ABSTRACT
Inter-cellular and inter-kingdom signaling systems of various levels of complexity regulate pathogenic and mutualistic interactions between bacteria, parasites, and fungi and animal and plant hosts. Inter-kingdom interactions between mutualistic bacteria such as rhizobia and legumes during nodulation and between fungi and plants during mycorrhizal associations, are characterized by the extensive exchange of molecular signals, which allow nitrogen and phosphate assimilation, respectively. A novel aspect of this signaling exchange is the existence of specific structures, the exosomes, that carry important molecules that shape the plant–pathogen interactions. Exosomes contain a wide array of molecules, such as lipids, proteins, messenger RNA, and microRNAs, that play important roles in cell-to-cell communication in animal and plant cells by affecting gene expression and other physiological activity in distant cells within the same organism (e.g., during cancer metastases and neuron injuries). In plant cells, it has been recently reported that exosomes go beyond organism boundaries and inhibit a pathogenic interaction in plants. Plant produce and send exosomes loaded with specific small miRNA which inhibit the pathogen infection, but the pathogen can also produce exosomes carrying pro-pathogenic proteins and microRNAs. Therefore, exosomes are the important bridge regulating the signal exchange. Exosomes are small membrane-bound vesicles derived from multivesicular bodies (MVBs), which carries selected cargos from the cytoplasm (protein, lipids, and microRNAs) and under certain circumstances, they fuse with the plasma membrane, releasing the small vesicles as cargo-carrying exosomes into the extracellular space during intercellular and inter-kingdom communication. Animal and plant proteomic studies have demonstrated that tetraspanin proteins are an integral part of exosome membranes, positioning tetraspanins as essential components for endosome organization, with key roles in membrane fusion, cell trafficking, and membrane recognition. We discuss the similarities and differences between animal tetraspanins and plant tetraspanins formed during plant–microbe interactions and their potential role in mutualistic communication.
KEYWORDS: Tetraspanin, exosomes, plant-microbe interaction, multivesicular bodies, nodulation, mycorrhization
Introduction
The multiple functions of tetraspanins
Tetraspanins constitute a diverse superfamily of transmembrane proteins, with 33 members identified in mammals and 17 in the model plant Arabidopsis thaliana. Although tetraspanin homologues have not yet been reported in yeast, bacteria, and archae, they are ubiquitous in eukaryotic cells and the first member of this family likely appeared around 570 million years ago.1,2 Several tetraspanin isoforms exist in deuterostomata, including fruitfly (Drosophila melanogaster), zebrafish (Danio rerio), and humans.3–5 Tetraspanins are involved in basic cell functions such as motility, fusion, and membrane and vesicular trafficking, and play key roles in diverse physiological processes including sperm–egg fusion, antigen presentation, and tissue differentiation.2,6–8 While knockout of the tetraspanins CD9- and CD81 in mice (Mus musculus) results in a clear defect in egg–sperm fusion, other tetraspanins are involved in virus and parasite interactions.2,6–10 More recently it has been reported that a single point mutation, L31S substitution in the third amino acid of TM1 has been associated with field-evolved resistance of cotton bollworm to transgenic Bt cotton.11Tetraspanins also regulate cellular invasiveness or metastases through associating with peptidases, matrix metalloproteinases, and urokinase plasminogen activator surface receptors in animal cells.12 In animals and plants, some tetraspanins have a broad tissue distribution, while others show a restricted expression; for instance, in plant cells, different tetraspanins are expressed in specialized tissues such as the reproductive tissue and meristems, during embryo development, and in specific cell niches, such as the quiescent center or the early initial cells that give rise to lateral roots.13–15
Tetraspanins appear to act as molecular organizers by forming homo- or heterodimers that localize to microdomains known as tetraspanin-enriched microdomains (TEMs). As components of TEMs, both animal and plant tetraspanins maintain a network of interactions with other membrane proteins, such as integrins, and signaling molecules, such as receptors, allowing the assembly of multi-molecular signaling platforms.12 Therefore, tetraspanins are dynamic master organizers within membranes that control the distribution and clustering of associated partner-proteins and thereby regulate cellular functions, such as signaling and adhesion.14,16 Their role in vesicular trafficking and as a component of exosomes, which mediate the transport of protein, lipids, and microRNAs during intercellular and interkingdom communication, open a new avenue of research in pathogenic and mutualistic interactions in plant and animal hosts.17
Tetraspanin structure
As its name implies, tetraspanins are proteins with four membrane-spanning domains. The amino- and carboxy-terminal tails and a small loop between transmembrane regions 2 (TM2) and 3 (TM3) are localized in the cytoplasmic compartment. Two loops, a small one (ECL1) located between TM1 and TM2, and a large one (ECL2) between TM3 and TM4, are extracellular (Figure 1).6,18 ECL2 can be subdivided into a conserved and variable region. The conserved region mediates dimerization, whereas the variable region is required for interactions with non-tetraspanin partner molecules. The highly conserved cysteine residues in the large ECL2 are essential tetraspanin signatures, and the polar residues conserved in the TM regions stabilize the tertiary structure.12
Figure 1.
