Abstract
Cyanogenic glycosides are defense compounds found in a wide range of plant species, including many crops. We demonstrate that the cyanogenic glucoside dhurrin, naturally found in sorghum, can be produced at high titers in Saccharomyces cerevisiae, constituting the first report of cyanogenic glycoside production in a microbe. Genetic modifications to increase the supply of the dhurrin precursor tyrosine enabled dhurrin production in excess of 80 mg/L. The dhurrin-producing yeast strain was used as a chassis to investigate previously uncharacterized enzymes identified close to the biosynthetic gene cluster containing the dhurrin pathway enzymes. This work shows the potential of heterologous expression in yeast to facilitate investigations of plant cyanogenic glycoside pathways.
Keywords: Dhurrin, Cyanogenic glycoside, Biosynthetic gene cluster, Plant natural product, Saccharomyces cerevisiae
Highlights
-
•
First production of cyanogenic glycosides in a microbe.
-
•
Strategies for optimizing production of cyanogenic glycosides.
-
•
Platform for rapidly characterizing the enzymes which constitute cyanogenic glycoside biosynthetic pathways.
1. Introduction
Plants produce a wide variety of natural products, including defense compounds which protect them against pathogens and herbivory. In crop plants, these compounds provide pest resistance but can also pose risks to human and animal health. One such class of compounds are cyanogenic glycosides, amino acid-derived plant natural products which defend against herbivory. When plant tissue is damaged, hydrolysis of cyanogenic glycosides by plant β-glycosidases produces an unstable cyanohydrin, which releases toxic cyanide on decomposition. Cyanogenic glycosides are found in a wide variety of plants, including many crop plants (Gleadow and Møller, 2014). In particular, the staple crop sorghum (Sorghum bicolor) produces a cyanogenic glycoside known as dhurrin. Cyanogenic glycosides enhance plant resistance to insects (Gleadow and Woodrow, 2002), and they can pose a risk to humans (Tokpohozin et al., 2016) and to livestock as well.
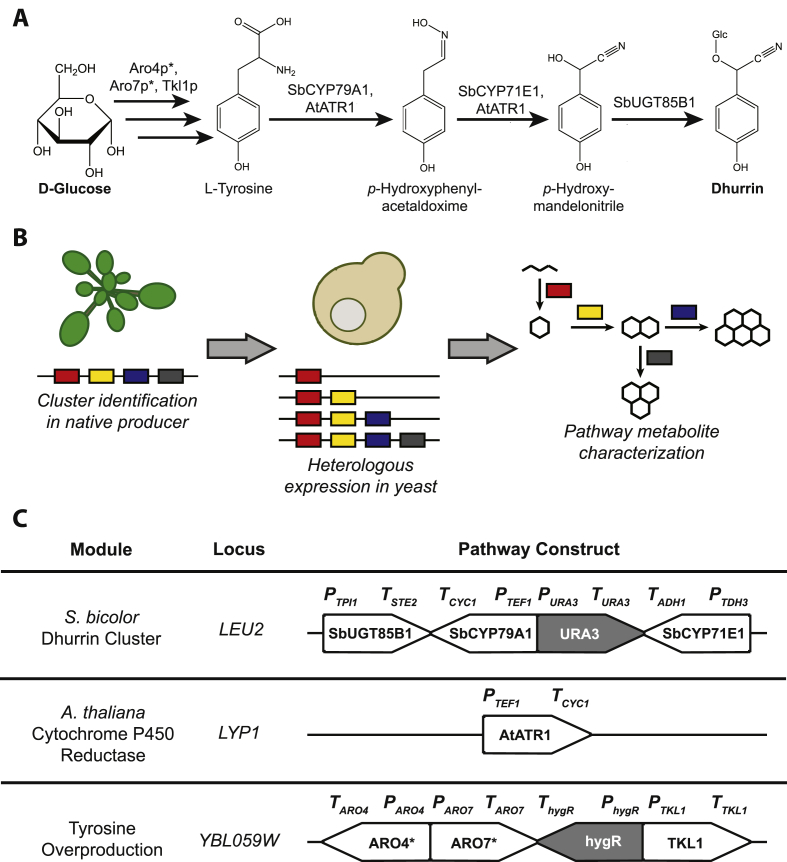
Dhurrin biosynthesis begins with the tyrosine N-monooxygenase SbCYP79A1 (Koch et al., 1995), which converts L-tyrosine into an oxime derivative, (E)-p-hydroxyphenylacetaldoxime. This intermediate is then converted into p-hydroxymandelonitrile by the 4-hydroxyphenylacetaldehyde oxime monooxygenase SbCYP71E1 (Bak et al., 1998), and lastly stabilized by glucosylation by the UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase SbUGT85B1 (Jones et al., 1999), yielding dhurrin (Fig. 1A). The genes encoding these three enzymes form a biosynthetic gene cluster, wherein CYP79A1, CYP71E1, and UGT85B1 appear adjacent to one another in the sorghum genome, along with a transporter which acts to sequester this potentially autotoxic compound in the plant cell vacuole (Darbani et al., 2016).
Fig. 1.
