ABSTRACT
Photomorphogenesis is an important developmental process that helps the seedlings adapt to external light conditions. B-Box proteins are a family of transcription factors that regulate photomorphogenic responses. BBX31 negatively regulates photomorphogenesis under visible light. In contrast, it promotes photomorphogenesis under UV-B and enhances tolerance to high doses of UV-B radiation. BBX31 and HY5 independently and oppositely regulate the ability of seedlings to adapt to varying light intensities. BBX31 also regulates primary root elongation under low intensities of white light. GC-MS and HPLC-based metabolite profiling identified differential accumulation of multiple primary and secondary metabolites in 35S:BBX31 that might enhance tolerance to UV-B.
KEYWORDS: B-box, BBX31, UV-B radiation, HY5, photomorphogenesis, light signaling, metabolites
Light acts as one of the most prominent environmental factors that regulates the development of newly emerged seedlings. In the presence of light, seedlings establish into autotrophic organisms by characteristically developing short hypocotyls and open cotyledons with chlorophyll accumulation – a phenomenon known as photomorphogenesis.1 Seedlings exhibit reduced hypocotyl elongation as a response not only to visible light but also to low doses of UV-B radiation.2 B-Box (BBX) proteins are a family of transcription factors that regulate light-dependent development.3,4 Most of the BBX proteins that are known to regulate photomorphogenesis act in a HY5-dependent manner.4 In a recent study, we reported that BBX31, a group V BBX protein in Arabidopsis, negatively regulates photomorphogenesis under visible light, whereas it acts as a positive regulator of photomorphogenesis under UV-B light.5 HY5 directly binds on the promoter of BBX31 and regulates its transcription.5,6 However, genetic evidences suggested that the regulation of photomorphogenesis by BBX31 was found to be independent of HY5. We found no evidence of physical interaction between BBX31 and HY5. Our RNA Sequencing data revealed that the genes regulated by BBX31 and HY5 are largely different.5 Here we discuss some additional features of BBX31-mediated cell elongation and UV-B protection.
BBX31 and HY5 oppositely and independently regulate hypocotyl elongation under varying light intensities
To compare the effects of the overexpression of BBX31 and the loss-of-function of HY5 in determining the ability of seedlings to adjust their hypocotyl growth to varying light intensities, we grew 35S:BBX31 and hy5-215 lines in increasing fluence of white light (10–90 μmolm−2s−1). Both these lines showed reduced inhibition of hypocotyl elongation upon increasing light intensity indicating that HY5 and BBX31 are positive and negative regulators of photomorphogenesis, respectively (Figure 1(a)). To understand if the two genes genetically interact to regulate light-mediated hypocotyl elongation, we analyzed the fluence response of 35S:BBX31/hy5-215 lines. These lines possessed longer hypocotyls compared to 35S:BBX31 and hy5-215 lines.5 35S:BBX31/hy5-215 lines completely failed to adapt to increasing intensities of white light, as their hypocotyls showed similar lengths in all the four fluence conditions provided (Figure 1(a)). The absence of significant change in the hypocotyl length of 35S:BBX31/hy5-215 in response to changing fluence suggests that BBX31 and HY5 might together constitute a pair of key regulators that independently regulate white light-mediated photomorphogenesis in Arabidopsis.
Figure 1.
