Abstract
Purpose
The study aims to isolate the culturable marine bacteria and to assess their potential as the bioremediation agent for petroleum hydrocarbons contamination in marine environment.
Methods
Bacteria isolates were obtained by repetitive streaks to obtain purified bacteria on Zobell marine agar plates before further analysis and culture through direct visualization on agar plates. Identification were conducted using 16S rDNA sequence which are compared using NCBI BLAST and, combined with phenotypic and phylogenetic data. The potential use of the selected bacteria was tested by culturing them with two carbon sources i.e., glucose and crude oil.
Result
Fifty-one culturable marine hydrocarbonoclastic bacteria were isolated from the Lombok Strait (LS-3, LS-13, LS-14, LS-15, LS-16 and LS-20) and Indian Ocean (IO-1, IO-6, IO-8, IO-19, IO-24 and IO-25). Twelve isolates were found to degrade crude oil efficiently at a >2% concentration and to grow with crude oil as their sole carbon and energy source. These 12 strains belong to the genus Bacillus, which is well known to produce surface active agents, and the oil displacement assay indicated the production of these agents by these strains. Within the genera Bacillus, five species (Bacillus flexus, B. methylotrophicus, B. aquimaris, B. horikoshii, and B. thioparans) were represented by the 12 identified strains.
Conclusion
Selected strains from the Lombok Strait and Indian Ocean were capable of degrading crude oil (2% v/v) by 43.9–71.9% over 14 days. These results are important for marine bioremediation in Indonesia, which often faces risks of oil spill contamination and disaster.
Keywords: Earth sciences, Environmental science, Microbiology
1. Introduction
Petroleum hydrocarbon contamination in many ecosystems is a major concern in Indonesia. At least 50 oil spills in Indonesian marine waters have been recorded from 1975 to 2017 (Darmayati, 2009). Research has been conducted to isolate cultivable and viable cells corresponding to strains capable of degrading polycyclic aromatic hydrocarbons (PAHs) compounds. Such research is considered a response to environmental pollution, e.g., oil spills, and is conducted in collaboration with the petroleum industry. Nogueira et al. (2015) suggested that the anthropogenic impacts of eutrophic conditions, harsh environments and other forms of pollution on mangroves can alter the relative abundance and community structure of microbes in sedimentary matrices.
In Indonesia, the application of bioremediation for oil and gas waste is underdeveloped. Feliatra (1999) reported the isolation of Acinetobacter, Arthrobacter, Micrococcus and Bacillus from the Malacca Strait. A similar study was performed in the Makassar Strait and Java Sea by Darmayati (2009). The enumeration, isolation and identification of oil-degrading bacteria by conventional methods showed that these bacteria, including Aeromonas sp., Pseudomonas sp., Bacillus sp., B. megaterium and Corynebacterium sp., were present in the marine environment. Biomolecular approaches have been used for the isolation and identification of oil-degrading bacteria in Indonesia since 2005. Darmayati et al. (2008) isolated 131 different hydrocarbonoclastic bacterial strains from Jakarta Bay and Seribu Island. Yuliani et al. (2012) also reported five marine bacterial isolates from Indonesian territory that had a high potential to degrade one group of compounds in crude oil, namely, PAHs. These compounds have gained public attention due to their toxicity and carcinogenicity. In a recent study, Syakti et al. (2013) isolated six PAH-degrading bacteria from mangrove habitats, including Bacillus aquimaris, B. megaterium, B. pumilus, Flexibacteraceae bacterium, Halobacillus trueperi, and Rhodobacteraceae bacterium. In addition, 18 PAHs were detected in five commercially valuable squid species from the Atlantic, Indian and Pacific Oceans, indicating the presence of PAHs in the water (Gomes et al., 2013). Many PAH-degrading bacteria are found in environments impacted by oil discharge.
Indonesia is a potential resource for megadiverse strains of bacteria that can potentially degrade PAHs because a major fraction of the world's crude oil is shipped along oil tanker routes that cross Indonesian water. In collaboration with the Institute for Marine Research and Observation, Ministry of Marine Affairs and Fisheries (BPOL-KKP), Republic of Indonesia, i.e., the Indian Ocean (IO) and Lombok Strait (LS), during a cruise using Baruna Jaya Research Vessels IV and VIII, the present study has been conducted. The aim of the study is to isolate bacteria from Indonesian sea water in the Lombok Strait and Indian Ocean particularly in Indonesian sector. The culturable isolates were then tested using a crude oil as contaminant model in order to assess their hydrocarbonoclastic potential.
2. Materials and methods
2.1. Sampling site
The isolated bacteria were originally collected from two Indonesian bodies of water, the IO and LS. The sampling points were monitored regularly in the framework of the project “The Java Upwelling Variation Observation” (JUVO) and SITE (Fig. 1). A global positioning system (GPS) and geographic information system (GIS) were used to target the different sites. Water samples were collected with a water sampler (Shiptex; Wildco Inc., USA) at a depth of 5 m.
Fig. 1.
Sampling locations for the in situ study. A. Indian Ocean. B. Lombok Strait.
2.2. Isolation and screening of hydrocarbonoclastic bacteria
From each sampling station, 3 subsamples (1 L each) were taken and filtered to eliminate particles and suspended matter. Then, 1 mL of filtered sea water was inoculated into 10 mL tubes containing 9 mL filtered and sterilized sea water, and 4 dilution series were prepared for each subsample. After a 10,000-fold dilution, 1 mL sea water was plated onto full-strength Zobell Marine Agar 2216 (HiMedia, VWR) consisting of the following (in grams per liter of distilled water): agar (15 g L-1), peptone (5 g L-1), yeast extract (1 g L-1), ferric citrate (0.1 g L-1), NaCl (19.45 g L-1), MgCl2 (8.8 g L-1), Na2SO4 (3.24 g L-1), CaCl2 (1.8 g L-1), KCl (0.55 g L-1), NaHCO3 (0.16 g L-1), KBr (0.08 g L-1), SrCl2 (0.034 g L-1), H3BO3 (0.022 g L-1), Na2SiO3 (0.004 g L-1), NaF (0.0024 g L-1), N2H4O3 (0.0016 g L-1), and Na2HPO4 (0.008 g L-1).
The final pH was adjusted to 7.6 ± 0.2 at 25 °C. Next, 55.25 g agar was suspended in 1000 mL distilled water, and the marine agar mixture was boiled to completely dissolve the medium prior to sterilization using an autoclave at 15 lb pressure (121 °C) for 15 min. After cooling to 45–50 °C, the agar was poured into sterile Petri plates, and plates inoculated with subsamples were incubated at 25 °C in a dark incubator room. Repetitive streaks were conducted to obtain purified bacteria on Zobell marine agar plates before further analysis and culture. The morphology of isolated bacterial colonies was determined by direct visualization on agar plates.
