cell lines did not demonstrate expression of CD133 (Fig. 1A, B and D). However, MDA‐MB‐468 cells did consistently express CD133 on their surface, at a level significantly higher than the other cell lines (Fig. 1C).
Human breast cancer cell lines contain sub‐populations of cells with enhanced ALDH activity
A complementary strategy for identifying cells with a stem/progenitor phenotype involves measurement of ALDH activity [24]. Human umbilical cord blood was assayed for ALDH activity in order to provide a control for setting up the flow cytometry protocol and for confirming that the assay was working appropriately (Fig. S2). The MDA‐MB‐435 cell line did not demonstrate any significant increase in ALDH activity (Fig. 2A). Interestingly, both the MDA‐MB‐231 and MDA‐MB‐468 cell lines showed two definitive subpopulations of cells: one that had ALDH activity at the level of the DEAB control, and one that had increased ALDH activity (Fig. 2B and C). The MCF‐7 cell line did not demonstrate any increased ALDH activity (Fig. 2D).
Figure 2.

Human breast cancer cell lines contain sub‐populations of cells with enhanced ALDH activity. (A–D) Flow cytometry analysis of ALDH activity. Cells were assayed with ALDEFLUOR® assay kit (StemCell Technologies) as per the manufacturer’s guidelines. As a negative control for all experiments, an aliquot of ALDEFLUOR®‐stained cells was immediately quenched with 1.5‐mM diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor. Cells were analysed using the green fluorescence channel (FL1) on a Beckman Coulter EPICS XL‐MCL flow cytometer. (A) MDA‐MB‐435 cells; (B) MDA‐MB‐231 cells; (C) MDA‐MB‐468 cells and (D) MCF‐7 cells. Left‐hand panels show representative dot plots of cells treated with the ALDH‐specific inhibitor DEAB (negative controls). Right‐hand panels show representative dot plots of ALDH activity. A minimum of 10,000 events were collected per sample. Cells which fell within the circled regions were considered to represent subpopulations of cells with enhanced ALDH activity relative to the rest of the cell population. The flow cytometry protocol was optimized and validated using human umbilical cord blood, which has a known population of stem/progenitor cells with enhanced ALDH activity (Fig. S2).
Strategy for isolation of stem‐like human breast cancer cells
MDA‐MB‐231 and MDA‐MB‐468 cell lines were chosen for further characterization and functional analysis because they had the most distinct subpopulations of cells with stem‐like characteristics (CSC marker expression, ALDH activity). Subsets of cells were isolated from MDA‐MB‐231 and MDA‐MB‐468 cell lines by FACS, using ALDH activity as the primary sort criteria and CD44+CD24− (MDA‐MB‐231 cells) or CD44+CD133+ (MDA‐MB‐468 cells) phenotype as the secondary sort criteria. The resulting cell subsets were designated as ALDHhiCD44+CD24− (MDA‐MB‐231) or ALDHhiCD44+CD133+ (MDA‐MB‐468) (‘stem‐like’) and ALDHlowCD44low/−CD24+ (MDA‐MB‐231) or ALDHlowCD44low/−CD133− (MDA‐MB‐468) (non ‘stem‐like’). Examples of the cell sorting strategy are represented in Fig. 3 (MDA‐MB‐231) and Fig. 4 (MDA‐MB‐468). From this point forward in the study, the non‐’stem‐like’ cells from both cell lines will be collectively referred to as respective ALDHlowCD44low/− subsets.
Figure 3.
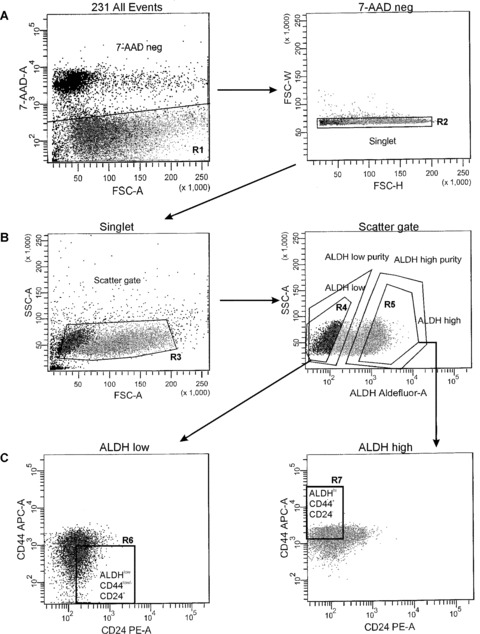
Strategy for isolation of stem‐like human breast cancer cells from the MDA‐MB‐231 cell line. Fluorescence‐activated cell sorting (FACS) was used to isolate ALDHhiCD44+ and ALDHlowCD44low/− human breast cancer cells for functional assays. MDA‐MB‐231 cells were concurrently labelled with 7‐AAD, fluorescent antibodies (CD44‐APC + CD24‐PE) and the ALDEFLUOR™ assay kit. Cell subsets were isolated using a four‐colour protocol on a FACS Vantage/Diva cell sorter, including ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ subsets (A–C): Representative schematic of a sequentially gated MDA‐MB‐231 cell line sort. (A) Cells were first selected for viability based on 7‐AAD negativity (R1, left panel) and for singlets (R2, right panel). (B) Cells were then selected based on light scatter (R3, left panel) and divided into ALDHlow (R4, bottom ∼20% of parent population) and ALDHhi (R5, top ∼20% of parent population) based on ALDH activity (right panel). (C) Finally, ALDHlow cells were further selected based on a CD44low/−CD24+ phenotype (R6, bottom 10% of parent population, gated on R1 + R2 + R3 + R4) (left panel), whereas ALDHhi cells were further selected based on expression of a CD44+CD24− phenotype (R7, top ∼10% of parent population, gated on R1 + R2 + R3 + R5) (right panel). The resulting cell subsets were designated as ALDHhiCD44+CD24− (R7, ‘stem‐like’) and ALDHlowCD44low/−CD24+ (R6, non ‘stem‐like’), and were collected for functional analysis of differences in malignant and metastatic behaviour in vitro and in vivo.
Figure 4.