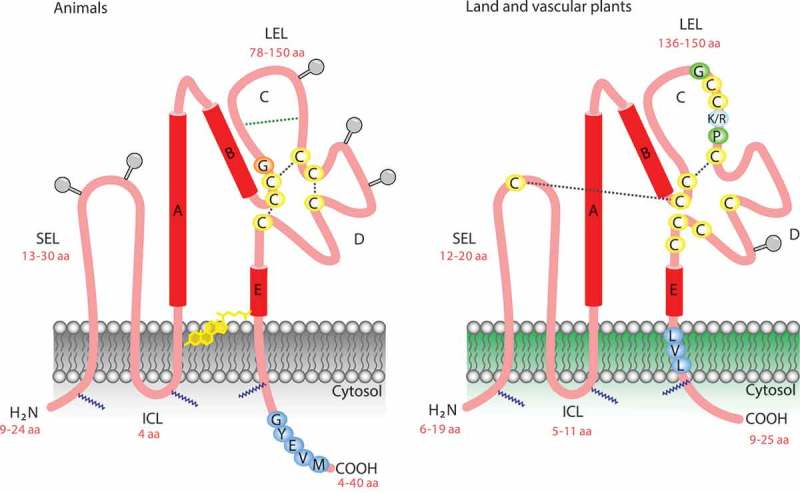
A general model describing the tetraspanin organization in the plasma membrane. Note the similarity and differences in the two extracellular loops. Plant tetraspanins contain a conserved cysteine that is not present in animal tetraspanin. (SEL) small extracellular loop, (LEL) large extracellular loop, (ICL) intracellular cytoplasmic loop, (GYEVM) endosome signal peptide, (GCCK/RPC) conserved sequence in plants, (LVL) putative plasmodesmata sorting.
Palmitoylation of intracellular and juxta-membrane cysteines might be required for initiating tetraspanin–tetraspanin web formation. This chemical modification also protects tetraspanins from lysosomal degradation, provides a link to cholesterol and gangliosides, and promotes cell–cell contact.6,19 In animal cells, palmitoylation of specific integrins also contributes to tetraspanin complex formation.20 Although bona fide integrins have not been identified in plant cells, other proteins such as NDR1 play similar functions.21 In plant cells, some tetraspanins possess a tyrosine-based sorting motif (Yxxϕ, with ϕ representing an amino acid with a bulky hydrophobic side chain) at the carboxyl end that delivers the tetraspanin to a specific intracellular compartment.17,22 However, internalization can also proceed through association with proteins with a sorting motif or by additional, not yet defined domains.12 The major extracellular loop ECL2 is a key region for tetraspanin function in animal and plant cells. Disulfide bonding between conserved cysteines in ECL2 produces a sub-loop structure in this region, which shows the greatest variability between tetraspanins.15,23 Some tetraspanins have an additional two-to-four cysteines within this sub-loop, which may also participate in disulfide bonding. The remainder of the ECL2 region is highly conserved and contains three alpha helices.2 The ECL2 loop appears to determine tetraspanin functional specificity and most specific monoclonal antibodies (mAb) bind to this region.2
Conservation of a single Cys residue at a specific point in the small loop is a unique feature of plant cell tetraspanins, suggesting that it plays a particular role that has not yet been determined. For instance, disulfide bonding between the big and small loops, which is important for tetraspanin–tetraspanin interactions, could stabilize the interaction in a redox-dependent fashion or affect the affinity for cholesterol or gangliosides present in the plasma membrane.24 In contrast to animal tetraspanins, plant tetraspanins have nine, rather than four, six, or eight, completely conserved Cys residues.14 In plants, missense mutations in these Cys residues in trn2–2 and trn2–3 alleles, or the exchange of an uncharged amino acid (Pro-164 in trn2–2 and Gly-177 in trn2–3) with a charged amino acid (Gluc) within the conserved EC2 region containing three Cys residues, could affect folding due a defect in disulfide bridges and thereby inhibit tetraspanin function.25 This is