ABSTRACT
How, if and in which cell types embryonic gene expression programs are elicited to induce tumor formation remains poorly understood. Through genomic analyses of regenerating, p53 deficient muscle stem cells we identified various oncogenomic amplifications, including but not limited to, the zygotic transcription factor Duxbl/DUXB to initiate tumorigenic transformation.
KEYWORDS: Cell of origin, muscle, stem cell, regeneration, genomic instability, rhabdomyosarcoma, zygotic gene activation, Duxbl, DUXB, DUX4, TP53, MET
A central challenge in cancer biology is the characterization of the ‘cancer-cell-of-origin’, i.e. the first cell that transforms and acquires sufficient capabilities to propagate into a tumor. Notably, it is commonly thought that progressive accumulation of genomic alterations and/or mutations are needed for neoplastic transformation.1 The hypothesis has been put forward that tissue-resident stem cells (SC) are more susceptible, due to their extended lifespan, to acquire oncogenomic lesions that would transform them to cancerous SCs. The inherent capacity of regenerative SCs to self-renew and proliferate, together with the observation that cancer often occurs in tissues with high regenerative capacity, supports this hypothesis.2
The adult skeletal muscle provides a paradigmatic example of tissue regeneration which is mediated by and dependent on rare Pax7 expressing muscle SCs (also known as satellite cells).3 Under physiological resting conditions, muscle SCs are predominantly quiescent but become activated upon regenerative cues such as an inflicting injury or during chronic regeneration in certain muscle diseases. Mice harboring an inactivating mutation in the gene dmd encoding for Dystrophin (known as mdx mice) mimic certain features of Duchenne muscular dystrophy including persistent and progressive muscle degeneration of mature myofibers. This elicits a constant regenerative pressure on the skeletal muscle and results in continuous activation of muscle SCs contributing to de novo myofiber formation.4 Recently, it was shown that germline inactivation of the tumor suppressor protein Trp53 (TP53 in humans, best known and hereafter referred to as p53) in chronically regenerating mdx mice develop fusion-negative rhabdomyosarcoma (RMS),5,6 a rare and aggressive childhood cancer and the most common soft-tissue sarcoma in children and adolescents.7 The cancer cell-of-origin under these settings is still unknown but we reasoned that muscle SCs could be a cellular origin of RMS due to the constant regenerative pressure on the muscle SC compartment in mdx mice. To test this we set up an inducible strategy coupling fluorescent lineage tracing and p53 deletion specifically in muscle SC in the mdx background (these mice are referred to as SCp53/mdx). In our model, all SCp53/mdx mice developed lineage-traced embryonic rhabdomyosarcomas (eRMS) in, or in immediate proximity to, the musculature of extremities clearly indicating the cancer cell of origin to be Pax7 expressing muscle SCs in these animals. Contrary, wild-type, mdx or SCp53 mice never developed tumors, which led us to conclude that muscle SC-specific loss of p53 in a regenerative environment is sufficient to generate RMS, or conversely, that a regenerative environment enables RMS formation upon muscle SC-specific loss of p53. Importantly, fluorescent lineage tracing of muscle SC through activation of a Rosa26Tomato reporter enabled FACS-based separation of tumor propagating cells (TPC) from the primary tumors, i.e.: lineage traced muscle SC successors that had transformed and were p53 deficient (TPCTOMpos) and non-lineage traced cells (TPCTOMneg) that essentially contained non-recombined, intact p53. Only TPCTOMpos but not TPCTOMneg developed secondary tumors when injected into immunocompromised mdx-nude mice further confirming that “genuine” RMS-forming cells originate from muscle SCs. These data additionally disclosed that the (non-traced) stromal cells in fact are not responsible for tumor initiation nor are they capable of forming tumors in secondary recipients (Figure 1(a)).
Figure 1.
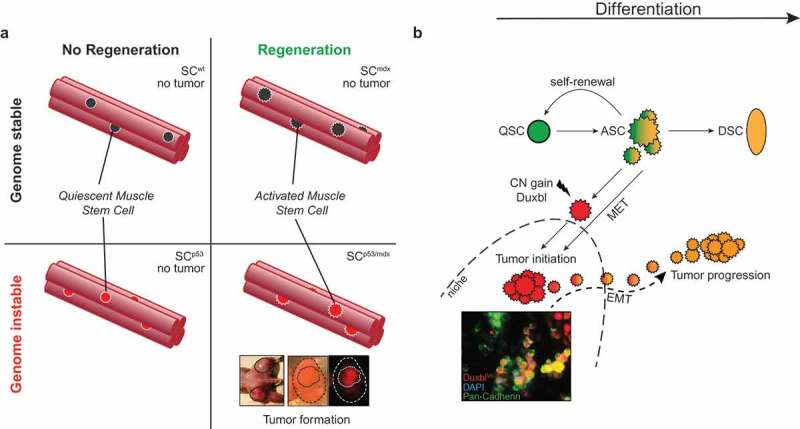
Interplay of stem-cell dependent tissue regeneration and genomic instability in tumor formation. (a) Relationship of regeneration and genomic instability in rhabdomyosarcoma (RMS) formation from muscle stem cells (SC). Both muscle regeneration and genomic instability are necessary to induce RMS tumors originating from muscle SCs. (b) Model of Duxbl/DUXB-mediated tumorigenesis. Healthy SCs contribute to muscle regeneration by differentiation of activated SCs upon injury. Copy number gain or expression of Duxbl/DUXB in activated SCs suppresses differentiation and promotes a gain of plasticity accompanied by epithelialization and initiation of tumorigenic colonies (inset). A secondary event likely involving EMT enables outgrowth of tumor cells from the tumor colony. ASC, activated stem cell; CN, copy number; DSC, differentiated stem cell; EMT, epithelial-to-mesenchymal transition; MET, mesenchymal-to-epithelial transition; QSC, quiescent stem cell.