A heterologous metabolic pathway for dhurrin biosynthesis in yeast. A: The proposed dhurrin biosynthetic pathway. B: Rationale for heterologous pathway reconstruction. Biosynthetic gene clusters identified in plant genomes can be rapidly reconstructed in model microbes such as yeast, enabling rapid characterization of cluster enzymes in a clean metabolite background. C: Modular strategy for pathway construction. Genes in gray are selection markers. Abbreviations: Glc, glucose; URA3, S. cerevisiae orotidine-5′-phosphate decarboxylase; AtATR1, Arabidopsis thaliana cytochrome P450 reductase; ARO4*, S. cerevisiae 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate synthase Q166K; ARO7*, S. cerevisiae chorismate mutase T226I; TKL1, S. cerevisiae transketolase; hygR, hygromycin B phosphotransferase.
Previous work involving heterologous reconstruction of the dhurrin pathway has focused on model plant systems. The pathway has been reconstructed in Arabidopsis thaliana (Tattersall et al., 2001; Kristensen et al., 2005), as well as in the chloroplast of Nicotiana tabacum (Gnanasekaran et al., 2016). Efforts have also been made to characterize parts of cyanogenic glucoside pathways in microbial systems. Microsomes prepared from yeast expressing individual P450s have been used to characterize enzymes in cyanogenic glycoside pathways in vitro (Forslund et al., 2004; Yamaguchi et al., 2014; Hamann and Møller, 2007). Strategies involving re-engineering the N-terminal domains of the dhurrin pathway P450s have also recently been applied to functionally co-express SbCYP79A1 and SbCYP71E1 in Escherichia coli (Vazquez-Albacete et al., 2017).
Reconstructing plant metabolic pathways in heterologous hosts provides a clean metabolite background, with minimal interference from endogenous pathways, in which to characterize the activities of pathway enzymes. In contrast to plants, which grow slowly and are difficult to genetically manipulate, their rapid growth and genetic tractability makes microbes convenient heterologous hosts. In baker's yeast (Saccharomyces cerevisiae), rapid genetic engineering is facilitated by efficient homology-guided repair, which enables integration of multiple genes into the genome simultaneously (Shao et al., 2009). More recently, the sequence-guided endonuclease Cas9 has been used to increase the efficiency of homologous integration in yeast by inducing double-stranded DNA breaks at the target site in the chromosome (Ryan et al., 2014). S. cerevisiae is a well-established host for plant pathways involving cytochrome P450 enzymes, which must interact with cytochrome P450 reductase partners (CPRs) at the endoplasmic reticulum, an organelle only found in eukaryotes. In recent years, S. cerevisiae has been shown to be an effective host for the production of plant natural products as complex as the opiates morphine (Galanie et al., 2015) and noscapine (Li et al., 2018), as well as the monoterpene indole alkaloid strictosidine (Brown et al., 2015). However, to our knowledge, no complete cyanogenic glycoside pathway has yet been reconstituted in a microbial system. We sought to characterize the dhurrin cluster through heterologous reconstruction in yeast (Fig. 1B).
Here, we present the first report of de novo cyanogenic glycoside production in a microbial host by co-expressing the dhurrin pathway enzymes with a plant cytochrome P450 reductase in yeast. We further increased titers of dhurrin by overexpressing selected host enzymes to boost the supply of the pathway precursor tyrosine. Additionally, we used the dhurrin-producing strain as a platform to investigate the function of previously uncharacterized putative enzymes located near the dhurrin cluster in the S. bicolor genome. This work demonstrates that the dhurrin pathway can be functionally reconstituted in S. cerevisiae, which therefore emerges as a microbial host organism for cyanogenic glycoside research.
2. Materials and methods
2.1. Plasmid construction
Lists of plasmids, primers, and gene sequences used in this study are provided in the Supplementary Material. Oligonucleotides were synthesized by Integrated DNA Technologies (IDT) and the Stanford Protein and Nucleic Acid Facility. Heterologous gene sequences were codon-optimized for expression in S. cerevisiae using GeneArt GeneOptimizer (Life Technologies) with default settings for S. cerevisiae and synthesized by Gen9, Inc. Expand High Fidelity PCR system (Sigma) was used for PCR amplifications according to manufacturer's instructions. Sequencing was performed by Elim Biopharmaceuticals, Inc.
To construct plasmids containing expression cassettes for sorghum genes (gene sequences bracketed by yeast promoters and terminators), sequences of sorghum genes were first obtained from the Phytozome plant genomics portal (Goodstein et al., 2012). The gene sequences were codon-optimized for yeast expression and synthesized. Codon-optimized sequences are provided in Table S1. To construct vectors for genomic integration, the synthesized genes were PCR-amplified and cloned into PCR-amplified plasmid backbones encoding promoters and terminators by Gibson assembly (Gibson et al., 2009). SbCYP71E1, SbCYP79A1, SbUGT85B1, Sobic.001G012000, and Sobic.001G012101 were inserted into pCS2656, pCS2657, pCS2661, pCS2663, and pCS2664, respectively, to yield pCS4182-6. Promoters and terminators used in these plasmids appear in Table S2. Oligonucleotides used to PCR-amplify genes and backbones, and for sequence verification, are listed in Table S3.
To construct plasmids targeting Cas9 cleavage activity to specified genomic loci, 20-bp guide RNAs in target loci were identified using the Benchling gRNA design tool (Hsu et al., 2013; Doench et al., 2016) (benchling.com). Plasmids pCS4187-9, containing a Cas9 expression cassette and transcription cassettes for gRNAs targeting loci LEU2 (5′-ggtaagagaaaggaagacga-3′), LYP1 (5′-gtatgtatgtttttgagtcg-3′), and YBR197C (5′-ttaacgaccagcattagtag-3′), respectively, were constructed via assembly PCR and Gibson assembly.
