ABSTRACT
Plant GH3 genes play pivotal roles in biotic stress through involving in hormonal homeostasis by conjugation to amino acids of the free-form of salicylic acid, jasmonic acid (JA) or indole-3-acetic acid. We recently showed that rice group I GH3 gene family, with four members, are the functional JA-Ile synthetases and positively mediated rice resistance to Xanthomonas oryzae pv. oryzae (Xoo). Here, we further found that these four genes are also positive regulators conferring resistance to Xanthomonas oryzae pv. oryzicola (Xoc), the devastating bacterial pathogen of rice. The transcript of these four genes were all activated upon Xoc invasion. The overexpressing plants showed less lesion length in comparison with wild type plant accompanying with higher pathogenesis-related genes accumulation, while the triple and quadruple suppressing plants showed susceptible to Xoc with less pathogenesis-related genes accumulation. Previous and present work demonstrate that rice group I GH3 family genes act as positive regulators in the resistance to Xoo and Xoc.
KEYWORDS: Rice, Xanthomonas oryzae pv. oryzicola, GH3, JA, JA-Ile
The GH3 proteins are encoded by a multigene family in various plant species with diverse members, involving in plant growth or development, biotic or abiotic stress by largely altering hormonal homeostasis.1 GH3 proteins can catalyze the formation of salicylic acid, jasmonic acid (JA), and indole-3-acetic acid (IAA) amido conjugates.2 The rice GH3 gene family contains 13 paralogs and divided into three groups. Group I harboring four members (GH3.3, GH3.5, GH3.6, and GH3.12) all have the JA-Ile synthetase activity by conversion of free JA into the active form JA-Ile, accompanying with mediated resistance to Xanthomonas oryzae pv. oryzae (Xoo) by regulating transcription pattern of JA-responsive genes.3 Group II contains eight members (GH3.1, GH3.2, GH3.4, GH3.8, GH3.9, GH3.10, GH3.11, and GH3.13), with which four of the group II members, GH3.1, GH3.2, GH3.8, and GH3.13, have the IAA-Asp synthetase activity by conversion of active IAA into inactive IAA-Asp. So far, only GH3.2 and GH3.8 had been reported as positive regulators conferring resistance to Xoo.4,5 Group III has one member (GH3.7) with function unknown.
We recently characterized four group I GH3 gene family members. These four GH3 proteins encode JA-Ile synthetase by catalyzing the conjugation of JA to Ile in vitro and in vivo.3 The overexpressing rice plants have less free JA accumulation and more active form JA-Ile concentration, accompanying with resistance to bacterial pathogen Xoo.3 To elucidate whether these four genes play roles of rice to another bacterial pathogen, Xanthomonas oryzae pv. oryzicola (Xoc), which causes bacterial leaf streak and also a devastating bacterial disease of rice,6 we firstly analyzed the expression patterns of these four genes after Xoc infection. We used qRT-PCR assay to examine the transcription level of the four genes in susceptible and resistant rice plants inoculated with Xoc strain RH3. WRKY45-2 OE is a transgenic plant showed broad-spectrum resistance to Xoc by overexpressing WRKY45-2, a central regulator of disease resistance,7 was used as resistant rice plant. The transcript level of four GH3 genes increased significantly in both resistant and susceptible plants after Xoc inoculation, while relatively higher transcript accumulation was observed in resistant plants compared within susceptible plants (Figure 1). In addition, WRKY45-2 OE plants have higher transcript level of GH3.3 and GH3.6, indicating the possibility that WRKY45 could directly regulate these two genes.8 These differential transcript patterns indicate that the four GH3 genes may be involved in the rice-Xoc interaction.
Figure 1.