intriguing, as crystallographic data and an analysis of the molecular dynamics of tetraspanin CD81 ECL2, showed the conformational plasticity of the EC2 region under different pH values and redox environments, suggesting that it acts as a molecular sensor of environmental change.15,26 It is possible that plant tetraspanins could have a similar response under pH and redox changes. Little is known about the C-terminal tail of tetraspanins, a divergent and short region (4 to 40 amino acids) in animals and plants that is longer in fungi.14,15,27 However, the C-terminal tail has been implicated in trafficking, as discussed below.28–30
Tetraspanin interactors: what we have learned from the animal field and how little is known in plant cells
In animal cells, the most prominent tetraspanin interactors are the integrins, growth factor receptors, G-protein-coupled receptors (GPCRs), several peptidases, transmembrane proteins associated with tumor progression (CD44), epithelial cell adhesion molecule (EPCAM), immunoglobulin (Ig), protein kinase C PKC), phosphatidylinositol 4-kinase (PI4KII), and phospholipase Cγ (PLCγ).12 By contrast, the interactors of plant tetraspanins are unknown and the literature is limited to the expression and subcellular localization of plant tetraspanins in a few cell models,4,14,15,31 and their role in recruiting ROS generating enzymes.32–34
In animal cells, TEM components have been dissociated using various detergents to examine interactions between tetraspanins and other molecules in the TEM. Different levels of interactions, ranging from strong associations to much weaker associations that are stable only in less hydrophobic detergents, such as Brij97, have been observed.23 Three types of primary tetraspanin interactions, types I–III, exist. Type I interactions involve direct protein–protein interactions, such as those occurring in tetraspanin homodimers, homotrimers, homotetramers, and some heterointeractions.23,35 Most tetraspanin–integrin and tetraspanin–tetraspanin interactions are type II interactions, which are less strong. Palmitoylation of tetraspanins, and possibly of their associating proteins, is essential for this type of interaction, which may be initiated in the Golgi apparatus.36 Weak type III interactions, such as those occurring between tetraspanin and several kinases, are also stabilized by palmitoylation. In addition to primary interactions, the functional activity of tetraspanins also depends on cholesterol and gangliosides, which enable higher-order tetraspanin complexes to form, which constitute the TEM.37 TEMs provide a signaling platform that recruits tetraspanin and its interactors. The palmitoylation of juxta-membrane cysteine residues of tetraspanins (Figure 1) is critical for the assembly of TEMs and is necessary for tetraspanin/tetraspanin interactions. Some integrins are also palmitoylated, and this modification appears to promote TEM formation. Cholesterol embedded in the membrane of the endosome is thought to stabilize TEMs.