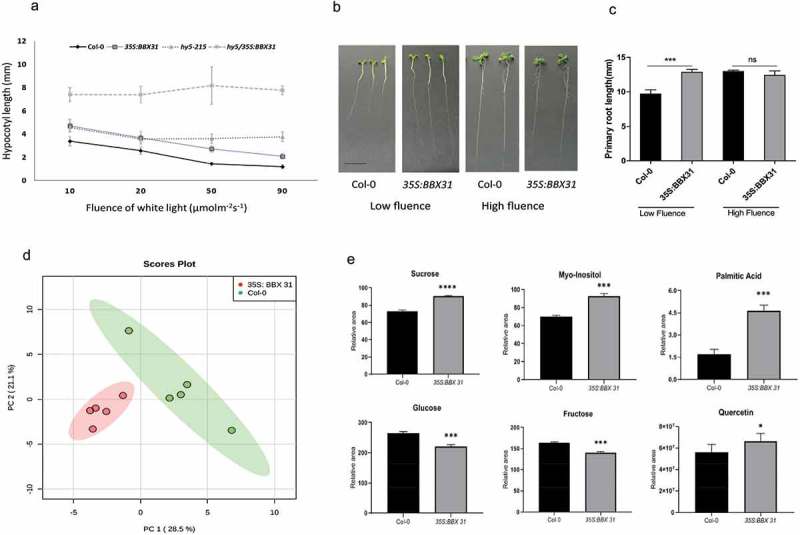
(a) hy5/35S:BBX31 exhibit constitutive elongated hypocotyl length over increasing fluence of white light. Line graph depicting hypocotyl lengths of Col-0, hy5, 35S:BBX31 and hy5/35S:BBX31 under different fluence of white light (b) Representative images of the seedlings grown in low white light (10 μmolm−2s−1) and high white light (90 μmolm−2s−1) for 7 days. (c) Quantification of primary root length of 7-day old seedlings of the indicated genotypes in low fluence white light (10 μmolm−2s−1) and high fluence white light (90 μmolm−2s−1). (d) Principal Component Analysis of metabolite profiles of Col-0 and 35S:BBX31 clustered separately depicting significant variations triggered by an early response of UV-B radiation. (e) Relative levels of key metabolites in Col-0 and 35S:BBX31 Arabidopsis seedlings on 24hr exposure to UV-B radiation exhibit significant variations due to metabolic readjustment. Error bars represent SD of atleast 3 replicates. Asterisks represent statistically significant differences (****p value < 0.0001; *** p value < 0.001, * p value < 0.01) as determined by Student’s t test.
BBX31 positively regulates primary root elongation under low-intensity white light
The BBX31 overexpressing lines exhibited longer hypocotyls owing to the longer constituting cells.5 We investigated if their primary roots showed any difference in length from the wild type. The 35S:BBX31 seedlings possessed significantly longer primary roots than Col-0 when grown under low fluence white light (10 μmolm−2s−1) (Figure 1(b, c)). However, under higher fluence (90 μmolm−2s−1) the primary root lengths of the wild type and 35S:BBX31 seedlings were similar (Figure 1(b, c)). Together these results suggest that BBX31 regulates primary root elongation specifically under low-intensity white light.
BBX31 regulates primary and secondary metabolites in early response to UV-B radiation
Seedlings overexpressing BBX31 survive better than wild-type when irradiated with high doses of UV-B.5 To understand the basis of this tolerance, we compared soluble metabolites obtained from 13-day-old Col-0 and 35S: BBX31 seedlings exposed to 24 h of UV-B radiation by GC-MS based metabolite profiling.7,8 Multivariate analysis using Principal component analysis revealed variations between Col-0 and 35S:BBX31 after exposure to UV-B radiation (Figure 1(d)). Prominent differences in accumulation patterns of sugars, sugar alcohols and fatty acids were observed. Levels of free sucrose, myoinositol, and palmitate enhanced significantly, while glucose and fructose levels decreased in UV-B treated 35S: BBX31 compared to wild-type (Figure 1(e)). These primary metabolites might act as signaling molecules to regulate protection against UV-B radiation. Myoinositol acts as a signaling molecule in maize leaves exposed to UV-B radiations.9 Sucrose is well known to adjust its levels under various environmental stress.10 Benzoic acid and glucopyranoside levels were elevated upon UV-B exposure both in Col-0 and 35S:BBX31 seedlings indicating their role in conferring UV-B stress tolerance. We found that UV-B radiation triggers the accumulation of phenolic compounds (gallic acid, coumaric acid, vanillic acid, and hydroxybenzoic acid) in 35S:BBX31.5 We also estimated levels of flavonoids in UV-exposed wild-type and BBX31 overexpressing seedlings using HPLC.11 We found significantly higher levels of quercetin in 35S:BBX31 compared to Col-0 (Figure 1(e)). Enhanced levels of phenols and flavonoids upon UV irradiation suggests that they might play an important role as UV-B absorbing molecules.