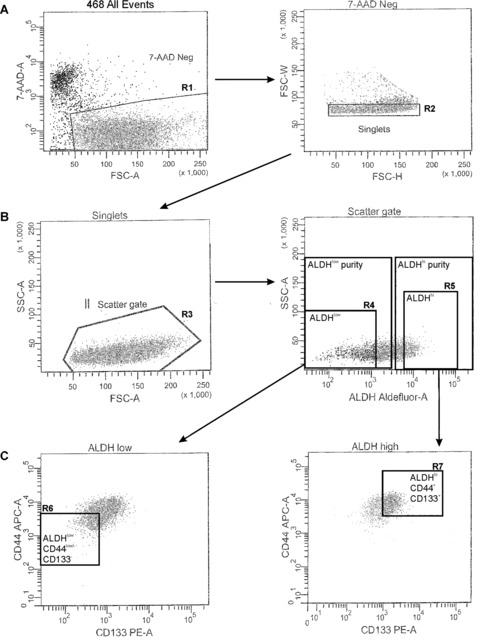
Strategy for isolation of stem‐like human breast cancer cells from the MDA‐MB‐468 cell line. Fluorescence‐activated cell sorting (FACS) was used to isolate ALDHhiCD44+ and ALDHlowCD44low/− human breast cancer cells for functional assays. MDA‐MB‐468 cells were concurrently labelled with 7‐AAD, fluorescent antibodies (CD44‐APC + CD133‐PE) and the ALDEFLUOR™ assay kit. Cell subsets were isolated using a four‐colour protocol on a FACS Vantage/Diva cell sorter, including ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− subsets (A–C): Representative schematic of a sequentially gated MDA‐MB‐468 cell line sort. (A) Cells were first selected for viability based on 7‐AAD negativity (R1, left panel) and for singlets (R2, right panel). (B) Cells were then selected based on light scatter (R3, left panel) and divided into ALDHlow (R4, bottom ∼20% of parent population) and ALDHhi (R5, top ∼20% of parent population) based on ALDH activity (right panel). (C) Finally, ALDHlow cells were further selected based on a CD44low/−CD133low/− phenotype (R6, bottom 10% of parent population, gated on R1 + R2 + R3 + R4) (left panel), whereas ALDHhi cells were further selected based on expression of a CD44+CD133+ phenotype (R7, top ∼10% of parent population, gated on R1 + R2 + R3 + R5) (right panel). The resulting cell subsets were designated as ALDHhiCD44+CD133+ (R7, ‘stem‐like’) and ALDHlowCD44low/−CD133− (R6, non ‘stem‐like’), and were collected for functional analysis of differences in malignant and metastatic behaviour in vitro and in vivo.
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced cell growth and colony formation in vitro
Differences in cell growth characteristics in vitro between sorted subpopulations for both cell lines were assessed (Fig. 5). ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells demonstrated increased growth in normal culture relative to respective ALDHlowCD44low/− cell subsets. Lag times (time to reach exponential growth phase) were also observed to be shorter for ALDHhiCD44+CD24− (MDA‐MB‐231 = 1 day) and ALDHhiCD44+CD133+ (MDA‐MB‐468 = 2 days) cells versus respective ALDHlowCD44low/− cell subsets (7 days for both cell lines) (Fig. 5A). Differences in colony‐forming ability and anchorage‐independent growth between sorted subpopulations were assessed using a soft agar assay (Fig. 5B and C). Both ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cell subsets formed significantly more colonies (P < 0.001) (Fig. 5B) and larger colonies (P < 0.05) (Fig. 5C) than respective ALDHlowCD44low/– cell subsets.
Figure 5.
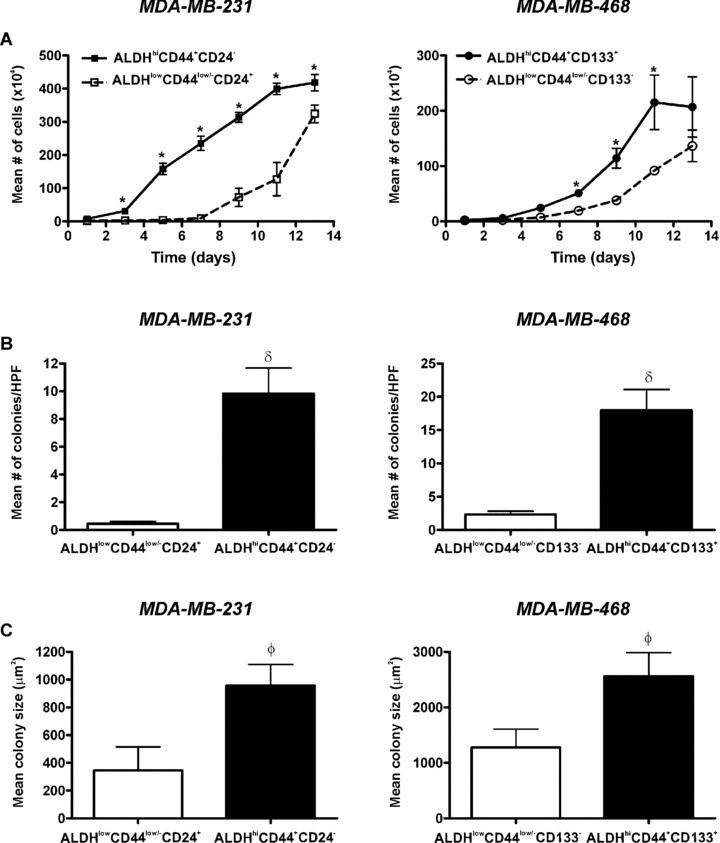
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced cell growth and colony formation in vitro. Cells were isolated by FACS as described in Figs 3 and 4 and subjected to in vitro assays for growth and colony formation. (A) Cell growth kinetics in normal (anchorage‐dependent) culture over time of ALDHhiCD44+CD24− (▪) versus ALDHlowCD44low/−CD24+ (□) cells isolated from the MDA‐MB‐231 cell line (left panel); and ALDHhiCD44+CD133+ (•) versus ALDHlowCD44low/− CD133− (○) cells isolated from the MDA‐MB‐468 cell line (right panel) (5.0 × 104 cells/60 mm plate; n = 3 plates/time‐point). Data are presented as the mean ± S.E.M. *= significantly different cell number than respective ALDHlowCD44low/− cell subsets (P < 0.05). (B, C) Colony forming ability of ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells (black bars) versus respective ALDHlowCD44low/− cell subsets (white bars) isolated from MDA‐MB‐231 (left panels) or MDA‐MB‐468 (right panels) cell lines. Cells (1.0 × 104 cells/60 mm plate, n= 4 plates/cell population) were grown in soft agar (0.6%) for 4 weeks. Five high‐powered fields of view (100×) were counted for each dish. (B) Mean number of colonies per plate and (C) Mean colony diameter (μm2). Data are presented as the mean ± S.E.M. δ= significantly different colony number than respective ALDHlowCD44low/− cell subsets (P < 0.001). The value φ= significantly different colony size than respective ALDHlowCD44low/− cell subsets (P < 0.05).