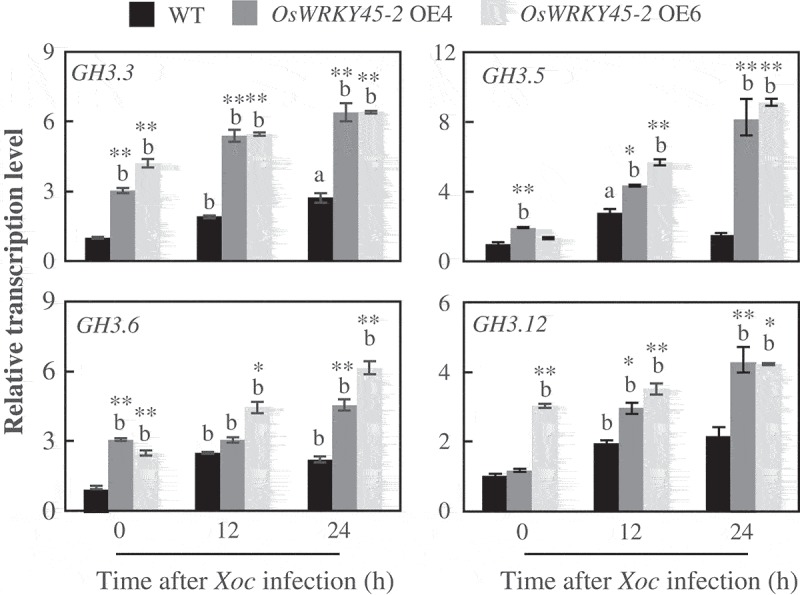
Expression patterns of group I GH3 family genes analyzed by qRT-PCR upon Xoc inoculation. The ‘a’ or ‘b’ indicates a significant difference was detected between non-infected and Xoc-infected plants of the same rice line at P < 0.05 or P < 0.01, respectively. Two (**) or one (*) asterisks indicate a significant difference was detected between WT and OsWRKY45-2 overexpressing of the same treatment at P < 0.01 or P < 0.05, respectively.
We then evaluate whether these four group I GH3 family genes play roles in resistance to Xoc. At the tillering stage, each three independently positive overexpressing transgenic rice plants with higher transcript levels were inoculated with Xoc strain RH3, and lesion lengths were calculated after 14 days inoculation.9 The transgenic plants showed enhanced resistance to Xoc strain RH3 with lesion length 3.2 ± 0.4 cm to 3.4 ± 0.2 cm for GH3.3, 2.8 ± 0.2 cm to 3.0 ± 0.2 cm for GH3.5, 3.7 ± 0.3 cm to 3.9 ± 0.2 cm for GH3.6, and 3.8 ± 0.2 cm to 3.9 ± 0.2 cm for GH3.12, compared to wild type with 4.6 ± 0.3 cm (Figure 2(a)). Simultaneously, the Xoc population was significantly lower in GH3.3-, GH3.5-, GH3.6-, and GH3.12-overexpressing plants than in wild type at different days post inoculation (Figure 2(b)).
Figure 2.
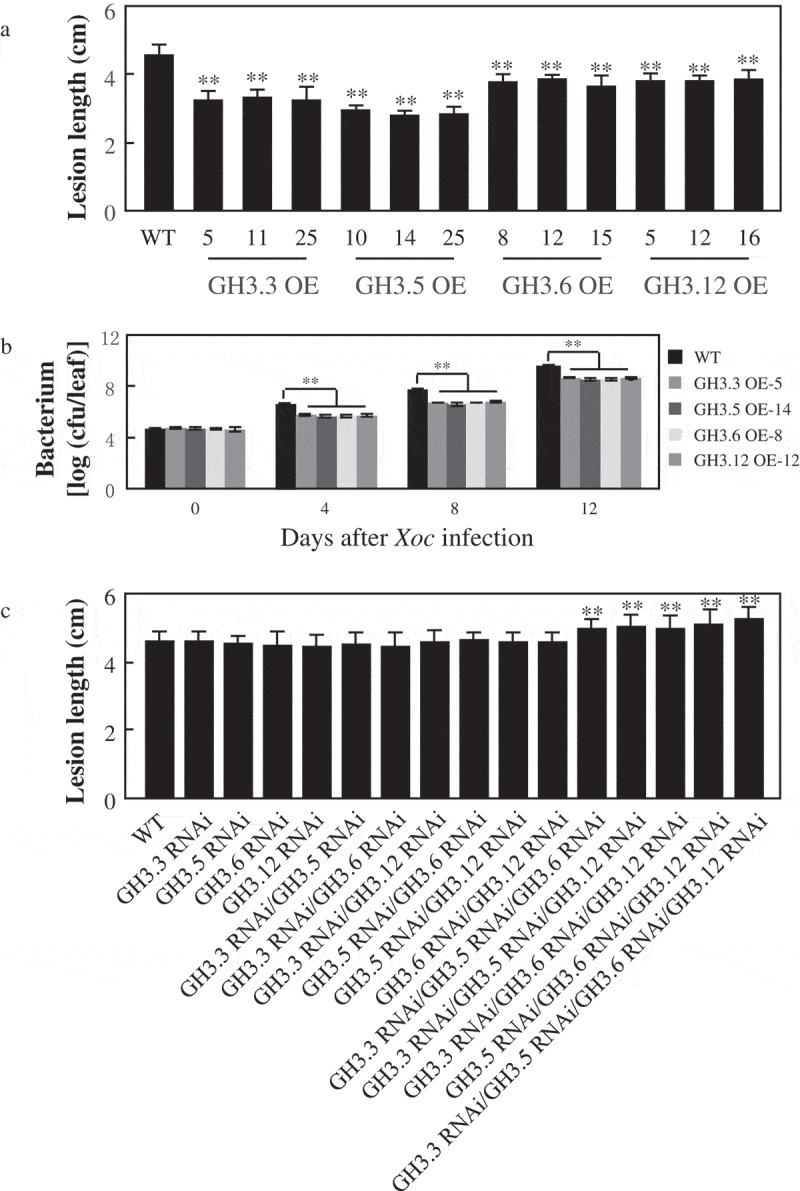
The group I GH3 family genes act as positive regulator in the resistance of rice to Xoc. Two (**) or one (*) asterisks indicate a significant difference was detected between WT (ZH11) and transgenic plants at P < 0.01 or P < 0.05, respectively. Data represent means (five to seven leaves from one plant) ± SD. (a) Enhanced resistance of GH3.3-overexpressing plants, GH3.5-overexpressing plants, GH3.6-overexpressing plants and GH3.12-overexpressing plants to Xoc strain RH3. (b) Growth of Xoc strain