Although TEMs share features with lipid rafts, they are independent and distinct from these structures. For instance, lipid rafts, but not TEMs, are disrupted by Triton X-100 at 4°C and the signature molecules of classical rafts, such as the glycosylphosphatidylinositol-anchored proteins and caveolin, do not associate with tetraspanins.12 Furthermore, TEMs can be purified in low-density sucrose gradient fractions and are enriched in certain lipids (e.g., cholesterol and ganglioside GM3), while lipid rafts have different biophysical properties, are more sensitive to cholesterol depletion, and contain different arrays of membrane proteins.6,7 Visualization of TEMs by fluorescence microscopy suggested that they are confined in discrete units at the nanometric scale for each class of tetraspanin. Recent single-molecule analyses of tetraspanin CD9 in animal cells shed light on tetraspanin dynamics in TEMs and the plasma membrane, redefining the tetraspanin web.7,38,39 These studies indicate that tetraspanins form stable interaction platforms that are distinct from lipid rafts and in permanent exchange with the rest of the cell membrane. In addition, CD9 mobility and partitioning into these platforms depends on palmitoylation and cholesterol.2
Although no direct interaction partners of tetraspanins have been identified in plant cells, the cellular distribution of tetraspanin in reproductive tissues has been described.15 Genetic data indicate that AtTET1 functions in a common pathway with TORNADO1 (TRN1), a leucine-rich-repeat protein that regulates patterning processes during Arabidopsis thaliana development, and double mutant analysis showed that these proteins are required for related, overlapping signaling events.14 Furthermore, AtTET1 functions together with WINDHOSE1 and 2 (WIH1/WIH2), two small peptides, to promote megasporogenesis.25,40,41 The short and divergent C-terminal tail of both animal and plant tetraspanins has been linked to targeting to intracellular locations and interaction with cytoskeletal or signaling molecules, including protein kinase C, integrins, and mu3A subunit AP-3.14,15,19 The phenotype of the Arabidopsis mutant trn2-4, which lacks 10 of 14 amino acids of the tetraspanin C-terminal tail, is as dramatic as that of the other trn2 alleles, suggesting that this region of plant tetraspanins is as critical as that described for animal tetraspanins.25 The C-terminal tail of the fungal tetraspanin Tps3 is larger than that of plant and animal tetraspanins and its specific interactors are unknown.27
Tetraspanins are key players in cancer development, with multiple antagonistic effects.42 For example, the tetraspanin CD82 is downregulated, while tetraspanins CD151 and tetraspanin 8 are induced during metastases and support tumor progression.12 Not all activities of tetraspanin rely on primary interactions with associating molecules; some biological phenomena can only be explained by considering the higher organization of tetraspanin complexes, which can interfere with the capacity of tetraspanins to recruit other molecules that negatively regulate a cellular event, such as the activation of a tyrosine phosphatase.43 However, recruitment of specific partner molecules in TEMs such as gangliosides could contribute to CD82-mediated metastasis-inhibiting activity.12 Tetraspanin CD9 also hampers the migration of metastasizing cells by interacting with the cytoskeleton. Indeed, overexpression of CD9 downregulates WAVE2 by a yet unknown mechanism,44 since WAVE is part of the ARP2 and ARP3 complex in actin polymerization, it links upstream signals to activation of the ARP2 and ARP3 complex (e.g., during lamellipodium and filipodium formation). Some tetraspanins have a Yxxϕ sorting motif, located close to the transmembrane region, that can disrupt the binding of the μ subunit of the AP2 adaptor complex, resulting in another cytoskeleton key regulatory component.12,17 Therefore, tetraspanins exert their function by modulating, stabilizing, or preventing the activities of their associated molecules, depending on the composition of the TEM.12,44,45
Role of tetraspanin in cell trafficking
Although tetraspanins were first identified as cell surface markers, it is now clear that some associate with the endosomal pathway, i.e., in early and late endosomes, multivesicular bodies [MVBs], clathrin-coated vesicles, lysosomes, and various types of secretory vesicles.46 Tetraspanins on the plasma membrane can be internalized via endocytosis and traffic to intracellular vesicles.47,48 Conversely, on cell stimulation, secretory vesicles containing tetraspanins fuse with the plasma membrane.46 Late endosomes and MVBs also fuse with the plasma membrane and release vesicles containing microRNA, proteins, and lipids known as exosomes that are enriched in certain tetraspanin proteins.49 Exosomes released from cells infected with intracellular pathogens, including mycobacteria, are potent stimulators of inflammation in uninfected cells.50 Thirteen of the 33 mammalian tetraspanins contain potential tyrosine-based sorting motifs, which have also been described in plants. However, in plants, additional motifs have been identified in tetraspanins, and the coat protein clathrin is necessary and sufficient for the uptake of cargo into clathrin-coated vesicles (CCVs). These motifs in the tetraspanins are based on the sequences YXXØ [D/E]XXXL[L/I] and FXNPXY, which are recognized by a family of adaptor protein (AP) complexes that determine the cellular localization of interacting proteins.2,17 The intracellular trafficking of tetraspanin CD63 has been well described.17,22 CD63 has a conserved GYEVM motif at its C-terminus, which interacts primarily with AP-2 complexes, linking tetraspanins to clathrin-mediated endocytosis pathways, and with AP-3 complexes for trafficking from endosomes to lysosomes. Furthermore, CD63 is also targeted to lysosomes directly from the trans-Golgi network.22 Tetraspanin CD63 interacts directly through its C-terminus with syntenin, which may compete with AP-2 and AP-3 to alter the trafficking of syntenin and therefore link its spatial localization with actual function.17,22 It has also been recently suggested that CD63 in animal cells is internalized from the plasma membrane by endocytosis of a specialized type of lipid raft known as caveolae, linking tetraspanin to the regulation of endocytosis.2,17,22