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced adhesion, migration and invasion in vitro
In vitro assays were used to compare sorted subpopulations from the perspective of differences in cell adhesion, migration and invasion (Fig. 6). ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells were observed to be significantly more adherent to fibronectin (MDA‐MB‐231) or vitronectin (MDA‐MB‐468) (Fig. 6A), significantly more migratory towards serum (Fig. 6B) and significantly more invasive through Matrigel (Fig. 6C) than respective ALDHlowCD44low/– cell subsets (P < 0.001).
Figure 6.
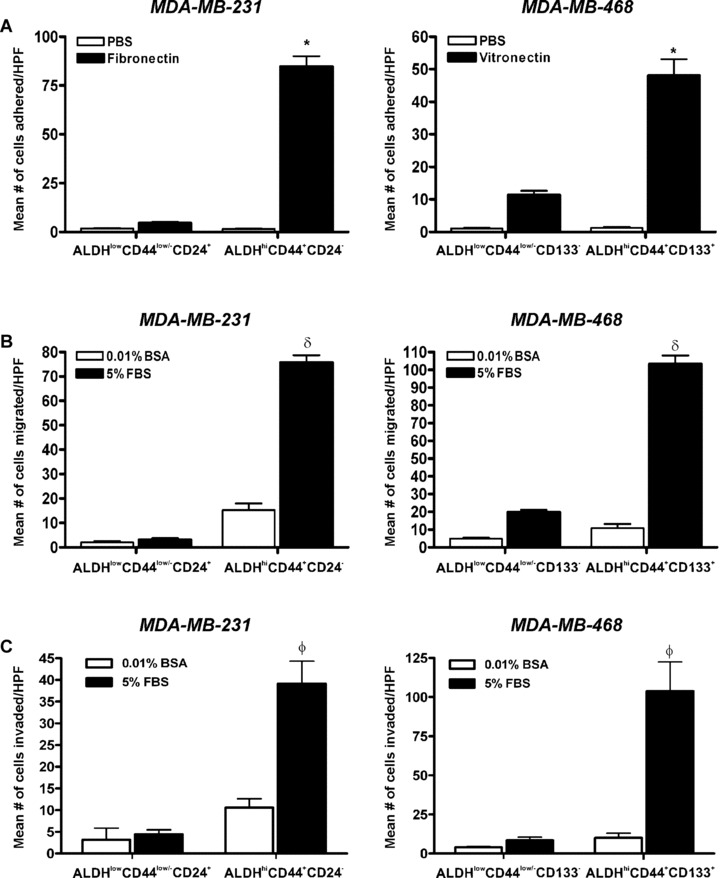
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced adhesion, migration and invasion in vitro. (A) Cell adhesion of ALDHhiCD44+CD24− versus ALDHlowCD44low/−CD24+ cells isolated from the MDA‐MB‐231 cell line (left panel); and ALDHhiCD44+CD133+ versus ALDHlowCD44low/− CD133− cells isolated from the MDA‐MB‐468 cell line (right panel). Cells were isolated by FACS as described in Figs 3 and 4 and plated onto sterile 96‐well non‐tissue culture plates treated with either 10 μg/ml of fibronectin (MDA‐MB‐231 cells), 5 μg/ml of vitronectin (MDA‐MB‐468 cells) (black bars) or PBS (negative control) (white bars), using 1 × 104 cells/well in triplicate wells for each sorted cell population. Cells were allowed to adhere for 5 hrs and adhered cells were quantified by manual counting of five high‐powered fields (HPF) per well. Data are presented as the mean ± S.E.M. *= significantly different than respective population of ALDHlowCD44low/− cells adhered to vitronectin or fibronectin (P < 0.001). (B) Cell migration and (C) Cell invasion of ALDHhiCD44+CD24− versus ALDHlowCD44low/−CD24+ cells isolated from the MDA‐MB‐231 cell line (left panels); and ALDHhiCD44+CD133+ versus ALDHlowCD44low/− CD133− cells isolated from the MDA‐MB‐468 cell line (right panels). Transwells (8 μm) were pre‐coated with either gelatin (migration assays; 6 μg/well) or Matrigel (invasion assays; 10 μg/well) and FACS‐isolated cells (3.5 × 104 cells/well; n= 3 for each cell population) were allowed to migrate or invade for 24 hrs towards chemoattractant media (5% foetal bovine serum) (black bars) or control media (0.01% bovine serum albumin) (white bars). Migrated or invaded cells were quantified by manual counting of five HPF per well. Data are presented as the mean ± S.E.M. δ= significantly different than respective population of ALDHlowCD44low/− cells migrating towards FBS (P < 0.001). φ= significantly different than respective population of ALDHlowCD44low/− cells invading through Matrigel towards FBS (P < 0.001).
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced tumorigenicity and metastasis in vivo
Standard experimental or spontaneous metastasis assays were used to compare the ability of sorted subpopulations to establish themselves and grow in vivo following cell injection into the tail vein (Fig. 7) or mammary fat pad (Fig. 8) of NOD/SCID‐IL2Rγ null mice. Following tail vein injection, metastatic tumour burden in the lung (percentage of lung occupied by tumour) was significantly higher in mice injected with ALDHhiCD44+CD24− (MDA‐MB‐231) or ALDHhiCD44+CD133+ (MDA‐MB‐468) cells versus respective ALDHlowCD44low/− cell subsets (Fig. 7A and B) (P < 0.05). Analysis of incidence of metastases (percentage of mice demonstrating metastatic growth) in the lung and extrapulmonary organs revealed that, although all subpopulations of cells were able to establish themselves in the lung (Fig. 7C), only ALDHhiCD44+ CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells were able to maintain that growth into larger metastases (Fig. 7A and B). Furthermore, metastases in extrapulmonary organs such as the pancreas, liver, spleen and/or kidney were only observed in mice injected with ALDHhiCD44+CD24− (MDA‐ MB‐231) or ALDHhiCD44+CD133+ (MDA‐MB‐468) cells, and not in mice injected with ALDHlowCD44low/− cell subsets (Fig. 7C).