Conclusion
In conclusion, BBX31 and HY5 antagonistically regulate photomorphogenesis. While BBX31 is a negative regulator, HY5 promotes photomorphogenesis. The two transcription factors regulate hypocotyl elongation independently and in opposite ways to help seedlings adapt to different light intensities. BBX31 also promotes primary root elongation. Under UV-B, readjustment of sugar, fatty acid, phenolic, and flavonoid metabolism occurs in Arabidopsis seedlings. Enhanced levels of the key intermediate primary and secondary metabolites in 35S:BBX31 confers additional UV-B protection.
Funding Statement
This work was supported by the Department of Biotechnology, Ministry of Science and Technology [BT/PR19193/BPA/118/195/2016]; Science and Engineering Research Board [ECR/2016/001176].
Acknowledgments
SD would like to thank Department of Biotechnology (Ramalingaswami Fellowship, IYBA, BT/PR19193/BPA/118/195/2016) and SERB (EMR/2016/000181), Government of India for funding. AY, PY, and ML acknowledge UGC, DBT, and MHRD-HTRA, Govt. of India, respectively, for their Ph.D. fellowships. SKM and ML acknowledge the Science and Engineering Research Board (SERB) Early career research funding (File No: ECR/2016/001176).
Disclosure of Potential Conflicts of Interest
Authors disclose no potential conflicts of interest.
References
- 1.Sullivan JA, Deng XW.. From seed to seed: the role of photoreceptors in Arabidopsis development. Dev Biol. 2003;260:289–297. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins GI. Photomorphogenic responses to ultraviolet-B light. Plant Cell Environ. 2017;40:2544–2557. doi: 10.1111/pce.12934. [DOI] [PubMed] [Google Scholar]
- 3.Gangappa SN, Botto JF. The BBX family of plant transcription factors. Trends Plant Sci. 2003;19:460–470. doi: 10.1016/j.tplants.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Vaishak KP, Yadukrishnan P, Bakshi S, Kushwaha AK, Ramachandran H, Job N, Babu D, Datta S. The B-box bridge between light and hormones in plants. J Photochem Photobio B. 2019;191:164–174. doi: 10.1016/j.jphotobiol.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 5.Yadav A, Bakshi S, Yadukrishnan P, Lingwan M, Dolde U, Wenkel S, Masakapalli SK, Datta S. The B-box-containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol. 2019:pp.01258.2018. doi: 10.1104/pp.18.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heng Y, Lin F, Jiang Y, Ding M, Yan T, Lan H, Zhou H, Zhao X, Xu D, Deng X. B-box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. Plant Physiol. 2019:pp.01244.2018. doi: 10.1104/pp.18.01244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 8.Masakapalli SK, Bryant FM, Kruger NJ, Ratcliffe RG. The metabolic flux phenotype of heterotrophic Arabidopsis cells reveals a flexible balance between the cytosolic and plastidic contributions to carbohydrate oxidation in response to phosphate limitation. Plant J. 2014;78(6):964–977. doi: 10.1111/tpj.12522. [DOI] [PubMed] [Google Scholar]
- 9.Paula C, Morrow DJ, Fernandes J, Walbot V. Rapid maize leaf and immature ear responses to UV-B radiation. Front Plant Sci. 2011;2:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maria B, Obata T, Mehterov N, Ivanov I, Petrov V, Toneva V, Fernie A, Gechev TS. Comparative metabolic profiling of Haberlea rhodopensis, Thellungiella halophyla, and Arabidopsis thaliana exposed to low temperature. Front Plant Sci. 2013;4:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoddami A, Wilkes MA, Roberts TH. Techniques for analysis of plant phenolic compounds. Molecules. 2013;18(2):2328–2375. doi: 10.3390/molecules18022328. [DOI] [PMC free article] [PubMed] [Google Scholar]