2.2.1. Oil displacement assay
Moreover, bacterial selection was performed by spreading enriched pure cultures diluted 10-fold onto agar plates coated with a layer of crude oil. Colonies producing clear zones were scored positive, picked from the plates, and purified again by repetitive streaking on agar plates. The inocula was incubated for 48 h at 25 °C. A positive reaction in terms of potential degradation was indicated by the formation of a clear zone around the colony or a change in color due to the transformation of crude oil based on strain selection. Fig. 2 shows the examples of the clear zone assays.
Fig. 2.
Example oil displacement assays. Positive results (A, B, C, D) and a negative result (E). A = IO-4, B=IO-25, C = LS-20, D = LS-13, E = IO-30.
2.3. Colony morphological characteristics
All visible masses of microorganisms are defined as a colony when they originate from a single mother cell. In this study, the key morphological characteristics of the bacterial colonies were described in terms of shape (including form, elevation and margin), texture and pigmentation. Form refers to the shape of the colony, i.e., coccus (spherical), rod (bacilli), or spiral (twisted). Elevation refers to the side profile, which can be flat, raised, umbonate, crateriform, convex, or pulvinate, while the margin or edge of the colony can be described as entire (smooth), irregular, undulated (wavy), filiform, etc.
The texture of the colony surface can appear shiny and smooth, dull, veined, rough or glistening. Some bacteria produce pigments, including green, red, yellow, white, and purple, and some are opaque (not transparent). The KOH test can quickly distinguish between gram-negative and gram-positive bacteria as a complement to Gram staining. KOH dissolves the thin peptidoglycan layer in the cell wall of gram-negative bacteria, resulting in cell wall lysis and the release of DNA material, which makes the solution viscous; this change is easily recognized when the solution is touched with a plastic loop. Gram-positive bacteria are not affected by KOH because they have a thicker peptidoglycan layer in the cell wall.
2.4. Media and enrichment of pure cultures of isolated bacteria
Selected bacteria were grown aerobically in ONR7 media containing 22.9 g NaCl, 3.99 g Na2SO4, 0.72 g KCl, 82 mg NaBr, 31 mg NaHCO3, 27 mg H3BO3, 2.6 g NaF, 0.27 g NH4Cl, 83 mg Na2HPO4, 1.3 g TAPSO, 11.18 g MgCl2, 1.46 g CaCl2, 24 mg SrCl2, and 2 mg FeCl2 and were enriched with crude oil as the sole carbon and energy source to a final concentration of 0.4 mg L-1 for two weeks. The flask was shaken on an orbital shaker at room temperature (25 ± 2 °C) at a speed of 150 rpm. After a two-week incubation, an aliquot of 5 mL of the enriched culture was transferred into a 250 mL conical flask containing 45 mL fresh ONR7+crude oil media. This step was repeated three times to enrich for crude oil-degrading bacteria.
2.5. Bacterial growth kinetics
This assay was conducted for only the selected strains from the IO to obtain data on bacterial growth kinetics. The IO strains were cultured using two different carbon sources, i.e., glucose and crude oil. Cultures in a 150 mL flask containing Zobell liquid media were incubated at 37 °C in a reciprocal shaker (150 rpm). Growth kinetics were observed starting at 0 min and continuing for 5 days at 6-h intervals using the dry weight method (Devianto and kardena, 2010). Bacterial cell division is presented using Lineweaver-Burk transformation.
2.6. Biodegradation potential
The petroleum hydrocarbon biodegradation test was conducted as described by (Latha and Kalaivani, 2012; Shahaby et al., 2015). Briefly, biodegradation testing was performed by inoculating the selected isolates into an Erlenmeyer flask with 50 mL liquid ONR7 media containing petroleum (up to 5% (v/v)). The control flask contained the same contents without bacterial isolates and was used to control for the loss from abiotic factors. Then, the Erlenmeyer flasks were incubated at room temperature on a reciprocal shaker (150 rpm) for 10 days.
The analysis was conducted by adding 2.5 mL of 3 N HCl to the flasks and then extracting the culture using 30 mL n-heptane for 24 h using a magnetic stirrer. After 24 h, the extract was decanted for 3 h to separate the organic and aqueous phases. The organic phase containing residual crude oil was then filtered by absorbing the remaining water with Na2SO4. The filtrate was evaporated using a rotary evaporator before being weighed. Aliquots for the biodegradation test were removed at 0, 1, 3, 5, 7, and 14 days.
2.7. DNA extraction and purification
DNA was extracted from bacterial samples with a DNA isolation kit as described by Precigou et al. (2001). The DNA was verified by 1% agarose gel electrophoresis in Tris-acetate-EDTA (TAE) buffer. DNA solutions were stored at -20 °C. Cell pellets were resuspended in 570 μL TE buffer (10 mM Tris–HCl and 1 mM EDTA, pH 8) by repeated pipetting. Next, we added 30 μL 10% sodium dodecyl sulfate and 3 μL 20 mg mL-1 proteinase K (recombinant PCR grade; Roche Diagnostics, Indianapolis, IN), and the mixture was incubated for 1 h at 37 °C. Then, 100 μL 5 M NaCl and 80 μL CTAB/NaCl solution (10% CTAB in 0.7 M NaCl) were added, and the mixture was incubated at 65 °C for 10 min.
An equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) was added, and the mixture was centrifuged for 5 min in a microcentrifuge. Supernatants were transferred to fresh tubes, washed with 1 volume chloroform/isoamyl alcohol (24:1) and centrifuged for 5 min. Supernatants were again transferred to fresh tubes, and nucleic acids were precipitated with 0.6 volumes isopropanol overnight at -20 °C. The precipitates were collected by centrifugation for 15 min at 4 °C and then washed with 70% ethanol. Dry pellets were dissolved in 100 μL of TE buffer.
2.8. 16S rRNA gene amplification and DNA sequencing
Genomic DNA was isolated by previously described methods. The 16S ribosomal RNA (rRNA) gene was amplified by PCR using the primers 27f (5′-AGTTTGATCCTGGCTCAG-3′) and 1492 (5′-ACGGCTACCTTGTTACGACTT-3′). The PCR mixture contained 9.5 μL H2O, 1 μL bacterial genomic DNA template (50 ng/μL), 1 μl each primer (15 pmol), and 12.5 μL PCR mix. Amplifications were performed with the following conditions: initial denaturation at 80 °C for 5 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s; and a final 7-min extension at 72 °C. Amplification products were analyzed by electrophoresis in 2% agarose gels and ethidium bromide staining according to Magarvey et al. (2004).