Figure 7.
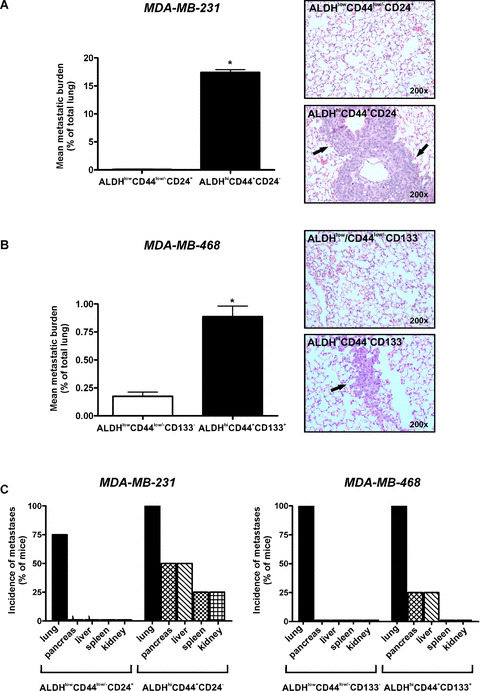
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced metastatic growth in vivo following tail vein injection. Cells were isolated by FACS as described in Figs 3 and 4 and injected into the lateral tail vein of female NOD/SCID‐IL2Rγ null mice using an established model of experimental metastasis (5 × 105 cells/mouse; 4 mice/cell population). At 5 weeks (MDA‐MB‐231) or 12 weeks (MDA‐MB‐468) after injection, mice were killed and assessed for metastatic burden in the lung and elsewhere. Tissue sections were subjected to haematoxylin and eosin staining (five random sections/tissue/mouse), and the incidence and extent of metastasis was determined in a blinded fashion. (A and B) Quantitative analysis of tumour burden (mean% of lung occupied by tumour) (left panels) and representative haematoxylin and eosin stained lung sections (right panels) following tail vein injection of (A) ALDHhiCD44+CD24− versus ALDHlowCD44low/−CD24+ cells isolated from the MDA‐MB‐231 cell line; and (B) ALDHhiCD44+CD133+ versus ALDHlowCD44low/−CD133− cells isolated from the MDA‐MB‐468 cell line. Arrowheads on haematoxylin and eosin images indicate regions of tumour within the lung. Data are presented as the mean ± S.E.M. *= significantly different than respective ALDHlowCD44low/− subsets (P < 0.05). (C) Incidence of metastatic growth in lung and extrapulmonary tissues following tail vein injection of ALDHhiCD44+CD24− versus ALDHlowCD44low/−CD24+ cells isolated from the MDA‐MB‐231 cell line (left panels); and ALDHhiCD44+CD133+ versus ALDHlowCD44low/−CD133− cells isolated from the MDA‐MB‐468 cell line (right panels).
Figure 8.
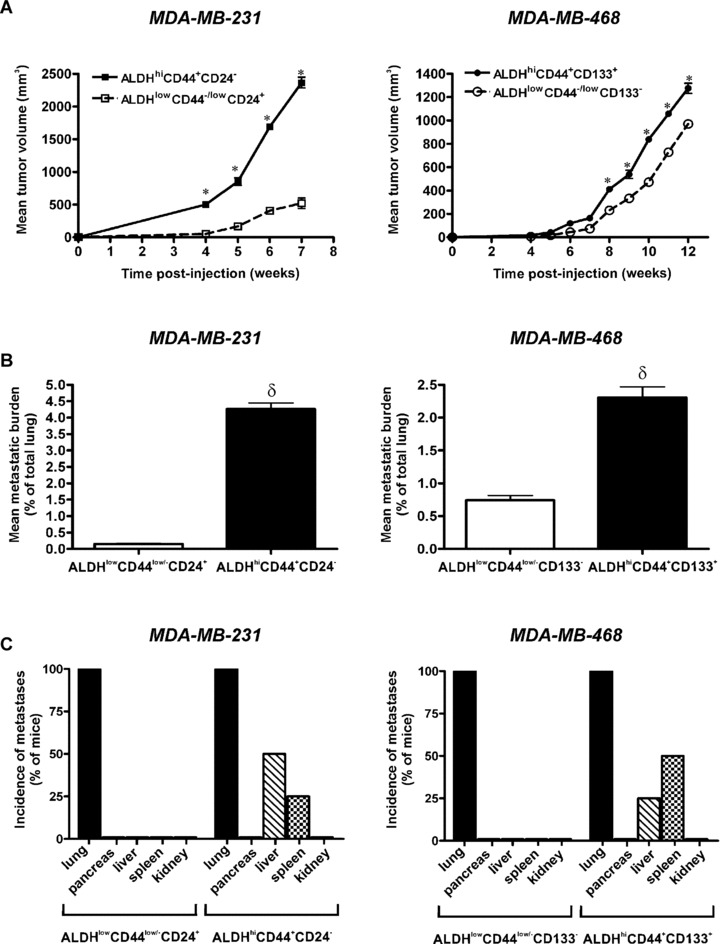
ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells demonstrate enhanced tumorigenicity and metastatic growth in vivo following mammary fat pad injection. Cells were isolated by FACS as described in Figs 3 and 4 and injected into the right thoracic mammary fat pad of female NOD/SCID‐IL2Rγ null mice using an established model of spontaneous metastasis (5 × 105 cells/mouse; 4 mice/cell population). At 7 weeks (MDA‐MB‐231) or 12 weeks (MDA‐MB‐468) after injection, mice were killed and assessed for metastatic burden in the lung and elsewhere. Tissue sections were subjected to haematoxylin and eosin staining (five random sections/tissue/mouse), and the incidence and extent of metastasis was determined in a blinded fashion. (A) Primary tumour growth kinetics following mammary fat pad injection of ALDHhiCD44+CD24− (▪) versus ALDHlowCD44low/−CD24+ (□) cells isolated from the MDA‐MB‐231 cell line (left panel); and ALDHhiCD44+CD133+ (○) versus ALDHlowCD44low/− CD133− (•) cells isolated from the MDA‐MB‐468 cell line (right panel). Data are presented as the mean ± S.E.M. *= significantly different tumour size than respective ALDHlowCD44low/− subsets at the same time‐point (P < 0.05). (B) Quantitative analysis of spontaneous lung metastasis tumour burden (mean% of lung occupied by tumour) following mammary fat pad injection of ALDHhiCD44+CD24− versus ALDHlowCD44low/−CD24+ cells isolated from the MDA‐MB‐231 cell line (left panel); and ALDHhiCD44+CD133+ versus ALDHlowCD44low/− CD133− cells isolated from the MDA‐MB‐468 cell line (right panel). Data are presented as the mean ± S.E.M. δ= significantly different than respective ALDHlowCD44low/− subsets (P < 0.05). (C) Incidence of spontaneous metastatic growth in lung and extrapulmonary tissues following mammary fat pad injection of ALDHhiCD44+CD24− versus ALDHlowCD44low/−CD24+ cells isolated from the MDA‐MB‐231 cell line (left panel); and ALDHhiCD44+CD133+ versus ALDHlowCD44low/− CD133− cells isolated from the MDA‐MB‐468 cell line (right panel).