Tetraspanins as key players in pathogenic interactions in animal and plant cells
Several infectious agents evolved mechanisms to exploit tetraspanins for pathogen entry, subsequent intracellular trafficking, replication, and even exit from the host cell. Tetraspanins have been associated with or shown to mediate infection by human hepatitis C (HCV), HIV, Plasmodium species that cause malaria, certain types of bacteria, and even prions.2 Some viruses (e.g., HIV, human T-cell leukemia virus [HTLV]) induce cell–cell fusion in a tetraspanin-dependent manner, resulting in the formation of giant cells or viral syncytia that contribute to spread of infection.2,51 In some cases, tetraspanins act as pathogen receptors; however, they can also work as ‘molecular organizers’, forming TEMs by the lateral association of tetraspanins with other tetraspanin and non-tetraspanin membrane proteins.2
In T cells themselves, tetraspanins in TEMs promote actin cytoskeleton associations that may lead to more efficient signaling or contribute to the formation of the immune synapse in T cells and it has been suggested that tetraspanins facilitate antigen presentation or antigen signaling.2 The localization of tetraspanins in TEMs has made it challenging to define the functions of individual family members, mainly because the antibodies that cross-link specific tetraspanins also perturb the organization and functions of interacting molecules, giving misleading information.23,43 However, the soluble recombinant EC2 domain is an alternative tool to investigate the role of specific tetraspanins.52 Recombinant EC2s appear to fold correctly and have biological activity in a number of systems.2,52 Nonetheless, knockouts clearly demonstrate that particular cellular functions (e.g., T-cell proliferation, sperm–egg fusion) can be affected by the modulation of a specific tetraspanin. Targeting of individual tetraspanins may provide a therapeutic means to modulate cellular processes without global detrimental effects on the organism, including plant cells.2
Plants have the capability to respond to abiotic and biotic stresses, including fungal pathogens. Fungi are responsible for major diseases of agriculturally important plants. For example, Magnaporthe grisea is the causative agent of the most devastating fungal disease (blast) of rice (Oryza sativa) worldwide, Botrytis cinerea is a necrotrophic pathogen affecting more than 200 different host plants, and Colletotrichum lindemuthianum is responsible for anthracnose diseases in a wide range of crops and ornamental plants. The ability of plant pathogenic fungi to infect their host plants depends on successful penetration into plant tissues. During the infection process, these fungi differentiate a specialized cell called the appressorium, in a process involving arrest of polarized growth, apex swelling, and cell wall reinforcement27,53,54 (Figure 2). Appressorium-mediated penetration occurs through the formation of a penetration peg at the base of the appressorium lying against the plant substratum, a region called the ‘appressorium pore’. The narrow penetration peg is able to perforate the host surface, which includes the plant cuticle and cell wall, allowing the fungus to penetrate into host tissues.27,53,54 The penetration peg emerges at a certain point of the base of the appressorium and requires spatial and temporal coordination of cellular functions, including reorganization of cytoskeletal elements; vesicular trafficking; exocytosis, including membrane and synthesis; assembly and remodeling of the cell wall.53,55,56 Fungal tetraspanins have emerged as key coordinators of the infection process in several pathogenic fungi (M. grisea, B. cinerea, and C. lindemuthianum).32,54,57,58 In these phytopathogenic fungi, Pls1 is essential for appressorium-mediated penetration into the host plant, since Pls1 null mutations result in appressoria that are unable to form functional penetration pegs.27,53,58 Using a transcriptional fusion between the PLS1 promoter and an eGFP reporter gene, clear expression is observed during the penetration of B. cinerea.54 These results suggest that processes involved in appressorium-mediated penetration are conserved among taxonomically unrelated fungal species.27,54 Since peg formation and penetration require the re-establishment of cell polarity at a focal point localized at the base of the appressorium, Pls1 tetraspanins might be required for the correct localization of the emergence site of the penetration peg. The finding that penetration peg was aborted or mislocalized in C. lindemuthianum clpls1 null mutants supports this interpretation.58 Therefore, functional studies of Pls1 tetraspanins in pathogenic fungi may reveal more general functions such as the generation of positional information during specific stages of fungal morphogenesis.