2.9. Molecular taxonomy
The obtained 16S rDNA sequences were compared to those in the NCBI database using the BLASTN algorithm (Gupta et al., 2015a, Gupta et al., 2015b). A phylogenetic tree was constructed using the neighbor-joining method with Kimura two-state parameters and pairwise-deletion model analyses, which were implemented using Molecular Evolutionary Genetic Analysis (MEGA) software version 5.0. The resultant tree topologies were evaluated by bootstrap analysis based on 1,000 replicates. The numbers at the nodes represent the percentage levels of bootstrap support (%).
3. Results and discussion
3.1. Hydrocarbon-degrading strain selection
Table 1 shows the cultivable bacterial isolates that could survive and grow when crude oil was used as a carbon source. Fifty-one isolates (20 from the LS and 31 from the IO) were tested successively using 1% (v/v) and 2% (v/v) crude oil provided by an oil and gas company in Indonesia. Through visual observation and oil displacement assay, 12 cultures were capable of dispersing the crude oil into small droplets to form an emulsion, which consequently changed the color of the medium. When 1% crude oil was used, all isolates showed remarkable colony growth, whereas 19.6% failed to grow when the concentration of crude oil was increased to 2%. Interestingly, all LS strains were able to grow with this concentration showing their readily adaptation to high concentrations of petroleum hydrocarbons. These strains were subjected to oil spreading tests in order to estimate their potential biosurfactant activity which undergo related with their hydrocarbonoclastic activity based on the clear zone diameter (cm) and oil displacement area (cm2).
Table 1.
Selected hydrocarbon-degrading strains from the Lombok Strait and Indian Ocean.
| Origin Isolate Code |
Prescreening |
Clear Zone Diameter (cm) | Oil Displacement Area (cm2) | ||
|---|---|---|---|---|---|
| 1% | 2% | Oil Spreading | |||
| LS-1 | √ | √ | - | - | - |
| LS-2 | √ | √ | - | - | - |
| LS-3 | √ | √ | √ | 1.98 | 12.32++ |
| LS-4 | √ | √ | - | - | - |
| LS-5 | √ | √ | - | - | - |
| LS-6 | √ | √ | - | - | - |
| LS-7 | √ | √ | - | - | - |
| LS-8 | √ | √ | - | - | - |
| LS-9 | √ | √ | - | - | - |
| LS-10 | √ | √ | - | - | - |
| LS-11 | √ | √ | - | - | - |
| LS-12 | √ | √ | - | - | - |
| LS-13 | √ | √ | √ | 2.05 | 13.21++ |
| LS-14 | √ | √ | √ | 1.94 | 11.83++ |
| LS-15 | √ | √ | √ | 1.82 | 10.41++ |
| LS-16 | √ | √ | √ | 0.99 | 3.08+ |
| LS-17 | √ | √ | - | - | - |
| LS-18 | √ | √ | - | - | - |
| LS-19 | √ | √ | - | - | - |
| LS-20 | √ | √ | √ | 3.54 | 39.39+++ |
| LS-21 | √ | √ | - | - | - |
| IO-1 | √ | √ | √ | 0.56 | 3.53+ |
| IO-2 | √ | √ | - | - | - |
| IO-3 | √ | √ | - | - | - |
| IO-4 | √ | √ | - | - | - |
| IO-5 | √ | √ | - | - | - |
| IO-6 | √ | √ | √ | 1.75 | 10.15++ |
| IO-7 | √ | √ | - | - | - |
| IO-8 | √ | √ | √ | 0.75 | 4.46+ |
| IO-9 | √ | √ | - | - | - |
| IO-10 | √ | - | - | - | - |
| IO-11 | √ | √ | - | - | - |
| IO-12 | √ | - | - | - | - |
| IO-13 | √ | √ | - | - | - |
| IO-14 | √ | - | - | - | - |
| IO-15 | √ | √ | - | - | - |
| IO-16 | √ | √ | - | - | - |
| IO-17 | √ | √ | - | - | - |
| IO-18 | √ | √ | - | - | - |
| IO-19 | √ | - | √ | 1.04 | 5.15++ |
| IO-20 | √ | - | - | - | - |
| IO-21 | √ | - | - | - | - |
| IO-22 | √ | √ | - | - | - |
| IO-23 | √ | √ | - | - | - |
| IO-24 | √ | √ | √ | 2.02 | 12.67++ |
| IO-25 | √ | √ | √ | 1.34 | 6.03++ |
| IO-26 | √ | √ | - | - | - |
| IO-27 | √ | - | - | - | - |
| IO-28 | √ | - | - | - | - |
| IO-29 | √ | - | - | - | - |
| IO-30 | √ | - | - | - | - |
| Control | - | - | - | - | - |
Six isolates from the LS formed a clear zone area, i.e., LS-3, LS-13, LS -14, LS -15, LS-16 and LS-20, with broad a diameter ranging from 0.99 cm to 2.05 cm and a surface area ranging from 3.08 cm2 to 39.39 cm2. Six isolates, i.e., IO-1, IO-6, IO-8, IO-19, IO-24 and IO-25, had the capacity to form a clear zone and an oil displacement area varying from 0.56 cm to 2.02 cm and from 3.53 cm2 to 12.67 cm2, respectively. Such criteria have been used previously to assess the hydrocarbon-degrading activity of isolated strains (Pacwa-Płociniczak et al., 2016; Mohanram et al., 2016). The importance of the clear zone diameter and oil displacement area might be directly related to the production of secondary metabolites, such as biosurfactant (Eubeler et al., 2010; Hidayati et al., 2011). The control, which contained no bacteria, did not generate a clear zone or oil displacement area.
Moreover, Thavasi et al. (2011) demonstrated that if biosurfactant is found within bacterial cultures, it can create a layer of oil in the oil spreading test and an oil-free clear zone. The clear zone diameter indicates the surfactant activity, which is also known as the oil displacement activity. Roy et al. (2018) suggested that the size of the oil displacement zone reflects surfactant activity; the larger the zone is, the higher the surfactant activity. This zone is measured in biosurfactant units, and one unit is defined as the amount of surfactant that creates an oil displacement zone of 1 cm2. In fact, biosurfactant lowers the viscosity of the concentrated oil trapped in the water (Deepika et al., 2016), which functions as an emulsified surfactant, i.e., compounds that can reduce the charge at the interface of two liquids (interfacial tention). Emulsion increases the dispersion of hydrocarbon compounds in the water and expands the contact between oil and bacteria (Thavasi et al., 2011).
Recently, Cai et al. (2019) reported on 37 marine bacteria capable of destabilizing the oil-water emulsion in oily waste water. Increased biosurfactant production increases the solubility of oil in water. After the initial screening was performed, twelve isolates were considered for further biodegradation tests to determine their potential to degrade petroleum hydrocarbons at higher concentrations (5% v/v) over 14 days.