In order to compare tumorigenicity and metastatic ability in a more clinically relevant model system, we next used a spontaneous model of metastasis involving orthotopic injection of breast cancer cells into the mammary fat pad. We observed that ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells demonstrated enhanced primary tumour growth relative to respective ALDHlowCD44low/− cell subsets (Fig. 8A). Spontaneous metastatic tumour burden in the lung was significantly higher in mice injected with ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells versus respective ALDHlowCD44low/− cell subsets (Fig. 8B) (P < 0.05). Interestingly, we did not observe a significant correlation between primary tumour size and metastatic burden in the lungs of individual mice, either in the ‘stem‐like’ populations (ALDHhiCD44+CD24− and ALDHhiCD44+CD133+) (R 2= 0.365) or in the respective ALDHlowCD44low/− populations (R 2= 0.530). Similar to the observations following tail vein injection, although all subpopulations of cells were able to establish themselves in the lung (Fig. 8C), only ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) had an enhanced capacity to maintain that growth into larger metastases (Fig. 8B), and to spontaneously metastasize to extrapulmonary organs such as the pancreas and spleen (Fig. 8C).
Discussion
The majority of breast cancer deaths occur as a result of metastatic disease rather than from the effects of the primary tumour. The inefficiency of the metastatic process, the inherently heterogeneous nature of solid tumours, and the influence of the tumour microenvironment dictate that only a small subset of cells can successfully navigate the metastatic cascade and eventually re‐initiate tumour growth to form life‐threatening metastases [1]. Although it has been speculated that this subset of cells may be CSCs, most studies to date have focused on the role of CSCs in primary tumour growth rather than on their potential contribution to metastatic behaviour. Furthermore, the challenges of obtaining large numbers of primary cells from patient samples combined with the technical complexity of studying metastasis of primary cells in vivo suggests that suitable alternative model systems need to be developed and validated in order to gain a greater insight into the role of stem‐like cells in metastasis.
In the present study, we demonstrated that commonly studied human breast cancer cell lines contain subpopulations of stem‐like cells based on both putative CSC marker expression (CD44/CD24 and CD133), and functional stem cell properties (enhanced ALDH activity). Furthermore, the ‘stem‐like’ cell content of the cell lines seemed to be associated with their aggressiveness (i.e. the more aggressive/metastatic the cell line, the greater the CD44+ and/or ALDHhi cell content). Our novel findings also demonstrated that ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) cells isolated from human breast cancer cell lines display significantly enhanced malignant/metastatic behaviour compared to respective ALDHlowCD44low/− subsets, including increased in vitro growth and colony forming ability in an anchorage‐independent environment and increased in vitro adhesion, migration and invasion. Moreover, when the different cell populations were isolated and injected into the tail vein or mammary fat pad of NOD/SCID‐IL2Rγ null mice, the ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) populations showed increased tumorigenicity, increased metastatic growth in the lungs and extrapulmonary metastases, whereas the respective ALDHlowCD44low/− populations showed minimal metastatic growth in vivo.
Our findings are supported by two recently published studies that also demonstrate a potential link between stem‐like cancer cells and successful metastatic behaviour [8, 41]. During the course of carrying out the present study, Yu et al. (2007) published findings demonstrating that stem‐like cells isolated from the SKBR3 human breast cancer cell line based on their mammosphere forming ability were more metastatic in vivo than unsorted parental SKBR3 cells [41]. In pancreatic cancer, Hermann et al. (2007) identified a subpopulation of cells within a CD133+ cell population that expressed the chemokine receptor CXCR4 [8]. In concert with its chemokine ligand SDF‐1, CXCR4 has been shown to play a key role in both normal stem cell migration and homing, as well as cancer cell migration and metastasis [42]. When the CXCR4+ pancreatic CSC subpopulation was eliminated in various highly metastatic pancreatic cancer cell lines, the cells could still efficiently form primary tumours but were no longer able to metastasize [8]. This suggests that the CSC population, at least in pancreatic cancer, is responsible for metastasis, but only because of a small subgroup of CD133+ CXCR4+ CSCs. Interestingly, both MDA‐MB‐231 and MDA‐MB‐468 cell lines demonstrate moderate and uniform expression of CXCR4 (Fig. S3), suggesting that CXCR4 may also be important for metastasis of stem‐like cells in breast cancer.
Conventionally, CSC populations have been isolated from solid human tumours based on cell surface expression of markers such as CD133 and CD44 [6, 7, 8, 9, 10, 11]. CD133 is a marker expressed by many types of normal stem cells, including neural and haematopoietic stem cells [9, 11], and has been shown to play a role in stem cell migration and asymmetric division [43, 44]. CD44 is a cell surface receptor for hyaluronic acid, and is involved in cell adhesion, migration and metastasis of cancer cells [23]. From a functional perspective, it is therefore not surprising that these markers would select for highly aggressive tumour cells. However, although defined cell surface markers have long been used to reliably isolate normal stem cells of various lineages, the inherent genetic instability of solid cancers suggests that it may be problematic to rely on cell surface marker expression alone to prospectively identify and isolate CSCs [20, 21, 22, 23]. Self‐protection is a key property of normal stem cells, and is of vital importance for protecting and maintaining the stem cell pool throughout the lifespan of an organism. ALDH activity is important for this self‐protection through its role in retinoic acid signalling and oxidation of intracellular aldehydes [24, 45], and thus provides a means of identifying and isolating stem‐like cancer cells based on a conserved stem/progenitor cell function.