Figure 2.
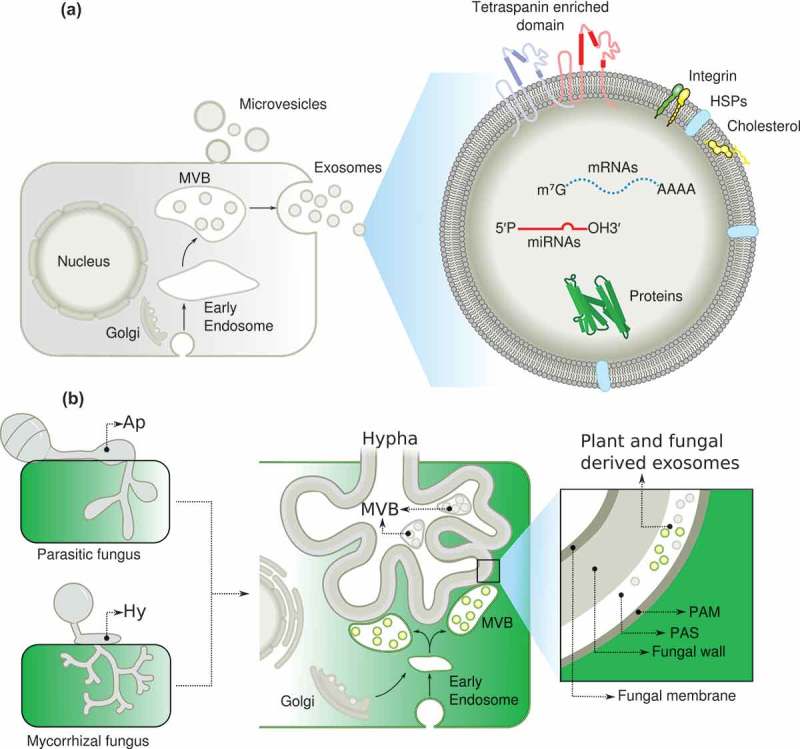
A model describing the role of exosomes in inter-kingdom communication. (a), Note the key role of exosomes in transporting molecules and tetraspanin in the exosome membranes as a marker. (b) Putative role of exosomes in mutualistic interactions. (MVB) multivesicular bodies, (Ap) appressorium, (Hy) hyphae, (PAM) periarbuscular membrane, (PAS) periarbuscular space, (HSPs) heat shock proteins.
Tetraspanins as regulators of localized ROS generation by NADPH oxidases (NOX)
The wide distribution of tetraspanins in eukaryotes and their notable absence in unicellular fungi suggest that tetraspanins co-emerged with multi-cellular organisms during evolution, and are thus associated with multicellularity.4 It is largely unknown how tetraspanins engage in multi-faceted functions at the cellular level. However, there is an emerging co-occurrence between tetraspanins and the mechanisms of ROS generation.33 In the nematode C. elegans, the rigid and flexible cuticular exoskeleton, which is composed of collagen, protects the internal tissues. Once secreted, this collagen is tyrosine cross-linked in a ROS-dependent manner, in a process assisted by BLI-3, a DUOX NADPH oxidase.59 This process also requires the participation of tetraspanin TSP-15, which recruits the NADPH oxidase.33,34,60 Inactivation of this tetraspanin or of BLI-3 produces similar phenotypes. This finding suggests a connection between the mechanism generating ROS and the tetraspanins. In fungi, tetraspanins have been related to NOX function based on the fact that mutants lacking either of these proteins show the same phenotypes. Furthermore, during plant infection with the pathogenic fungus M. grisea or B. cinerea, the tetraspanin PLS1 specify the infection sites, and the tetraspanin and ROS generated by NADPH oxidase coordinate ROS production at the infection site. Furthermore, in the pathogenic C. lindemuthianum, a tetraspanin is required in the appressorium, which is a swollen not growing structure, but in order to progress the fungal infection a re-initiation of the polarity is required.32–34,58,61 Furthermore, in Claviceps purpurea, Nox2 and Pls1 are also important for host–pathogen interaction.62
These data point to a strong connection between tetraspanins and the ROS generating machinery, both in animal and plant cells, suggesting that tetraspanins regulate localized ROS production.33,34,60,63 Localized ROS production is also required during Casparian strip lignification in plants, where CASP, a protein that recruits NADPH oxidase, plays a role similar to tetraspanin, by bringing together NADPH oxidase and peroxidase and ensuring the localized activation of the oxidase.64 Tetraspanins have also been found at the tip of growing pollen tubes, a region that requires NADPH oxidase activity for the generation of ROS, key players in the regulation of polar growth.65,66 Moreover, tetraspanins accumulate at the site of female gametophyte differentiation, suggesting an active role during pollen fertilization, a well-described ROS-dependent process that regulates programmed cell death.15 It is tempting to speculate that tetraspanins recruit the ROS-generating machinery in a site-localized manner.