We found that p53-deficient muscle SCs show increasing rates of DNA double-strand breaks which prompted us to search for recurrent genomic alterations in purified TPCTOMpos that would point to causal mutations playing a role in tumor formation. Surprisingly however, the mutational load in TPCTOMpos was astonishingly low indicating that a progressive accumulation of mutations in muscle SCs is likely not the cause of tumorigenic transformation. Instead, and in almost every individually analyzed specimen, we identified discrete genomic copy number (CN) amplifications many of which harbor genes of known mutational targets of eRMS including Yap1, Cdk4, Met, Jun and others. A subset of tumors revealed a more subtle but recurrent amplification of the poorly described genomic locus 14qA3. This region harbors the Duxbl gene which is synteny to its putative human ortholog DUXB. Duxbl/DUXB belongs to the homeobox-containing Dux family of transcription factors with human DUX4 as its founding member. Notably, DUX4, or the murine ortholog Dux, was recently shown to be responsible for driving gene expression signatures known as zygotic gene activation (ZGA) at the cleavage stage of totipotent zygotes.8,9 These observations led us to test whether Dux transcription factors might act at a putative interface of stem cell potency and tumor formation. To this end, we overexpressed Duxbl in wildtype muscle SCs. Strikingly, this impaired myogenic differentiation and resulted in the emergence of immortalized and morphologically rounded clones (referred to as SCDuxbl) prone to spontaneously form epithelial-like spherical aggregates, which is reminiscent of mesenchymal-to-epithelial (MET) transition. Intriguingly, we found that SCDuxbl expressed the pluripotency factors Sox2 and Klf4 both of which are required for inducing the essential process of MET during reprogramming of somatic cells to induced pluripotent stem cells (iPSCs).10 Transplantation of SCDuxbl led to tumor formation upon subcutaneous transplantation in immunocompromised mice. Interestingly however, SCDuxbl contributed to myofiber formation when injected directly into the strongly pro-differentiating environment of skeletal muscle. Thus, depending on the local microenvironment SCDuxbl can adopt different cell fates. We concluded that overexpression of Duxbl confers a cellular plasticity via a MET-like process but a niche with constant low differentiation pressure is required to facilitate tumor initiation and colonization which could be provided in conditions of chronic injury (Figure 1(b)). The Dux-factor/ZGA axis appears to play an important role beyond RMS since we found this axis to be reactivated in over different 30 types of human cancer. From our study a unifying theme emerges indicating how genomic instability can trigger reactivation of zygotic gene networks through a low number of oncogenomic lesions in regenerating SCs, which in turn can elicit (re-)acquisition of SC plasticity features acting at a three-sided interface of SC potency, cancer formation and regenerative potential.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Visvader JE. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 2.Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Günther S, Kim J, Kostin S, Lepper C, Fan C-M, Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13(5):590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldrin L, Zammit PS, Morgan JE. Satellite cells from dystrophic muscle retain regenerative capacity. Stem Cell Res. 2015;14(1):20–29. doi: 10.1016/j.scr.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camboni M, Hammond S, Martin LT, Martin PT. Induction of a regenerative microenvironment in skeletal muscle is sufficient to induce embryonal rhabdomyosarcoma in p53-deficient mice. J Pathol. 2012;226(1):40–49. doi: 10.1002/path.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21(9):2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 7.El Demellawy D, McGowan-Jordan J, de Nanassy J, Chernetsova E, Nasr A. Update on molecular findings in rhabdomyosarcoma. Pathology. 2017;49(3):238–246. doi: 10.1016/j.pathol.2016.12.345. [DOI] [PubMed] [Google Scholar]
- 8.Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49(6):925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madissoon E, Jouhilahti E-M, Vesterlund L, Töhönen V, Krjutškov K, Petropoulos S, Einarsdottir E, Linnarsson S, Lanner F, Månsson R, et al. Characterization and target genes of nine human PRD-like homeobox domain genes expressed exclusively in early embryos. Sci Rep. 2016;6:1. doi: 10.1038/srep28995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]