When we analysed ALDH activity in human breast cancer cell lines, the pattern of ALDH activity in different cell lines often corresponded to their observed pattern of CSC marker expression. For example, in the case of the moderately metastatic MDA‐MB‐231 and weakly metastatic MDA‐MB‐468 cell lines (where subpopulations were evident in the marker expression analysis), two distinct subpopulations were also observed with respect to ALDH activity. Similarly, non‐metastatic MCF‐7 cells did not contain a detectable CD44+CD24− or CD133+ subpopulation and showed minimal increase in ALDH activity. There is evidence to support the idea that isolating CSC‐like cells by both marker expression and ALDH activity is most accurate. When Ginestier et al. (2007) isolated stem‐like cells from primary human breast tumours using both ALDH activity and CD44+CD24−, the ALDHhiCD44+CD24− population was more tumorigenic than populations identified by either marker expression or ALDH activity alone. Interestingly, the overlap between CSC marker expression (CD44+CD24−) and high ALDH activity in primary tumours was observed to be quite small (∼1%) [32]. In the present study, we investigated two distinct subpopulations of ‘stem‐like’ cells; ALDHhiCD44+CD24− cells isolated from the moderately metastatic MDA‐MB‐231 cell line and ALDHhiCD44+CD133+ cells isolated from the weakly metastatic MDA‐MB‐468 cell line. Although both populations behaved in a similar manner from the point of view of having significantly enhanced malignant and metastatic ability relative to their respective ALDHlowCD44low/− subsets, the in vitro and in vivo differences in growth‐related functional behaviour of ALDHhiCD44+CD133+ and ALDHlowCD44low/−CD133− subsets were more subtle than those between ALDHhiCD44+CD24− and ALDHlowCD44low/−CD24+ subsets. These may simply be cell‐line specific differences, or they may be related to the differential use of CD133 versus CD24 to refine the ALDH isolation procedure. This is supported by a recent study by Shmelkov et al. (2008), which demonstrated that CD133 may not be restricted to stem cells and that both CD133+ and CD133− metastatic colon cancer cells can initiate tumours in vivo[46]. This, combined with the findings of our study and those of Ginestier et al. (2007) [32], suggests that there is heterogeneity even within the CSC population of breast tumours, and that ALDHhiCD44+ cells (and ALDHhiCD44+CD24− cells in particular) may represent the most aggressive and malignant population within the CSC pool.
When the in vitro proliferative capacity of ALDHhiCD44+CD24− (MDA‐MB‐231) and ALDHhiCD44+CD133+ (MDA‐MB‐468) versus ALDHlowCD44low/− populations was compared in this study, it was observed that the ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ populations demonstrated enhanced growth. Similarly, in vivo studies demonstrated that ALDHhiCD44+CD24− and ALDHhiCD44+ CD133+ showed enhanced primary tumour growth in the mammary fat pad relative to respective ALDHlowCD44low/–populations. Interestingly, although all cell subsets could establish themselves in the lung as very small micrometastases, again only the ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ cells had a significantly enhanced capacity for metastatic growth resulting in increased tumour burden in the lung and metastases to extrapulmonary organs such as spleen and pancreas. In the context of stem cell properties, these results were a bit surprising, because it is well established that the proliferation rate of primitive stem cells is lower than that of committed cells [43, 47, 48, 49]. However, ALDH has been shown to identify rapidly dividing cells that represent a progenitor cell population in human umbilical cord blood and bone marrow ([50] and our unpublished data), and this may also be true in the context of breast cancer. The studies described here suggest that both ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ breast cancer cells may in fact demonstrate a more proliferative progenitor‐like phenotype (rather than a quiescent stem cell phenotype), although further studies are required to investigate this idea in more detail.
In summary, the novel findings presented here indicate that ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ stem‐like cancer cells isolated from MDA‐MB‐231 and MDA‐MB‐468 human breast cancer cell lines demonstrate enhanced malignant/metastatic behaviour in vitro and in vivo, and suggest that breast cancer cell lines may provide a suitable model system for starting to investigate the role of stem‐like cells in metastasis. Furthermore, our study represents the first report that selection and isolation of stem‐like breast cancer cells on the basis of ALDH activity can enhance for functional cell properties that contribute to metastasis, including in vitro adhesion, migration and invasion, and in vivo growth in primary and secondary organ sites. Further elucidation of the mechanisms by which ALDHhiCD44+CD24− and ALDHhiCD44+CD133+ populations successfully metastasize and the translation of this knowledge into the clinic could have potentially important implications for the management and treatment of breast cancer.
Supporting information
Fig. S1 Compiled quantitative analysis of CD44+CD24‐ surface marker expression in human breast cancer cell lines. Human breast cancer cells (MDA‐MB‐435, MDA‐MB‐231, MDA‐MB‐468, MCF‐7) were incubated with an anti‐CD44 antibody (clone IM7) conjugated to fluorescein isothiocyanate (FITC) and an anti‐CD24 antibody (clone ML5) conjugated to phycoerytherin (PE), or appropriate FITC and PE‐conjugated IgG isotype controls. Representative dot plots from the flow cytometry analysis can be observed in Fig. 1. The percentage of cells with a CD44+CD24‐ phenotype was used to assess CD44/CD24 cell surface expression in each cell line. Data are presented as the mean ± S.E.M. * ∇ significantly larger CD44+CD24‐ population relative to all other cell lines (P < 0.01). γ ∇ significantly larger CD44+CD24‐ population relative to MDA‐MB‐468 and MCF‐7 cell lines (P < 0.01).
Fig. S2 Optimization and validation of the ALDEFLUOR® assay kit and flow cytometry protocol using human umbilical cord blood. Cord blood was assayed with ALDEFLUOR® assay kit (StemCell Technologies) as per the manufacturer's guidelines. As a negative control for all experiments, an aliquot of ALDEFLUOR®‐stained cord blood was immediately quenched with 1.5‐mM diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor. Samples were analysed using the green fluorescence channel (FL1) on a Beckman Coulter EPICS XL‐MCL flow cytometer, using the protocol set‐up and data acquisition template recommended by the manufacturer. (A) Representative dot plots of cord blood samples treated with the ALDH‐specific inhibitor DEAB (negative control). (B) Representative dot plots of ALDH activity in cord blood. Cells which fell within the circled regions represent subpopulations of cells with enhanced ALDH activity relative to the rest of the cell population.