To construct a plasmid for expression of SbCYP71E1 in yeast, the SbCYP71E1 sequence and ADH1 terminator from pCS4182 were PCR-amplified and introduced into the Gateway pDONR221 vector using BP Clonase II Enzyme Mix (Thermo Fisher) to produce pCS4190. The insert sequence was subsequently recombined into pAG414-ccdB expression vectors from the S. cerevisiae Advanced Gateway Destination Vector Kit (Alberti et al., 2007) using LR Clonase II (Thermo Fisher) to produce pCS4191 (expressing SbCYP71E1 under pGAL1) and pCS4192 (expressing SbCYP71E1 under pGPD). Oligonucleotides used to PCR-amplify SbCYP71E1-ADH1t and sequence-verify constructs appear in Table S4.
2.2. Strain construction
Yeast strains used in this study are listed in Table S5. All yeast strains are haploid and are derived from CEN.PK2 (MATα URA3-52; TRP1-289; LEU2-3/112; HIS3Δ1; MAL2-8C; SUC2).
For overexpression of native yeast genes involved in tyrosine biosynthesis, the genes ARO4Q166K, ARO7T226I, and TKL1, and hphNTI were PCR-amplified from the yeast strain CSY105817, and integrated by the DNA assembler method (Shao et al., 2009) into the YBL059W locus of CEN.PK2. Transformed colonies were selected by growth on a selective medium containing 200 mg/L hygromycin B (Sigma). Tyrosine overproduction was verified as described in section 2.3 below.
All other integrations were carried out by co-electroporating the DNA fragment(s) to be integrated together with a Cas9 plasmid which carries a G418 resistance selection marker, Cas9 coding sequence, and a guide RNA (gRNA) directing Cas9 cleavage at a target integration site (Ryan et al., 2014). Forty base pairs of homology were added using PCR primers at each end of each DNA fragment to guide genomic integration. In all electroporations, 200 ng of each DNA fragment and of the selected Cas9 plasmid were used.
To integrate single genes, promoter-gene-terminator cassette DNA fragments were co-electroporated with Cas9 plasmids targeting sites in the genomic locus of interest. Integrants were selected by plating on yeast extract peptone dextrose (YPD; BD Diagnostics) with 400 mg/L G418 sulfate (Calbiochem) to select for the Cas9 plasmid. Integrants were confirmed by yeast colony PCR, and the Cas9 plasmid was cured by regrowth in YPD with replica plating to YPD + G418 plates to identify colonies without G418 resistance.
Multigene integrations to build variants of the dhurrin pathway were carried out following the DNA assembler approach. A URA3 selection marker was PCR-amplified from pCS1748 (Liang et al., 2012) and included in the assembly, and successful integrants were selected by plating on a yeast nitrogen base (YNB; BD Diagnostics) medium containing 2% dextrose and lacking uracil (YNB − U). Successful integrants were identified by PCR of junctions between each assembly fragment. Oligonucleotides used to PCR-amplify expression cassettes for integration and verify constructs by colony PCR appear in Table S4.
2.3. Characterizing dhurrin pathway metabolite production
Yeast were cultured in synthetic-complete YNB +2% dextrose (unless stated otherwise) at 30 °C. In culture plate experiments, yeast were grown in 500 μL cultures in 96-well culture plates, after inoculation with 5 μL of seed cultures grown in 500 μL to saturation. In shake-flask experiments, yeast were cultured in 125-mL baffled shake flasks, after inoculation at OD 0.05 from 5 mL seed cultures grown in culture tubes. Optical density was measured using a Nanodrop spectrophotometer.
To measure the concentrations of metabolites that accumulated in the culture broth, yeast cells were removed by centrifugation, and the culture medium was diluted tenfold in water. Samples were analyzed using reverse-phase LC-MS on an Agilent 6420 triple-quad LC–MS, using an Agilent EclipsePlus C18 column (2.1 × 50 mm, 1.8 μm) and positive ionization. Isocratic 5% CH3CN (v/v in water + 50 μM NaCl, 0.1% formic acid) was used to separate metabolites over 4 min, with a flow rate of 0.4 mL min−1. The addition of 50 μM NaCl to the aqueous phase produced a Na+ adduct of dhurrin, which was used for detection as previously reported (Takos et al., 2010). The MS detection method for tyrosine used the mass transition 182.1 to 91.1 as the quantifier and 182.1 to 136.0 as the qualifier. For dhurrin, 334.1 to 145.0 was used as the quantifier and 334.1 to 184.9 as the qualifier. For 4-hydroxybenzylaldehyde, 123.1 to 77.0 was used as the quantifier and 123.1 to 95.0 was used as the qualifier. The transitions 152.1 to 119.8 and 152.1 to 134.5 were used to monitor production of p-hydroxyphenylacetaldoxime.
2.4. Strain characterization: untargeted metabolomics
Yeast were cultured in 500 μL YNB +2% dextrose media at 30 °C. Cultures were inoculated at OD 0.05 from seed cultures grown under the same conditions to saturation. For experiments on the complete dhurrin pathway, a YNB dropout medium lacking uracil was used; for experiments on truncated dhurrin pathways, a YNB dropout medium lacking uracil and tryptophan was used. After 24 h of growth, 200 μL aliquots of culture media were flash-frozen, lyophilized overnight, and reconstituted in 100 μL methanol +0.1% formic acid for LC-MS analysis on an Agilent 6520 quantitative time-of-flight (qTOF) mass spectrometer. Five samples were analyzed for each condition.