RH3 in leaves of GH3.3-, GH3.5-, GH3.6-, GH3.12-overexpression plants (T1 generation). Data represent mean (nine leaves from three plants) ± SD. Two (**) asterisks indicate a significant difference was detected between WT and transgenic plants at P < 0.01. (c) The response of GH3.3, GH3.5, GH3.6, and GH3.12 single, double, triple, and quadruple suppressing plants to Xoc strain RH3.
We next evaluate the response of suppressing plants to Xoc. After inoculation of GH3.3-, GH3.5-, GH3.6-, and GH3.12-suppressing plants with Xoc strain RH3 at tillering stage,9 the individual suppressing plants had comparable lesion length in comparison with wild type plants. The double suppressing plants also have the similar lesion length compared with wild type. While the triple suppressing plants with lesion length 5.0 ± 0.3 cm to 5.2 ± 0.4 cm, and the quadruple suppressing plants with lesion length 5.3 ± 0.4 cm, showed longer lesion length compared with wild type with 4.6 ± 0.3 cm (Figure 2(c)). Taken together, these results demonstrate that GH3.3, GH3.5, GH3.6, and GH3.12, with functional redundancy of the same clade of group I GH3 family, positively regulate rice resistance to Xoc.
To further verify the function of group I GH3 family genes conferring resistance to Xoc in rice, we determined the transcription pattern of pathogenesis-related (PR) genes, OsPR1a, OsPR8, and OsPR10, which have been validated as marker genes during rice-Xoc interaction.10 The qRT-PCR analyses showed that these three genes were significantly activated in the GH3.3-, GH3.5-, GH3.6-, and GH3.12-overexpressing plants (Figure 3), while depressed in the GH3.3, GH3.5, GH3.6, and GH3.12 triple and quadruple suppressing plants, but not in the GH3.3, GH3.5, GH3.6, and GH3.12 single and double suppressing plants (Figure 3), compared with wild type plants.
Figure 3.