Fig. S3 MDA‐MB‐231 and MDA‐MB‐468 human breast cancer cell lines express CXCR4. (A‐‐B) Representative flow cytometry histograms of (A) MDA‐MB‐231 cells and (B) MDA‐MB‐468 cells. Cells were incubated with an anti‐CXCR4 antibody (clone 12G5) conjugated to phycoerytherin (PE) (black profiles) or a PE‐conjugated IgG isotype control (white profiles). A minimum of 10,000 events were collected per sample.
Please note: Wiley‐Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item
Acknowledgements
We thank Michael Keeney, Kristin Chadwick and Krysta Levac for their invaluable advice and technical help with the flow cytometry/FACS experiments. We also thank Heather Broughton for her work in maintaining the mouse colony, and George Ormond for his assistance with data analysis. This work was supported by grants from the London Regional Cancer Program (A.L.A.); Krembil Foundation, Juvenile Diabetes Research Foundation and Ontario Research and Development Fund (D.A.H). A.K.C. is the recipient of a Research Fellowship from the Canadian Breast Cancer Foundation (Ontario Chapter), and a Canadian Institutes for Health Research Strategic Training Scholarship/Translational Breast Cancer Scholarship through the London Regional Cancer Program. A.L.A. is supported by the Imperial Oil Foundation.
References
- 1. Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer . 2002; 2: 563–72. [DOI] [PubMed] [Google Scholar]
- 2. Luzzi KJ, MacDonald IC, Schmidt EE, et al . Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol . 1998; 153: 865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weiss L. Metastatic inefficiency. Adv Cancer Res . 1990; 54: 159–211. [DOI] [PubMed] [Google Scholar]
- 4. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med . 1997; 3: 730–7. [DOI] [PubMed] [Google Scholar]
- 5. Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self‐renewal capacity. Nat Immunol . 2004; 5: 738–43. [DOI] [PubMed] [Google Scholar]
- 6. Al‐Hajj M, Wicha MS, Benito‐Hernandez A, et al . Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA . 2003; 100: 3983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collins AT, Berry PA, Hyde C, et al . Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res . 2005; 65: 10946–51. [DOI] [PubMed] [Google Scholar]
- 8. Hermann PC, Huber SL, Herrler T, et al . Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell . 2007; 1: 313–23. [DOI] [PubMed] [Google Scholar]
- 9. O’Brien CA, Pollett A, Gallinger S, et al . A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature . 2007; 445: 106–10. [DOI] [PubMed] [Google Scholar]
- 10. Schatton T, Murphy GF, Frank NY, et al . Identification of cells initiating human melanomas. Nature . 2008; 451: 345–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Singh SK, Hawkins C, Clarke ID, et al . Identification of human brain tumour initiating cells. Nature . 2004; 432: 396–401. [DOI] [PubMed] [Google Scholar]
- 12. Li F, Tiede B, Massague J, et al . Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res . 2007; 17: 3–14. [DOI] [PubMed] [Google Scholar]
- 13. Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J Cell Mol Med . 2008; 12: 374–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer‐initiating cells to radiation. J Natl Cancer Inst . 2006; 98: 1777–85. [DOI] [PubMed] [Google Scholar]
- 15. Ponti D, Costa A, Zaffaroni N, et al . Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res . 2005; 65: 5506–11. [DOI] [PubMed] [Google Scholar]
- 16. Sheridan C, Kishimoto H, Fuchs RK, et al . CD44+/CD24‐ breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res . 2006; 8: R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abraham BK, Fritz P, McClellan M, et al . Prevalence of CD44+/CD24‐/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res . 2005; 11: 1154–9. [PubMed] [Google Scholar]
- 18. Balic M, Lin H, Young L, et al . Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res . 2006; 12: 5615–21. [DOI] [PubMed] [Google Scholar]
- 19. Liu R, Wang X, Chen GY, et al . The prognostic role of a gene signature from tumorigenic breast‐cancer cells. N Engl J Med . 2007; 356: 217–26. [DOI] [PubMed] [Google Scholar]
- 20. Clarke MF, Dick JE, Dirks PB, et al . Cancer Stem Cells–Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res . 2006; 66: 9339–44. [DOI] [PubMed] [Google Scholar]
- 21. Fillmore C, Kuperwasser C. Human breast cancer stem cell markers CD44 and CD24: enriching for cells with functional properties in mice or in man Breast Cancer Res . 2007; 9: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Polyak K. Breast cancer stem cells: a case of mistaken identity Stem Cell Rev . 2007; 3: 107–9. [DOI] [PubMed] [Google Scholar]
- 23. Shipitsin M, Campbell LL, Argani P, et al . Molecular definition of breast tumor heterogeneity. Cancer Cell . 2007; 11: 259–73. [DOI] [PubMed] [Google Scholar]
- 24. Storms RW, Trujillo AP, Springer JB, et al . Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA . 1999; 96: 9118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Appel B, Eisen JS. Retinoids run rampant: multiple roles during spinal cord and motor neuron development. Neuron . 2003; 40: 461–4. [DOI] [PubMed] [Google Scholar]
- 26. Hess DA, Craft TP, Wirthlin L, et al . Widespread non‐hematopoietic tissue distribution by transplanted human progenitor cells with high aldehyde dehydrogenase activity. Stem Cells . 2008; 26: 611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hess DA, Meyerrose TE, Wirthlin L, et al . Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood . 2004; 104: 1648–55. [DOI] [PubMed] [Google Scholar]
- 28. Hess DA, Wirthlin L, Craft TP, et al . Selection based on CD133 and high aldehyde dehydrogenase activity isolates long‐term reconstituting human hematopoietic stem cells. Blood . 2006; 107: 2162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cheung AM, Wan TS, Leung JC, et al . Aldehyde dehydrogenase activity in leuk‐emic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia . 2007; 21: 1423–30. [DOI] [PubMed] [Google Scholar]
- 30. Matsui W, Wang Q, Barber JP, et al . Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res . 2008; 68: 190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pearce DJ, Taussig D, Simpson C, et al . Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells . 2005; 23: 752–60. [DOI] [PubMed] [Google Scholar]