Data files from qTOF-MS experiments were exported to mzXML format and analyzed with custom R scripts, using the XCMS package (Smith et al., 2006) to identify differentially produced compounds. When comparing two sample sets, a compound was considered to be differentially produced if a signal above 10,000 was observed in at least one sample, the p-value of a T-test comparing the differentially expressed conditions was less than 0.001, and the fold change was greater than 10.
3. Results and discussion
3.1. Production of dhurrin in S. cerevisiae
To reconstruct the dhurrin pathway in yeast, a dhurrin pathway module encoding expression of SbCYP79A1, SbCYP71E1, and SbUGT85B1 was integrated into the LEU2 locus of the wild-type yeast strain (CEN.PK2) (Fig. 1C), yielding CSY1212. However, dhurrin was produced at levels of less than 3 mg/L after 96 h of growth in a synthetic-complete medium (Fig. 2A). The first two steps of the dhurrin pathway are catalyzed by cytochrome P450 enzymes, which interact with cytochrome P450 reductases (CPRs) at the endoplasmic reticulum in order to function. The Arabidopsis thaliana CPR AtATR1 has previously been shown to couple with diverse heterologous plant P450s in yeast, leading these enzymes to function more effectively than when relying solely on the native yeast CPR (Trenchard et al., 2015; Li and Smolke, 2016). A plant CPR module encoding the expression of AtATR1 was amplified from pCS1056 (Hawkins and Smolke, 2008) and integrated into the LYP1 locus of CSY1212, yielding CSY1213 (Fig. 1C). This strain produced over 50 mg/L dhurrin after 96 h of growth in a synthetic-complete medium (Fig. 2A). The identity of the dhurrin product was confirmed by comparing its fragmentation spectrum to that of an authentic standard (Figure S1).
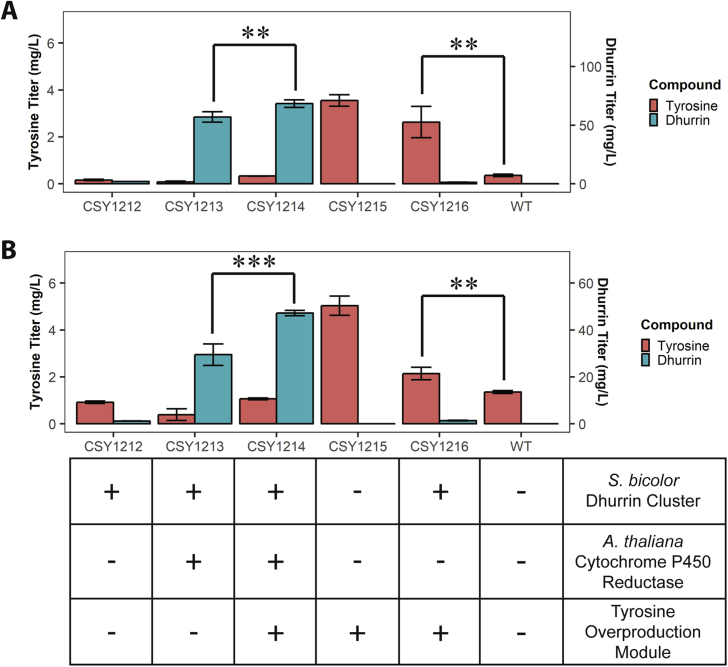
Fig. 2.
Effect of strain enhancements on dhurrin and tyrosine production levels. A: Tyrosine and dhurrin titers after 96 h growth of engineered yeast strains in plate culture, with 40 mg/L tyrosine initially present in the medium. B: Tyrosine and dhurrin titers without tyrosine initially present in the medium; otherwise as in A. The table below panels A and B schematically depicts the genotypes of tested strains. **: p < 0.05, one-tailed t-test; ***: p < 0.01, one-tailed t-test.
We attempted to increase dhurrin titers by ensuring an adequate supply of L-tyrosine, the amino acid precursor of dhurrin. We leveraged previous work optimizing plant pathways with tyrosine as a precursor (Galanie et al., 2015; Trenchard et al., 2015) to identify a set of yeast enzymes involved in tyrosine biosynthesis as targets for overexpression and optimization. A tyrosine overproduction module encoding overexpression of three enzymes - a feedback-inhibited mutant of the yeast enzyme 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate (DAHP) synthase (ARO4Q166K), chorismate mutase (ARO7), and transketolase (TKL1) - was integrated into the YBL059W locus of CSY1213, yielding CSY1214 (Fig. 1C). We additionally integrated this tyrosine overproduction module into the YBL059W locus of wild-type CEN.PK2, yielding CSY1215. Lastly, we integrated the dhurrin module into CSY1215, yielding CSY1216.