3.2. Growth kinetics of selected isolates
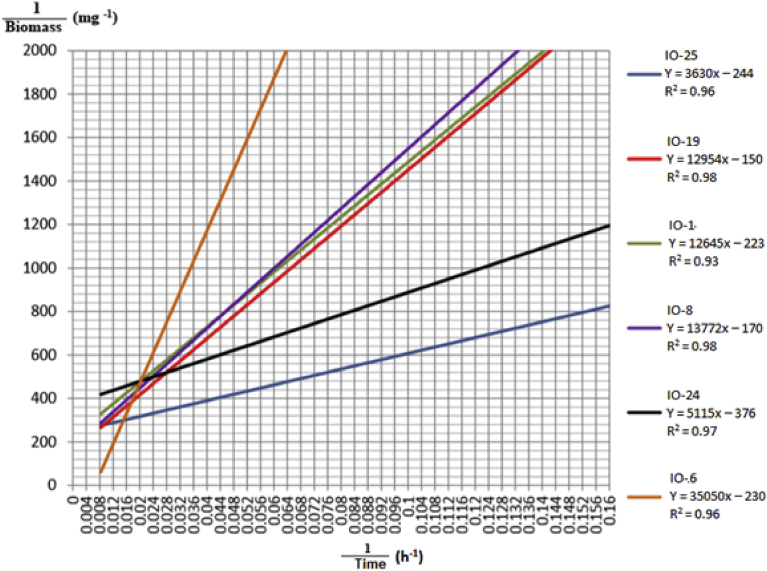
Growth kinetics were ascertained for only selected strains from the IO. The results showed (Fig. 3) that the generation time varied for each of the 6 strains using glucose as the carbon source. Bacterial cell division is presented using Lineweaver-Burk transformation. The generation times were as follows: IO-25, 13 h; IO-19, 14 h; IO-1 and IO-8, 21 h; IO-24, 19 h; and IO-6, 26 h. According to Syakti et al. (2013), the monosaccharide glucose is more easily metabolized by bacteria than other carbon sources, such as petroleum hydrocarbons. Prince et al. (2003) stated that the generation time differed among different generations of bacteria by a few minutes, hours or days, depending on the growth rate and environmental conditions such as temperature, pH, and nutrient availability, which all influence bacteria growth.
Fig. 3.
Bacterial growth kinetics of IO isolates on glucose as a carbon source using Lineweaver-Burk transformation.
We expected that the use of petroleum hydrocarbon as the carbon source would slow cell division. However, as shown in Fig. 4, the generation times were as follows: IO-25, 52 h (over four time as long); IO-19, 70 min; IO-1, 49 h; IO-24, 40 min; IO-24, 31 min; IO-8, 29 h and IO-6, 18h. These data reveal that each bacterial isolate is capable of utilizing petroleum as a carbon source for growth (Salleh et al., 2003). The bacterial cell division results confirmed that the growth kinetics of bacteria are slower with crude oil than with glucose as their energy and carbon source.
Fig. 4.
Bacterial growth kinetics of IO isolates on crude oil (2% v/v) as a carbon source using Lineweaver-Burk transformation.
3.3. Biodegradation test
The results showed that 12 of the 51 total bacterial isolates had the potential to degrade crude oil. For the LS isolates, LS-20 had the greatest potential (71.9%), followed by LS-13 (32.88%), LS-3 (30.24%), LS-14 (24.91%), LS-15 (23.35%) and LS-16 (16.64%). The IO isolates showed higher removal rates over the ten-day experiment, with the best performance by IO-25 (70.26%), followed by IO-8 (67.71%), IO-19 (65.52%), IO-1 (60.92%), IO-6 (59.45%) and IO-24 (26.14%). Fig. 5 shows the bioremoval of crude oil by each selected isolate over time, with the inclusion of an abiotic control. In general, petroleum degradation progressed steadily in the first three days, indicating that the isolates had already adapted to crude oil as their sole carbon and energy source.
Fig. 5.
Crude oil remaining (%) after degradation by selected strains adapted to 2% crude oil. Biodegradation tests were conducted in triplicate.
For instance, LS-20 showed to decrease the oil extent from 1 g to 0.22 g in the end of experimentation. With the remaining extent of oil that was 0.49 g on the seventh day, we could stated that the average percentage decrease in petroleum per day was 5.02%. The LS-3, LS-13 and LS-14 isolates showed a range of degradation activity, with the amount remaining varying from 10.3%-14.5% (day 3), 18.5%–24.8% (day 5), 35.7%–39.8% (day 7) and 53.5%–61.4% (day 14). The degradation activity of isolate LS-15 steadily decreased from 9.2% (day 3) to 18.7% (day 5), 41.6% (day 7), and 51.99% (day 14). The smallest decrease in concentration was observed for isolate LS-16, which at days 3–7 showed decreases in oil concentration of 8.3% and 37.7%, respectively. The arithmetical average decrease was 3.22% per day. Among the IO isolates, the best performance was noted for IO-25. IO-25 degraded 10.3% of the oil over the first three days and then degraded 24.7%, 55.8% and 70.3% of the oil at days 5, 7 and 14, respectively. IO-8, IO-19 and IO-24 showed relatively similar performances, ranging from 11.3% to 14.7% oil degradation over the first three days, 21.2%–26.4% by day 5, 35.7%–39.4% by day 7 and 63.9%–67.7% by day 14. Moreover, IO-6 showed decreased progress from day three (7.1%) to day five (15.5%), but degradation increased after day seven. The least degradation was observed for IO-24, which degraded crude oil by 43. 9% over the 14-day time course.
From Table 3, the biodegradation reaction order (n) of all the strains is rounded up to 1. For instance, during the logarithmic phase (from day 2 to day 14), the rates of hydrocarbon removal followed first-order kinetics, and the maximum and minimum slopes of the line represent the first-order kinetic constant k (Table 3). Similar results were found in previous studies on the bioremediation of petroleum-contaminated sediment (Birsch et al., 2017; Thessen and North, 2017). The reaction rate can also be calculated integrally by using the actual reaction order, and this kinetics approach to determining the biodegradation rate of crude oil for each bacteria yielded significantly different values, with IO-25, IO-24, and LS-15 most likely able to degrade the crude oil faster than the others.
Table 3.