- 32. Ginestier C, Hur MH, Charafe‐Jauffret E, et al . ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell . 2007; 1: 555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welch DR. Technical considerations for studying cancer metastasis in vivo . Clin Exp Metastasis . 1997; 15: 272–306. [DOI] [PubMed] [Google Scholar]
- 34. Price JE, Polyzos A, Zhang RD, Daniels LM. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res . 1990; 50: 717–21. [PubMed] [Google Scholar]
- 35. Rae JM, Creighton CJ, Meck JM, et al . MDA‐MB‐435 cells are derived from M14 melanoma cells–a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat . 2007; 104: 13–9. [DOI] [PubMed] [Google Scholar]
- 36. Rae JM, Ramus SJ, Waltham M, et al . Common origins of MDA‐MB‐435 cells from various sources with those shown to have melanoma properties. Clin Exp Metastasis . 2004; 21: 543–52. [DOI] [PubMed] [Google Scholar]
- 37. Allan AL, George R, Vantyghem SA, et al . Role of the integrin‐binding protein osteopontin in lymphatic metastasis of breast cancer. Am J Pathol . 2006; 169: 233–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schulze EB, Hedley BD, Goodale D, et al . The thrombin inhibitor Argatroban reduces breast cancer malignancy and metastasis via osteopontin‐dependent and osteopontin‐independent mechanisms. Breast Cancer Res Treat . 2007. [DOI] [PubMed] [Google Scholar]
- 39. Furger KA, Allan AL, Wilson SM, et al . Beta(3) integrin expression increases breast carcinoma cell responsiveness to the malignancy‐enhancing effects of osteopontin. Mol Cancer Res . 2003; 1: 810–9. [PubMed] [Google Scholar]
- 40. Ishikawa F, Yasukawa M, Lyons B, et al . Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood . 2005; 106: 1565–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu F, Yao H, Zhu P, et al . let‐7 regulates self renewal and tumorigenicity of breast cancer cells. Cell . 2007; 131: 1109–23. [DOI] [PubMed] [Google Scholar]
- 42. Kucia M, Reca R, Miekus K, et al . Trafficking of normal stem cells and metastasis of cancer stem cells involve similar mechanisms: pivotal role of the SDF‐1‐CXCR4 axis. Stem Cells . 2005; 23: 879–94. [DOI] [PubMed] [Google Scholar]
- 43. Beckmann J, Scheitza S, Wernet P, et al . Asymmetric cell division within the human hematopoietic stem and progenitor cell compartment: identification of asymmetrically segregating proteins. Blood . 2007; 109: 5494–501. [DOI] [PubMed] [Google Scholar]
- 44. Giebel B, Corbeil D, Beckmann J, et al . Segregation of lipid raft markers including CD133 in polarized human hematopoietic stem and progenitor cells. Blood . 2004; 104: 2332–8. [DOI] [PubMed] [Google Scholar]
- 45. Sladek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol . 2003; 17: 7–23. [DOI] [PubMed] [Google Scholar]
- 46. Shmelkov SV, Butler JM, Hooper AT, et al . CD133 expression is not restricted to stem cells, and both CD133 and CD133 metastatic colon cancer cells initiate tumors. J Clin Invest . 2008; 118: 2111–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brummendorf TH, Dragowska W, Zijlmans J, et al . Asymmetric cell divisions sustain long‐term hematopoiesis from single‐sorted human fetal liver cells. J Exp Med . 1998; 188: 1117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mayani H, Dragowska W, Lansdorp PM. Lineage commitment in human hemopoiesis involves asymmetric cell division of multipotent progenitors and does not appear to be influenced by cytokines. J Cell Physiol . 1993; 157: 579–86. [DOI] [PubMed] [Google Scholar]
- 49. Punzel M, Zhang T, Liu D, et al . Functional analysis of initial cell divisions defines the subsequent fate of individual human CD34(+)CD38(−) cells. Exp Hematol . 2002; 30: 464–72. [DOI] [PubMed] [Google Scholar]
- 50. Nagano M, Yamashita T, Hamada H, et al . Identification of functional endothelial progenitor cells suitable for the treatment of ischemic tissue using human umbilical cord blood. Blood . 2007; 110: 151–60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Compiled quantitative analysis of CD44+CD24‐ surface marker expression in human breast cancer cell lines. Human breast cancer cells (MDA‐MB‐435, MDA‐MB‐231, MDA‐MB‐468, MCF‐7) were incubated with an anti‐CD44 antibody (clone IM7) conjugated to fluorescein isothiocyanate (FITC) and an anti‐CD24 antibody (clone ML5) conjugated to phycoerytherin (PE), or appropriate FITC and PE‐conjugated IgG isotype controls. Representative dot plots from the flow cytometry analysis can be observed in Fig. 1. The percentage of cells with a CD44+CD24‐ phenotype was used to assess CD44/CD24 cell surface expression in each cell line. Data are presented as the mean ± S.E.M. * ∇ significantly larger CD44+CD24‐ population relative to all other cell lines (P < 0.01). γ ∇ significantly larger CD44+CD24‐ population relative to MDA‐MB‐468 and MCF‐7 cell lines (P < 0.01).
Fig. S2 Optimization and validation of the ALDEFLUOR® assay kit and flow cytometry protocol using human umbilical cord blood. Cord blood was assayed with ALDEFLUOR® assay kit (StemCell Technologies) as per the manufacturer's guidelines. As a negative control for all experiments, an aliquot of ALDEFLUOR®‐stained cord blood was immediately quenched with 1.5‐mM diethylaminobenzaldehyde (DEAB), a specific ALDH inhibitor. Samples were analysed using the green fluorescence channel (FL1) on a Beckman Coulter EPICS XL‐MCL flow cytometer, using the protocol set‐up and data acquisition template recommended by the manufacturer. (A) Representative dot plots of cord blood samples treated with the ALDH‐specific inhibitor DEAB (negative control). (B) Representative dot plots of ALDH activity in cord blood. Cells which fell within the circled regions represent subpopulations of cells with enhanced ALDH activity relative to the rest of the cell population.
Fig. S3 MDA‐MB‐231 and MDA‐MB‐468 human breast cancer cell lines express CXCR4. (A‐‐B) Representative flow cytometry histograms of (A) MDA‐MB‐231 cells and (B) MDA‐MB‐468 cells. Cells were incubated with an anti‐CXCR4 antibody (clone 12G5) conjugated to phycoerytherin (PE) (black profiles) or a PE‐conjugated IgG isotype control (white profiles). A minimum of 10,000 events were collected per sample.
Please note: Wiley‐Blackwell Publishing are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Supporting info item