We analyzed these strains, along with wild-type CEN.PK2, for tyrosine levels and dhurrin production after 96 h of growth in a synthetic complete medium (Fig. 2A) and in a medium lacking tyrosine (Fig. 2B). In both media, strains harboring the tyrosine overproduction module but lacking the dhurrin module or the plant CPR module (CSY1215, CSY1216) exhibited significantly higher tyrosine levels than other strains tested. In the synthetic complete medium, 3.6 ± 0.2 mg/L tyrosine was detected in CSY1215 and 2.6 ± 0.7 mg/L in CSY1216, versus 0.4 ± 0.1 mg/L in CEN.PK2 and less in other strains tested. In the medium lacking tyrosine, 5.0 ± 0.4 mg/L tyrosine was detected in CSY1215 and 2.1 ± 0.3 mg/L in CSY1216, versus 1.4 ± 0.1 mg/L in CEN.PK2 and less in other strains tested. The strains lacking the plant CPR module (CSY1212, CSY1215, and CSY1216) produced only small amounts of dhurrin (less than 3 mg/L in all cases). In addition, the strain harboring the dhurrin pathway module and the plant CPR module but lacking the tyrosine overproduction module (CSY1213) produced significantly less dhurrin than CSY1214. In the synthetic-complete medium, 57 ± 4 mg/L dhurrin was detected in CSY1213 versus 68 ± 3 mg/L in CSY1214. In the medium lacking tyrosine, 29 ± 5 mg/L dhurrin was detected in CSY1213 versus 47 ± 1 mg/L in CSY1214.
3.2. Characterizing rates of dhurrin production and cell growth
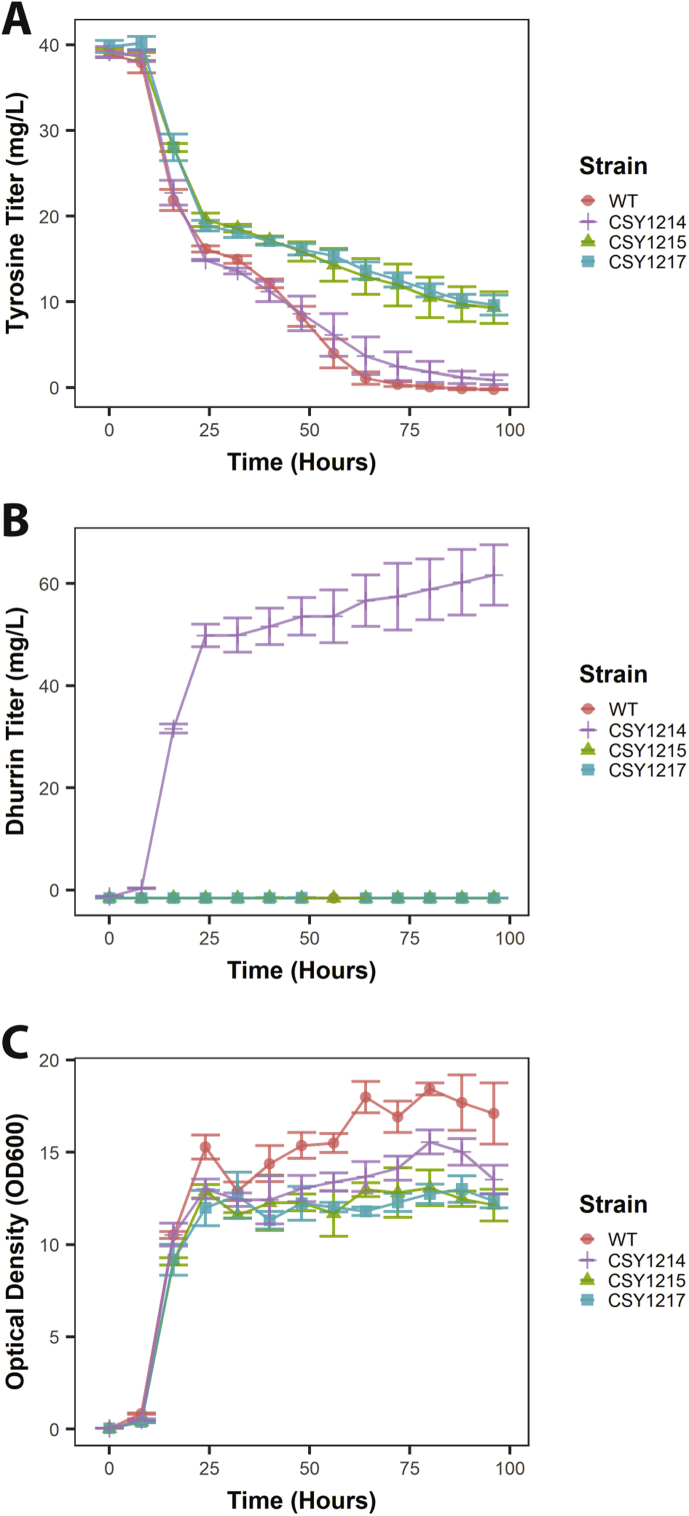
To test the effect of the tyrosine overexpression and AtATR1 modules in the absence of the dhurrin pathway, we integrated our AtATR1 expression cassette at the LYP1 locus of CSY1215, yielding CSY1217. The wild-type strain (CEN.PK2), complete dhurrin-producing strain with tyrosine overproduction module (CSY1214), wild-type strain with tyrosine overproduction module (CSY1215), and marker-only control strain (CSY1217) were characterized for tyrosine levels, dhurrin production, and optical density (OD) over a four-day time course experiment in shake flasks. Tyrosine was initially present in the growth medium at approximately 40 mg/L, but was rapidly depleted in CEN.PK2 to 16.2 ± 0.4 mg/L after 24 h (Fig. 3A). Tyrosine titers for CSY1214 were similar to those in wild-type CEN.PK2 throughout the experiment (Fig. 3A). In contrast, tyrosine was present at substantially higher concentrations for CSY1215 and CSY1217 throughout the experiment – after 96 h, less than 1 mg/L tyrosine was present in CEN.PK2 and CSY1214 as compared to over 9 mg/L tyrosine in both CSY1215 and CSY1217. This result is likely due to additional tyrosine production resulting from the presence of the tyrosine overproduction module and the lack of dhurrin production in these strains.