Kinetics of crude oil biodegradation by the selected strains.
| Strains | Maximum Slope |
Minimum Slope |
n | k | Biodegradation Kinetics* |
|---|---|---|---|---|---|
| LS-3 | -11.734 | -11.578 | 1.002 | -0.066 | Ct−0.02 = C0−0.02 - kt |
| LS-13 | -12.053 | -11.494 | 0.989 | -0.070 | Ct0.11 = C00.11 - kt |
| LS-14 | -11.263 | -11.002 | 1.033 | -0.059 | Ct−0.33 = C0−0.33 - kt |
| LS-15 | -11.039 | -8.583 | 0.989 | -0.054 | Ct0.11 = C00.11 - kt |
| LS-16 | -9.999 | -9.977 | 0.986 | -0.017 | Ct0.04 = C00.04 - kt |
| LS-20 | -14.624 | -14.557 | 0.993 | -0.094 | Ct0.07 = C00.07 - kt |
| IO-1 | -12.300 | -12.143 | 0.998 | -0.071 | Ct0.02 = C00.02 - kt |
| IO-6 | -11.507 | -11.206 | 0.996 | -0.066 | Ct0.04 = C00.04 - kt |
| IO-8 | -11.757 | -11.469 | 1.024 | -0.064 | Ct−0.024 = C0−0.024 - kt |
| IO-19 | -12.949 | -12.695 | 1.009 | -0.079 | Ct−0.09 = C0−0.09 - kt |
| IO-24 | -9.648 | -9.431 | 0.974 | -0.044 | Ct0.26 = C00.26 - kt |
| IO-25 | -15.506 | -15.088 | 1.028 | -0.097 | Ct0.28 = C00.28 - kt |
For most of the experiments, the crude oil initially coalesced and formed a separate layer on the surface of the liquid culture media. After one day, due to mechanical shaking, the oil gradually formed droplets in culture. The formation of dispersed oil and emulsion was observed after day three, suggesting the presence of biosurfactant produced by the bacteria (Hidayati et al., 2011; Syakti et al., 2013). This surfactant will further increase the rate of transfer of hydrocarbon compounds into microorganisms (Yoon et al., 2003).
The extent of biodegradation depends on several factors, including the target compound. According to Sierra-García et al. (2014), aliphatic hydrocarbons are more easily degraded than aromatic hydrocarbons. Generally, short-chain hydrocarbons are degraded before more complex hydrocarbons. Another factor is the bacteria; Shekhar et al. (2015) and Yadav et al. (2016) stated that the microorganism's ability to produce biosurfactant is related to the production of enzymes capable of breaking down complex organic compounds. The process of biodegradation is also influenced by abiotic factors, including pH, temperature, and nutrient and oxygen availability. These abiotic components can be adjusted to optimize bacterial growth, resulting in faster biodegradation (Chakraborty et al., 2012; Dzionek et al., 2016).
3.4. Identification of culturable marine hydrocarbonoclastic bacteria
Table 2 shows that the LS isolates (LS-3, LS -13, LS-14, LS-15, LS-16, and LS-20) have six different morphologies, including smooth (entire) and undulate shape characteristics. Colonies were found to be several colors, including yellow, opaque, white, and purple, with two types of elevation, i.e., convex and raised. In terms of the surface, colonies were glistening and smooth. Our data suggest that the isolates with similar morphology likely belong to the same genera. Regarding the IO isolates, there were three colony colors, including bright yellow, bright white and brownish white. IO-1 was bright yellow, IO-3 and IO-25 were bright white, and the IO-8, IO-19, and IO-24 colonies were opaque. All isolates formed circular colonies with edges. The elevation of the colonies from all isolates was either convex (IO-1, IO-8 and IO-19) or raised (IO-3, IO-24, and IO-25). The observed morphological characteristics indicate possible relationships among the bacterial isolates.
Table 2.
Morphological characteristics and colony shape of the selected marine hydrocarbonoclastic strains. * All isolates are gram positive.
| Origin Isolate Code |
Cell Form |
Color | Elevation | Margin | Surface | Gram* +/- |
|---|---|---|---|---|---|---|
| LS-3 | coccus | opaque | convex | entire | glistening | + |
| LS-13 | rod | yellow | raised | undulate | glistening | + |
| LS-14 | coccus | opaque | raised | entire | glistening | + |
| LS-15 | coccus | white | raised | entire | glistening | + |
| LS-16 | coccus | purple | raised | undulate | glistening | + |
| LS-20 | coccus | white | convex | entire | glistening | + |
| IO-1 | small coccus | bright yellow | convex | entire | glistening | + |
| IO-6 | coccus | bright white | raised | entire | smooth | + |
| IO-8 | coccus | opaque | convex | entire | smooth | + |
| IO-19 | rod | opaque | convex | entire | glistening | + |
| IO-24 | coccus | opaque | raised | entire | glistening | + |
| IO-25 | rod | bright white | raised | entire | glistening | + |
All isolates were categorized as gram-positive bacteria based on a KOH (3%) test while other studies reported in Southern Ocean found the culturable marine bacteria were dominated by gram-negative (Gupta et al., 2015a, 2015b).
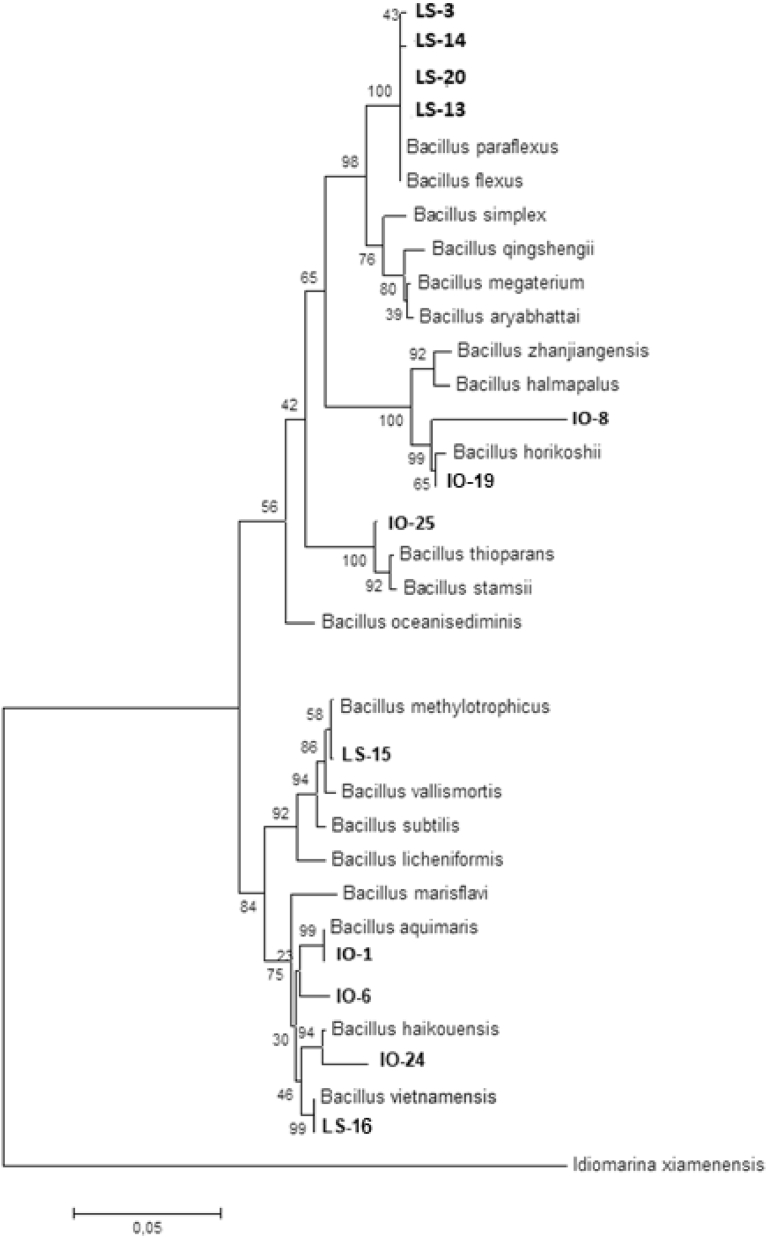
Twelve selected isolates were recultured for purification and identification. After purification, DNA extraction and sequencing, each DNA sequence was analyzed using GenBank, and nucleotide homology is shown in Table 3. Fig. 6 shows based on molecular identification and BLAST, the isolates LS-3, LS-13, LS-14 and LS-20 had similarity with B. flexus (98%–99%), LS-15 with B. methylotrophicus (99%), and LS-16 with B. vietnamensis (96%) and B. aquimaris (96%). Concerning the IO isolates, we identified IO-4 as B. aquimaris (98%), IO-21 as B. thioparans (100%), and IO-23 and IO-39 as B. horikoshii (99% and 98%, respectively). The identity of two other isolates (IO-6 and IO-2) could not be determined due to poor DNA and RNA quantification and purity (see Table 4).