Exosomes as signaling shuttles, from inter-cellular communication to inter-kingdom communication
Inter-cellular and inter-kingdom signaling require specific molecules that can diffuse and be recognized by a different cell. Exosomes have emerged as a ubiquitous mechanism for transferring information between cells and organisms across all three kingdoms of life.67,68 Initially described in animal cells as a general signaling pathway that allows communication between tumor and distal tissue during metastases, exosomes have also been shown to function in immune surveillance in plant cells during host–pathogen interactions,69–72 and we propose that exosomes could also contribute to the regulation of mutualistic interactions.
Exosomes containing proteins, lipids, and RNAs produced by fungal pathogens can promote plant infection by transmitting pathogen-related molecules that could escape the host immune system or inhibit immune responses by favoring immune cell apoptosis. Furthermore, exosomes secreted by the plant host can have anti-infection roles by inducing immune responses that inhibit pathogen proliferation or infection. Therefore, exosomes might act as regulatory “bridges” during plant–pathogen interactions.70
How are exosomes produced and how are they related to tetraspanins? The general view is that exosomes are derived from multivesicular bodies (MVBs), which form when the endosomal membrane undergoes invagination, taking up proteins, lipids, inhibitors of apoptosis, and microRNAs from the cytosol.73,74 The resulting vesicles inside the endosomes are termed MVBs. MVB either fuse with lysosomes or the plasma membrane, releasing their intraluminal vesicles as exosomes. There are other similar structures such as the ectosomes and microvesicles, which are assembled and released directly from the plasma membrane and therefore represent a different size and origin. The molecular composition of exosomes reflects their origin from intraluminal vesicles and includes several tetraspanins both in animals and plants.71 It is important to note that exosomal proteins maintain their activity. Another notable feature is phosphatidylserine content on the outer membrane leaflet of the exosome in animal cells, which facilitates or triggers exosome uptake by phosphatidylserine-binding proteins (such as scavenger receptors, integrins, and complement receptors). The finding that exosomes can contain mRNAs and microRNAs that can be transferred to target cells where they can be translated or mediate RNA silencing, is a critical aspect of intercellular communication during the immune response, cell-to-cell spread of infectious agents, and tumor progression.75–79 This process is specific to the target cell, such that RNA is transcribed in one cell, but travels long distances to affect another cell. Notably, the relative abundance of proteins, mRNA, and microRNA differs between exosomes and the cytoplasm of donor cells, suggesting a certain degree of specificity in the segregation of these components and therefore in the specificity of this type of communication. This implies that cargo is actively sorted into multivesicular bodies, through mechanisms such as monoubiquitylation, localization in cholesterol-rich membrane microdomains, or higher-order oligomerization. Although the contribution of tetraspanins to the composition of exosomes is well established, their impact on the functions of exosomes is unknown. However, it seems likely that tetraspanins are involved in the cross-talk between distant cells.12 Specific tetraspanins and their interacting proteins, might contribute to exosome function and define the target cell.80 Future studies providing the molecular mechanism to select the cargo and target cells by the exosomes will provide important information to manipulate the formation and targeting of exosomes.2,81,82
Exosomes in inter-kingdom communication during plant–pathogen interactions and future directions in mutualistic interactions