Fig. 3.
Metabolite levels and cell growth trends over time from yeast strains engineered for dhurrin biosynthesis. Tyrosine concentrations (A), dhurrin concentrations (B), and cell densities (optical density at 600 nm) (C) obtained during time course growth experiment.
Trends in dhurrin production over time from CSY1214 tracked closely with growth rate. While most of the productivity occurred during the early exponential growth stage, reaching titers of 50 ± 2 mg/L after 24 h of growth, the metabolite continued to accumulate throughout the experiment to a final titer of 62 ± 6 mg/L after 96 h (Fig. 3B). All engineered yeast strains exhibited a growth defect relative to CEN.PK2 – the wild-type strain reached over 17 OD600, while the engineered strains only reached from 12 to 13 OD600. However, the dhurrin-producing strain CSY1214 grew similarly to CSY1215 and CSY1217 (Fig. 3C). These results suggest that although tyrosine overproduction may impose an increased metabolic burden on engineered cells, dhurrin does not inhibit yeast growth at the titers produced in this experiment.
3.3. Characterizing intermediate accumulation in complete and partial dhurrin pathway strains
We sought to detect intermediates of the dhurrin pathway in order to determine whether inefficiencies or bottlenecks existed in the pathway converting tyrosine to dhurrin. We integrated a URA3 expression cassette in the LEU2 locus of the strain CSY1217 (which contains the tyrosine overexpression and AtATR1 modules), using the approach used to construct CSY1214; this yielded the marker-only control strain CSY1218. Similarly, we constructed CSY1219, a strain which was derived from CSY1217 but only expressed the URA3 expression cassette and the first enzyme in the pathway, SbCYP79A1, from the LEU2 locus. These strains were then transformed with either pCS4192, a plasmid which expresses SbCYP71E1, or pAG414-ccdB-GPD, a selection marker-only control. To measure production of p-hydroxyphenylacetaldoxime, the product of SbCYP79A1, in the absence of an authentic standard, we used the CFM-ID MS fragment prediction tool (Allen et al., 2014) and previously reported spectra for detection of the related compound phenylacetaldoxime (Yamaguchi et al., 2014) to predict MRM transitions for this intermediate. As a proxy for production of p-hydroxymandelonitrile, we measured this unstable compound's degradation product, p-hydroxybenzylaldehyde (Yamaguchi et al., 2014), using an authentic standard.
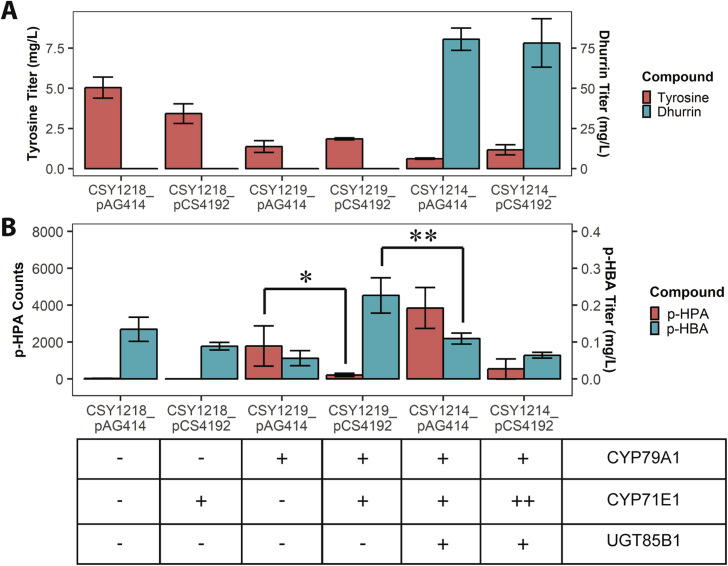
The strains were cultured for 48 h in plate culture and analyzed for production of tyrosine, dhurrin, p-hydroxyphenylacetaldoxime, and p-hydroxybenzylaldehyde (Fig. 4A and B). In contrast to previous experiments, we cultured these strains in a medium lacking uracil and tryptophan to ensure plasmid stability. We found that culturing the strains in this medium increased dhurrin titers, to 81 ± 7 mg/L in CSY1214 transformed with pAG414.
Fig. 4.
Precursor, product, and intermediate titers in complete and partial dhurrin pathway strains. A: Tyrosine and dhurrin titers after 48 h growth in plate culture. B:p-hydroxyphenylacetaldoxime (“p-HPA”) ion counts and p-hydroxybenzylaldehyde (“p-HBA”) titers after 48 h growth in plate culture. “++” in strain key indicates two expressed copies of the enzyme – one via genomic integration and one via low-copy plasmid. The table below panels A and B schematically depicts the genotypes of tested strains. *: p < 0.1, one-tailed t-test; **: p < 0.05, one-tailed t-test.