Fig. 6.
Phylogenetic tree. The history of evolution was inferred using the neighbor-joining method. The percentage of trees that replicate the associated taxa are clustered together in the bootstrap (1,000 replicates) and displayed at the branches. Evolutionary distances were calculated using Kimura's method. This analysis was performed using a 16-nucleotide sequence. All positions containing gaps or missing data were eliminated. A total of 511 positions are in the final dataset. Evolution analysis was performed using MEGA5.
Table 4.
Nucleotide homology and identification of hydrocarbonoclastic marine bacteria.
| Origin Isolate Code |
Identification | Homology | GenBank Accession Number |
|---|---|---|---|
| LS-3 | Bacillus flexus | 99% | NR_113800.1 |
| LS-13 | Bacillus flexus | 98% | NR_113800.1 |
| LS-14 | Bacillus flexus | 99% | NR_113800.1 |
| LS-15 | Bacillus methylotrophicus | 99% | NR_116240.1 |
| LS-16 | Bacillus aquimaris | 96% | NR_025241.1 |
| LS-20 | Bacillus flexus | 99% | NR_113800.1 |
| IO-1 | Bacillus aquimaris | 98% | NR_025241.1 |
| IO-6∗ | - | - | - |
| IO-8 | Bacillus horikoshii | 98% | NR_040852.1 |
| IO-19 | Bacillus horikoshii | 99% | NR_040852.1 |
| IO-24∗ | - | - | - |
| IO-25 | Bacillus thioparans | 100% | NR_043762.1 |
DNA sequencing data for strains IO-6 and IO-24 were poor, and these strains were thus not identified.
According to Drancourt et al. (2000) and Agogué et al. (2005), homology greater than 97% indicates the same species, homology within 93%–97% indicates the same genus but a different species, and homology below 93% likely indicates a new species. Isolates LS-3, LS-13, LS-14, LS-20 and LS-15 had nucleotide homology greater than 97%, but they are unlikely to be the same strains. LS-16 is likely in the same genus as the other isolates but is not B. vietnamensis or B. aquimaris because the nucleotide homology was 93–97%. The neighboring relationships among the bacteria are shown in a phylogenetic tree (Fig. 6). Several previous studies have successfully isolated biosurfactant-producing bacteria. For instance, Hassanshahnian (2014) isolated 7 strains belonging to Shewanella alga, S. upenei, Vibrio furnissii, Gallaecimonas pentaromativorans, Brevibacterium epidermidis, Psychrobacter namhaensis and Pseudomonas fluorescens from a petrochemicals plant in the Persian Gulf. In another study, Nkem et al. (2016) isolated two indigenous hydrocarbon-degrading bacteria, Acinetobacter baumannii and Cellulosimicrobium cellulans, from a tar ball found on Rhu Sepuluh beach, Terengganu, Malaysia.
Fig. 3 shows that the bacterial isolates LS-3, LS-13, LS-14 and LS-20 are 98%–99% similar to B. flexus, gram positive, rod shaped, motile facultative alkaliphile that produces white colonies (Tambekar et al., 2015). Wang and Shao (2012) demonstrated that B. flexus isolated from the Atlantic Ocean was able to degrade paraffin, an alkane. Isolate LS-15 corresponded to B. methylotrophicus with 99% similarity; B. methylotrophicus are gram-positive bacteria that form endospores, are aerobic and motile, and produce tan white colonies. The physico-chemical parameters used to culture B. methylotrophicus, including temperature (30 °C) and pH (8), have been described in a previous study (Madhaiyan et al., 2010). Regarding the hydrocarbonoclastic potential, Chandankere et al. (2014) successfully isolated B. methylotrophicus, which produces biosurfactant for the degradation of crude oil.
Among the IO isolates, the strain B. aquimaris (IO-1) is gram positive and rod shaped and grows in a broad range of temperatures. This strain has been found in both the LS and IO. B. aquimaris is capable of forming spores to survive in less favorable environmental conditions and can produce a variety of proteins useful for bioremediation (Yoon et al., 2003; Syakti et al., 2013, 2017). Fig. 4 shows that the isolates IO-8 and IO-19 have 98% and 99% homology with B. horikoshii, which are facultative anaerobic gram-positive bacteria. B. horikoshii can grow under alkaline conditions (pH 8–9.5) (Madhaiyan et al., 2010) and has potential for protease enzyme production for applications in the food, leather, bioremediation, detergent and pharmaceutical industries (Lu and Yi, 2009; Vermelho et al., 2012). The isolate IO-25 was identified with 100% sequence homology as B. thioparans, which can degrade petroleum. A previous study conducted by Rodríguez-Tirado et al. (2012) found that B. thioparans produces an emulsifying agent that is useful for the bioremediation of petroleum hydrocarbons, including heavy metals. Despite promising degradation capabilities during the 14-day experiment, we could not identify IO-6 (59.5%) and IO-24 (70.3%) due to the lack of a comparable gene sequence using BLAST. However, their morphological characteristics, test results, Gram staining and adaptation to 2% crude oil contamination suggest that these bacteria reside within the genus Bacillus.