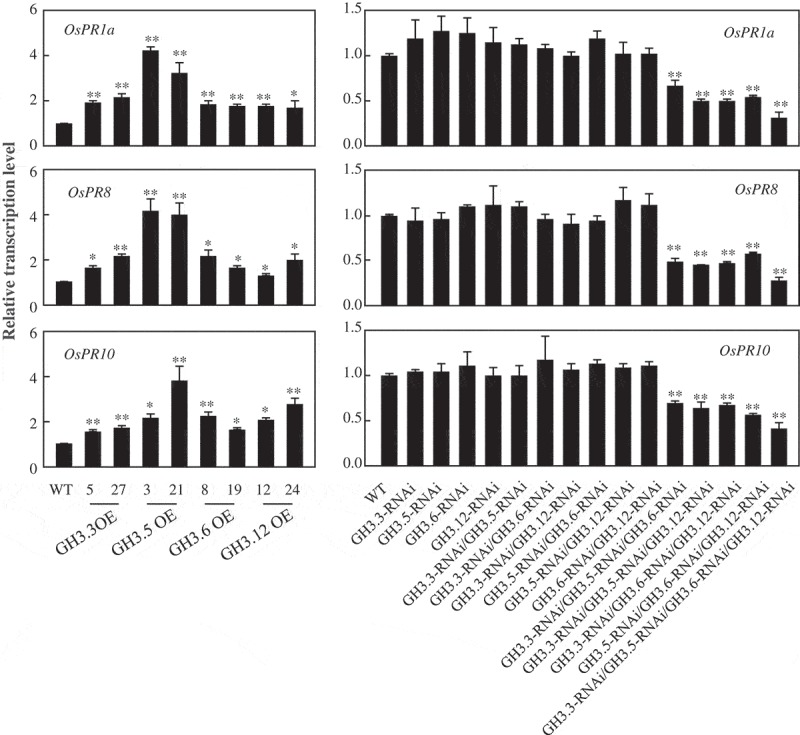
Relative transcript level of pathogen-responsive gene OsPR1a, OsPR8, and OsPR10 in GH3.3-, GH3.5-, GH3.6-, GH3.12-overexpressing plants and GH3.3, GH3.5, GH3.6, GH3.12 single, double, triple, and quadruple suppressing plants. Two (**) or one (*) asterisks indicate a significant difference was detected between WT and transgenic plants at P < 0.01 or P < 0.05, respectively.
In conclusion, our previous and present work indicate that the rice group I GH3 family genes containing GH3.3, GH3.5, GH3.6, and GH3.12 four members act as positive regulators in the resistance to Xoo and Xoc, the two devastating bacterial pathogens of rice. Exploring the haplotype of the four genes with relative higher transcript level provides an alternative strategy for disease resistance improvement of rice.11
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (31822042, 31871946).
Abbreviations
- JA
jasmonic acid
- IAA
indole-3-acetic acid
- Xoo
Xanthomonas oryzae pv. oryzae
- Xoc
Xanthomonas oryzae pv. oryzicola
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Fu J, Yu H, Li X, Xiao J, Wang S.. Rice GH3 gene family: regulators of growth and development. Plant Signal Behav. 2011;6:570–574. doi: 10.4161/psb.6.4.14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Staswick PE, Tiryaki I, Rowe M.. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic and indole-3-acetic acids in an assay for adenylation. Plant Cell. 2002;14:1405–1415. doi: 10.1105/tpc.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui S, Hao M, Liu H, Xiao J, Li X, Yuan M, Wang S. The group I GH3 family genes encoding JA-Ile synthetase act as positive regulator in the resistance of rice to Xanthomonas oryzae pv. oryzae. Biochem Biophys Res Commun. 2019;508:1062–1066. doi: 10.1016/j.bbrc.2018.12.057. [DOI] [PubMed] [Google Scholar]
- 4.Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell. 2008;20:228–240. doi: 10.1105/tpc.107.055657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 2011;155:589–602. doi: 10.1104/pp.110.163774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niño-Liu DO, Ronald PC, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. 2006;7:303–324. doi: 10.1111/j.1364-3703.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 7.Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. A pair allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 2009;151:936–948. doi: 10.1104/pp.109.145623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19:2064–2076. doi: 10.1105/tpc.106.046250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, Dong X. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545:491–494. doi: 10.1038/nature22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ju Y, Tian H, Zhang R, Zuo L, Jin G, Xu Q, Ding X, Li X, Chu Z. Overexpression of OsHSP18.0-Cl enhances resistance to bacterial leaf streak in rice. Rice. 2017;10:12. doi: 10.1186/s12284-017-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