The results indicate that the dhurrin-producing yeast strains consumed pathway intermediates efficiently. In strains lacking CYP79A1, MS fragment ions corresponding to p-hydroxyphenylacetaldoxime were observed only at trace levels. Significantly more p-hydroxyphenylacetaldoxime was observed in the strain which expressed SbCYP79A1 (CSY1219) transformed with the control plasmid (pAG414-ccdB-GPD) than with the plasmid expressing SbCYP71E1 (pCS4192) (1800 ± 1100 ions versus 220 ± 90 ions) (Fig. 4B). While p-hydroxybenzylaldehyde was observed in all yeast strains tested, significantly more of this compound was produced when co-expressing only CYP79A1 and CYP71E1 in CSY1219 transformed with pCS4192 relative to the complete dhurrin pathway construct CSY1214 transformed with pAG414 (0.23 ± 0.05 mg/L versus 0.11 ± 0.02 mg/L). (Fig. 4B).
3.4. Characterizing putative S. bicolor enzymes using dhurrin-producing chassis strains
We used the dhurrin-producing platform yeast strain to examine uncharacterized enzymes encoding genes located near the dhurrin biosynthetic gene cluster in the S. bicolor genome for putative activities associated with the dhurrin pathway. The open reading frames Sobic.001G012000 (Sb120) and Sobic.001G012100 (Sb121) are both located within 20 kb of the biosynthetic gene cluster encoding the dhurrin pathway in the sorghum genome (Figure S2); Sb120 is predicted to be a member of the CYP71E1 family of cytochrome P450s and Sb121 is predicted to be a dehydrogenase. We first examined whether Sb120 can function analogously to SbCYP71E1 in the dhurrin pathway. Following the approach used to construct the dhurrin-producing strain CSY1214, we constructed CSY1220, which expresses Sb120 instead of SbCYP71E1. Analysis of this strain indicated that it produced no dhurrin (Figure S3), indicating that Sb120 is not a functional analogue to SbCYP71E1.
We next examined potential roles for Sb120 and Sb121 in the dhurrin pathway through untargeted metabolomics. We integrated Sb120 and Sb121 expression cassettes separately into the YBR197C locus of our dhurrin-producing strain CSY1214, yielding CSY1221 and CSY1222, respectively. Strains CSY1214, CSY1221, CSY1222, and the marker-only control strain CSY1218 were cultured for 48 h in five 500 μL samples each of a medium lacking uracil. Culture media were assayed using liquid chromatography coupled to quantitative time-of-flight mass spectrometry (qTOF-MS). qTOF-MS results from CSY1214 were compared to data from CSY1218, CSY1221, and CSY1222 in order to identify ions present at significantly different abundances in the compared strains. From this analysis, four potentially differentially-produced candidate ions were identified (Figure S4). However, on closer inspection, none of the candidate ions were specific to Sb120 or Sb121 expression, suggesting that these signals did not represent enzyme activity on dhurrin (Figure S5).
Finally, we examined whether Sb120 and Sb121 could exhibit activity on intermediates in the dhurrin pathway. We integrated Sb120 or Sb121 into the YBR197C locus of the CYP79A1-expressing strain CSY1219, yielding CSY1223 and CSY1224, respectively. We transformed these strains with either the CYP71E1-expressing plasmid pCS4192 or the selection marker control plasmid pAG414-ccdB-GPD. These strains were cultured and assayed by qTOF-MS as previously described to determine the effect of expressing Sb120 or Sb121 in the presence of SbCYP79A1 or in the presence of SbCYP71E1 and SbCYP79A1. No candidate ions specific to Sb120 or Sb121 expression were identified for either enzyme in any background tested (Figure S6; Figure S7). Taken together, the results indicate that if these genes encode functional enzymes, they do not act on dhurrin or on dhurrin pathway intermediates.
4. Conclusion
In summary, we have demonstrated the production of the cyanogenic glycoside dhurrin in yeast through functional expression of enzymes from a S. bicolor biosynthetic gene cluster. We found that production of dhurrin was significantly enhanced by overexpression of native yeast enzymes producing the dhurrin precursor tyrosine, and that the A. thaliana CPR AtATR1 is capable of coupling with S. bicolor P450 enzymes in yeast to increase dhurrin production. While we were unable to demonstrate activity for the previously uncharacterized enzymes Sb120 and Sb121 in the dhurrin gene cluster, our untargeted metabolomics workflow may be applied to characterize other putative cyanogenic glucoside-producing enzymes in heterologous hosts.
Cyanogenic glycoside pathways have independently evolved in a wide range of plants. As initiatives like the 1000 Plant Genomes Project produce more plant genomes and automated pipelines for identifying putative plant biosynthetic pathways are developed (Medema and Osbourn, 2016), more candidate cyanogenic glycoside clusters are likely to be identified. Our work demonstrates that enzymes from classes commonly implicated in cyanogenic glycoside production can be functionally expressed in yeast. The capability to rapidly test these clusters in a genetically tractable microbial host free of potentially confounding plant enzyme activities will be a valuable tool for future studies of cyanogenic glycoside biosynthesis.