4. Conclusion
In conclusion, among 51 isolates from Indonesia, i.e., the LS and IO, 12 had hydrocarbonoclastic properties. Using biomolecular methods, five distinct species of hydrocarbonoclastic marine bacteria and two species likely belonging to the Bacillus genera were identified. This study identified bacterial strains that can potentially remediate the marine environment. The strain B. flexus LS-20 and B. thioparans IO-25 may have good potential as bioremediation agents. Prior to the next application, cultivation conditions, i.e., optimum medium composition, temperature, pH, salinity, dissolved oxygen level, and light intensity, should be precisely studied to obtain optimal performance. Our results reinforce the potential of isolating valuable bioremediation agents from Indonesian marine environments to mitigate oil contamination on the Indonesian coast. Additionally, our results suggest that microbial biodegradation is a natural process involved in the remediation of marine environments.
Declarations
Author contribution statement
Agung Dhamar Syakti: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Priyati Lestari, Satya Simanora, Lilik Kartika Sari, Febrianti Lestari, Fadliyah Idris, Teguh Agustiadi, Syafsir Akhlus, Nuning Vita Hidayati, Riyanti Riyanti: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
Agung D. Syakti was supported by a Competitive Grant (HIKOM) from Ristekdikti for 2018 (1637/UN23.14/PN.01.00/2018).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We thank the Institute for Marine Research and Observation (BPOL)–Ministry of Marine Affairs and Fisheries of The Republic of Indonesia for cruises, sampling, the use of R/V Baruna Jaya IV (Indian Ocean) and R/V Baruna Jaya VIII (Lombok Strait) and excellent technical assistance. The study was supported by a Competitive Grant (HIKOM) from Ristekdikti for 2018 (1637/UN23.14/PN.01.00/2018) awarded to Dr. A.D. Syakti.
References
- Agogué H., Casamayor E.O., Bourrain M., Obernosterer I., Joux F., Herndl G.J., Lebaron P. A survey on bacteria inhabiting the sea surface microlayer of coastal ecosystems. FEMS Microbiol. Ecol. 2005;54:269–280. doi: 10.1016/j.femsec.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Birch H., Hammershoj R., Comber M., Mayer P. Biodegradation of hydrocarbon mixtures in surface waters at environmentally relevant levels – effect of inoculum origin on kinetics and sequence of degradation. Chemosphere. 2017;184:400–407. doi: 10.1016/j.chemosphere.2017.05.169. [DOI] [PubMed] [Google Scholar]
- Cai Q., Zhu Z., Chen B., Zhang B. Oil-in-water emulsion breaking marine bacteria for demulsifying oily wastewater. Water Res. 2019;149:292–301. doi: 10.1016/j.watres.2018.11.023. [DOI] [PubMed] [Google Scholar]
- Chakraborty R., Wu C.H., Hazen T.C. Systems biology approach to bioremediation. Curr. Opin. Biotechnol. 2012;23:483–490. doi: 10.1016/j.copbio.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Chandankere R., Yao J., Cai M., Masakorala K., Jain A.K., Choi M.M.F. Properties and characterization of biosurfactant in crude oil biodegradation by bacterium Bacillus methylotrophicus USTBa. Fuel. 2014;122:140–148. [Google Scholar]
- Darmayati Y. Development of oil bioremediation research on marine environment in Indonesia. J. Coast. Dev. 2009;12(3):105–110. [Google Scholar]
- Darmayati Y., Harayama S., Yamazoe A., Hatmanti A., Sulistiani, Nuchsin R., Kunarso D. Hydrocarbonoclastic bacteria from Jakarta bay and Seribu islands. Mar. Res. Indones. 2008;33(1):55–64. [Google Scholar]
- Deepika K.V., Kalam S., Ramu Sridhar P., Podile A.R., Bramhachari P.V. Optimization of rhamnolipid biosurfactant production by mangrove sediment bacterium Pseudomonas aeruginosa KVD-HR42 using response surface methodology. Biocatal. Agr. Biotechnol. 2016;5:38–47. [Google Scholar]
- Devianto L.A., Kardena E. Pengaruh Glukosa terhadap Produksi Biosurfaktan oleh Azotobacter vinelandii dan Pengaruh Biosurfaktan Terhadap Biodegradasi TPH oleh Konsorsium Bakteri Petrofilik. Biotechnology. 2010;8(1):1–10. [Google Scholar]
- Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J. Clin. Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzionek A., Wojcieszyńska D., Guzik U. Natural carriers in bioremediation: a review. Electron. J. Biotechnol. 2016;23:28–36. [Google Scholar]
- Eubeler J.P., Bernhard M., Knepper T.P. Environmental biodegradation of synthetic polymers II. Biodegradation of different polymer groups. TrAC - Trends in Anal. Chem. 2010;29:84–100. [Google Scholar]
- Feliatra. Identifikasi bakteri patogen (Vibrio sp.) di perairan Nongsa Batam Propinsi Riau. J. Nat. Indones. 1999;11(1):8–33. [Google Scholar]
- Gomes F., Oliveira M., Ramalhosa M.J., Delerue-Matos C., Morais S. Polycyclic aromatic hydrocarbons in commercial squids from different geographical origins: levels and risks for human consumption. Food Chem. Toxicol. 2013;59(2013):46–54. doi: 10.1016/j.fct.2013.05.034. [DOI] [PubMed] [Google Scholar]
- Gupta P., Agrawal H.K., Bandopadhyay R. Distribution pattern of bacteria in the two geographic poles and Southern Ocean from the reported 16S rDNA sequences. Curr. Sci. 2015;108:1926–1930. [Google Scholar]
- Gupta P., Balaji R., Parani M., Chandra T.S., Shukla P., Bandopadhyay R. Phylogenetic analysis and biological characteristic tests of marine bacteria isolated from Southern Ocean (Indian sector) water. Acta Oceanol. Sin. 2015;34:73–82. [Google Scholar]
- Hassanshahnian M. Isolation and characterization of biosurfactant producing bacteria from Persian Gulf (Bushehr provenance) Mar. Pollut. Bull. 2014;86(1-2):361–366. doi: 10.1016/j.marpolbul.2014.06.043. [DOI] [PubMed] [Google Scholar]
- Hidayati N.V., Hilmi E., Haris A., Effendi H., Guiliano M., Doumenq P., Syakti A.D. Fluorene removal by biosurfactants producing Bacillus megaterium. Waste Biomass Valorization. 2011;2:415–422. [Google Scholar]
- Latha R., Kalaivani R. Bacterial degradation of crude oil by gravimetric analysis. Analysis. 2012;3:2789–2795. [Google Scholar]