Acknowledgements
We thank Prashanth Srinivasan, Shireen Rudina, and Calvin Schmidt for valuable comments on the manuscript. We thank Aaron Cravens for providing Cas9 plasmids pCS4187 and pCS4188, targeting LEU2 and LYP1, respectively, and Sijin Li for constructing the tyrosine overexpression module. We thank Professor Beth Sattely for use of the qTOF-MS instrumentation to perform metabolomics experiments. This work was supported by the National Institutes of Health (grants to C.D.S.), the Stanford Bio-X Institute, and the Siebel Scholars Foundation (graduate student fellowships to B.K.).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00092.
Author contributions
B.K. and C.D.S. conceived of the project and designed the experiments. B.K. and C.D.S. wrote the manuscript. B.K. performed all experiments.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alberti S., Gitler A.D., Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913–919. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F., Pon A., Wilson M., Greiner R., Wishart D. CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res. 2014;42:W94–W99. doi: 10.1093/nar/gku436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Kahn R.A., Nielsen H.L., Moller B.L., Halkier B.A. Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogen. Plant Mol. Biol. 1998;36:393–405. doi: 10.1023/a:1005915507497. [DOI] [PubMed] [Google Scholar]
- Brown S., Clastre M., Courdavault V., O'Connor S.E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. U.S.A. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbani B. The biosynthetic gene cluster for the cyanogenic glucoside dhurrin in Sorghum bicolor contains its co-expressed vacuolar MATE transporter. Sci. Rep. 2016;6:37079. doi: 10.1038/srep37079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 2004;135:71–84. doi: 10.1104/pp.103.038059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gleadow R.M., Møller B.L. Cyanogenic glycosides: synthesis, physiology, and phenotypic plasticity. Annu. Rev. Plant Biol. 2014;65:155–185. doi: 10.1146/annurev-arplant-050213-040027. [DOI] [PubMed] [Google Scholar]
- Gleadow R.M., Woodrow I.E. Constraints on effectiveness of cyanogenic glycosides in herbivore defense. J. Chem. Ecol. 2002;28:1301–1313. doi: 10.1023/a:1016298100201. [DOI] [PubMed] [Google Scholar]
- Gnanasekaran T. Transfer of the cytochrome P450-dependent dhurrin pathway from Sorghum bicolor into Nicotiana tabacum chloroplasts for light-driven synthesis. J. Exp. Bot. 2016;67:2495–2506. doi: 10.1093/jxb/erw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D.M. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T., Møller B.L. Improved cloning and expression of cytochrome P450s and cytochrome P450 reductase in yeast. Protein Expr. Purif. 2007;56:121–127. doi: 10.1016/j.pep.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Hawkins K.M., Smolke C.D. Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 2008;4:564–573. doi: 10.1038/nchembio.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.R., Moller B.L., Hoj P.B. The UDP-glucose:p-hydroxymandelonitrile-O-glucosyltransferase that catalyzes the last step in synthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor. Isolation, cloning, heterologous expression, and substrate specificity. J. Biol. Chem. 1999;274:35483–35491. doi: 10.1074/jbc.274.50.35483. [DOI] [PubMed] [Google Scholar]
- Koch B.M., Sibbesen O., Halkier B.A., Svendsen I., Møller B.L. The primary sequence of cytochrome P450tyr, the multifunctional N-hydroxylase catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 1995;323:177–186. doi: 10.1006/abbi.1995.0024. [DOI] [PubMed] [Google Scholar]
- Kristensen C. Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1779–1784. doi: 10.1073/pnas.0409233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Smolke C.D. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 2016;7:12137. doi: 10.1038/ncomms12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E3922–E3931. doi: 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.C., Chang A.L., Kennedy A.B., Smolke C.D. A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity. Nucleic Acids Res. 2012;40:e154. doi: 10.1093/nar/gks636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M.H., Osbourn A. Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat. Prod. Rep. 2016;33:951–962. doi: 10.1039/c6np00035e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O.W. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3 doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Zhao H., Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., Want E.J., O'Maille G., Abagyan R., Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Takos A. Genetic screening identifies cyanogenesis-deficient mutants of Lotus japonicus and reveals enzymatic specificity in hydroxynitrile glucoside metabolism. Plant Cell. 2010;22:1605–1619. doi: 10.1105/tpc.109.073502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall D.B. Resistance to an herbivore through engineered cyanogenic glucoside synthesis. Science. 2001;293:1826–1828. doi: 10.1126/science.1062249. [DOI] [PubMed] [Google Scholar]
- Tokpohozin S.E., Fischer S., Sacher B., Becker T. β-d-Glucosidase as ‘key enzyme’ for sorghum cyanogenic glucoside (dhurrin) removal and beer bioflavouring. Food Chem. Toxicol. 2016;97:217–223. doi: 10.1016/j.fct.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Trenchard I.J., Siddiqui M.S., Thodey K., Smolke C.D. De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng. 2015;31:74–83. doi: 10.1016/j.ymben.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Albacete D. An expression tag toolbox for microbial production of membrane bound plant cytochromes P450. Biotechnol. Bioeng. 2017;114:751–760. doi: 10.1002/bit.26203. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Yamamoto K., Asano Y. Identification and characterization of CYP79D16 and CYP71AN24 catalyzing the first and second steps in L-phenylalanine-derived cyanogenic glycoside biosynthesis in the Japanese apricot, Prunus mume Sieb. et Zucc. Plant Mol. Biol. 2014;86:215–223. doi: 10.1007/s11103-014-0225-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.