- Lu Y., Yi R. Bacillus horikoshii, a tetrodotoxin-producing bacterium isolated from the liver of puffer fish. Ann. Microbiol. 2009;59:453–458. [Google Scholar]
- Madhaiyan M., Poonguzhali S., Kwon S.W., Sa T.M. Bacillus methylotrophicus sp. nov., a methanol-utilizing, plant-growth-promoting bacterium isolated from rice rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2010;60:2490–2495. doi: 10.1099/ijs.0.015487-0. [DOI] [PubMed] [Google Scholar]
- Mohanram R., Jagtap C., Kumar P. Isolation, screening, and characterization of surface-active agent-producing, oil-degrading marine bacteria of Mumbai Harbor. Mar. Pollut. Bull. 2016;105:131–138. doi: 10.1016/j.marpolbul.2016.02.040. [DOI] [PubMed] [Google Scholar]
- Nkem B.M., Halimoon N., Yussof F. Md., Wan Johari W.,L., Zakaria M.P., Medipally S.R., Kannan N. Isolation, identification and diesel-oil biodegradation capacities of indigenous hydrocarbon-degrading strains of Cellulosimicrobium cellulans and Acinetobacter baumannii from tarball at Terengganu beach, Malaysia. Mar. Pollut. Bull. 2016;107(1):261–268. doi: 10.1016/j.marpolbul.2016.03.060. [DOI] [PubMed] [Google Scholar]
- Nogueira V.L.R., ocha L.L., Colares G.B., Angelim A.L., Normando L.R.O., Cantao M.E., Agnez-Lima L.F., Andreote F.D., Melo V.M.M. Microbiomes and potential metabolic pathways of pristine and anthropized Brazilian mangroves. Reg. Stud. Marine Sci. 2015;2:56–64. [Google Scholar]
- Pacwa-Płociniczak M., Płociniczak T., Iwan J., Żarska M., Chorążewski M., Dzida M., Piotrowska-Seget Z. Isolation of hydrocarbon-degrading and biosurfactant-producing bacteria and assessment their plant growth-promoting traits. J. Environ. Manag. 2016;168:175–184. doi: 10.1016/j.jenvman.2015.11.058. [DOI] [PubMed] [Google Scholar]
- Precigou S., Goulas P., Duran R. Rapid and specific identification of nitrile hydratase (NHase)-encoding genes in soil samples by polymerase chain reaction. FEMS Microbiol. Lett. 2001;204(1):155–161. doi: 10.1111/j.1574-6968.2001.tb10879.x. 2001. [DOI] [PubMed] [Google Scholar]
- Prince R.C., Lessard R.R., Clark R. Bioremediation of marine oil spills. Oil Gas Sci. Technol. 2003;58(4):463–468. [Google Scholar]
- Rodríguez-Tirado V., Green-Ruiz C., Gómez-Gil B. Cu and Pb biosorption on Bacillus thioparans strain U3 in aqueous solution: kinetic and equilibrium studies. Chem. Eng. J. 2012;181–182:352–359. [Google Scholar]
- Roy A., Dutta A., Pal S., Gupta A., Sarkar J., Chatterjee A., Saha A., Sarkar P., Sar P., Kazy S.K. Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Biores. Technol. 2018;253:22–32. doi: 10.1016/j.biortech.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Salleh A.B., Ghazali F.M., Rahman R.N.Z.A., Basri M. Bioremediation of petroleum hydrocarbon pollution. Indian J. Biotechnol. 2003;2:411–425. [Google Scholar]
- Shahaby A.F., Alharthi A.A., Tarras A.E.El. Bioremediation of petroleum oil by potential biosurfactant-producing bacteria using gravimetric assay. Int.J.Curr.Microbiol.App.Sci. 2015;4:390–403. [Google Scholar]
- Shekhar S., Sundaramanickam A., Balasubramanian T. Biosurfactant producing microbes and their potential applications: a review. Crit. Rev. Environ. Sci. Technol. 2015;45:1522–1554. [Google Scholar]
- Sierra-García I.N., Alvarez J.C., De Vasconcellos S.P., De Souza A.P., Dos Santos Neto E.V., De Oliveira V.M. New hydrocarbon degradation pathways in the microbial metagenome from brazilian petroleum reservoirs. PLoS One. 2014;9 doi: 10.1371/journal.pone.0090087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syakti A.D., Yani M., Hidayati N.V., Siregar A.S., Doumenq P., Sudiana I.M. The bioremediation potential of hydrocarbonoclastic bacteria isolated from a mangrove contaminated by petroleum hydrocarbons on the cilacap coast, Indonesia. Bioremed. J. 2013;17:11–20. [Google Scholar]
- Syakti A.D., Arofah N., Purnomowati R., Salim A. Sludge valorization from biofloc-based Aquaculture systems for bioremediation of crude oil-contaminated sediment. Waste Biomass Valorization. 2017;8:561–572. [Google Scholar]
- Tambekar D.H., Tambekar S.D., Borkar P.R., Laddha Y.P. Molecular characterization and detoxification of phenol by haloalkaliphilic bacterium Bacillus flexus. Int. J. Adv. Biotechnol. Res. 2015;6:232–237. [Google Scholar]
- Thavasi R., Jayalakshmi S., Banat I.M. Effect of biosurfactant and fertilizer on biodegradation of crude oil by marine isolates of Bacillus megaterium, Corynebacterium kutscheri and Pseudomonas aeruginosa. Biores. Technol. 2011;102:772–778. doi: 10.1016/j.biortech.2010.08.099. [DOI] [PubMed] [Google Scholar]
- Thessen A.N., North E.W. Calculating in situ degradation rates of hydrocarbon compounds in deep waters of the Gulf of Mexico. Mar. Pollut. Bull. 2017;122(1-2):77–84. doi: 10.1016/j.marpolbul.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Vermelho A.B., Supuran C.T., Guisan J.M. Microbial enzyme: applications in industry and in bioremediation. Enzym. Res. 2012;2012:980681. doi: 10.1155/2012/980681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Shao Z. Diversity of flavin-binding monooxygenase genes (almA) in marine bacteria capable of degradation long-chain alkanes. FEMS Microbiol. Ecol. 2012;80:523–533. doi: 10.1111/j.1574-6941.2012.01322.x. [DOI] [PubMed] [Google Scholar]
- Yadav A.K., Manna S., Pandiyan K., Singh A., Kumar M., Chakdar H., Kashyap P.L., Srivastava A.K. Isolation and characterization of biosurfactant producing Bacillus sp. from diesel fuel-contaminated site. Microbiology. 2016;85:56–62. [Google Scholar]
- Yoon J.H., Kim I.G., Kang K.H., Oh T.K., Park Y.H. Bacillus marisflavi sp. nov. and Bacillus aquimaris sp. nov., isolated from sea water of a tidal flat of the Yellow Sea in Korea. Int. J. Syst. Evol. Microbiol. 2003;53:1297–1303. doi: 10.1099/ijs.0.02365-0. [DOI] [PubMed] [Google Scholar]
- Yuliani H., Sahlan M., Hermansyah H., Wijanarko A. Selection and identification of polyaromatic hydrocarbon degrading bacteria. World Appl. Sci. J. 2012;20(8):1133–1138. [Google Scholar]