Eukaryotic regulatory small RNAs (sRNAs) that induce RNA interference (RNAi) are involved in a plethora of biological processes, including host immunity and pathogen virulence for successful infection.83–85 In plants, diverse classes of sRNAs contribute to the regulation of host innate immunity. These immune-regulatory sRNAs operate through distinct RNAi pathways that trigger transcriptional or post-transcriptional gene silencing. Remarkably, the influence of regulatory sRNAs is not limited to the individual organism in which they are generated and it can sometimes extend to interacting species from different kingdoms.71,86 Similarly, many pathogen-derived sRNAs regulate pathogen virulence, where they trigger gene silencing in the interacting organism, a phenomenon called cross-kingdom RNAi.85,86 Aggressive fungal pathogens such as Botrytis and Verticillium spp. that cause severe crop losses worldwide secrete effector proteins that suppress host immune responses.86 Notably, the plant fungal pathogen Botrytis cinerea delivers and translocates small RNA effectors into host cells, where they hijack the plant RNAi machinery and suppress host immunity.85,86 Upon B. cinerea infection, expression of the microRNA Bc-siR37 is specifically induced in the fungus, downregulating at least eight predicted Arabidopsis target genes and thereby enhancing disease susceptibility to B. cinerea.86 Furthermore, knockout of the target genes of Bc-siR37 enhances disease susceptibility, further indicating that these genes promote plant defense against B. cinerea.86 In summary, by regulating the expression of specific target genes in the plant host, it is possible to control multiple fungal diseases simultaneously. Therefore, cross-kingdom RNAi opens up a vastly unexplored area of research on mobile sRNAs in the host–pathogen interactions.71,78,85 The big question is how is cross-kingdom communication carried out? Exosomes are produced in all domains of life, but little is known about exosomes in plants despite their implication in plant defense.71,87–89 This work lays the foundation for the concept of cell-to-cell communication through exosomes in plants.79 This idea is also supported in sunflower (Helianthus annuus), where exosomes in extracellular fluids of seedlings are enriched in cell wall remodeling enzymes and defense proteins which can be taken up by the phytopathogenic fungus Sclerotinia sclerotiorum, and affect fungal growth and even killing fungal cells.88
In animal cells, sRNAs can be transported by extracellular vesicles, specific transmembrane proteins, high-density lipoprotein complexes, or gap junctions.90 The gastrointestinal parasite Heligmosomoides polygyrus also secretes exosomes, thereby transporting microRNAs (miRNAs) into mammalian cells and suppressing host immunity.76 By contrast, the mechanism by which sRNAs are transported from hosts to interacting pathogens is unknown. In plant cells, it is well known that sRNAs travel through plasmodesmata, and thus systemically through the vasculature; however, cross-kingdom/organism RNA interference (RNAi) mediated by exosomes opens up a new area of research.71 Because exosomes transfer miRNAs between animal cells as part of a long-distance signaling system and also in parasites interacting with mammalian host cells, the recent finding that plants and pathogenic fungi employ a similar mechanism involving exosome-mediated sRNA transfer indicates that exosomes constitute a general mechanism for intercellular and inter-kingdom signaling. This is a bi-directional communication with exosomes generated in the host to control the pathogen, but also with exosomes generated in the pathogen to promote the infection. Thus, studies should examine how exosomes travel across the boundaries between organisms of different taxonomic kingdoms.71 In Arabidopsis, the cuticle is an important barrier against pathogens. However, in cuticle mutants, higher ROS production is able to sustain constitutive resistance against B. cinerea due to an increase in the cuticle permeability, which facilitates the diffusion and recognition of the cuticle monomers that induce the innate response. However, increased cuticle permeability could also facilitate exosome transport and diffusion in the permeable cuticle, allowing for neutralization of the pathogen through the exchange of miRNAs.91–93
Mammalian tetraspanins, such as CD63, are specific exosome markers.77 Arabidopsis has 17 TETRASPANIN (TET)-like genes, but only closely related TET8 and TET9, which are structurally similar to mammalian CD63, are induced by B. cinerea infection, at the fungal infection sites and as part of the exosomes.15,94 Since mammalian exosomes are derived from MVBs, it is not surprising that the Arabidopsis thaliana MVB marker Rab5-like GTPase ARA695 is also enriched at infection sites, and is partially co-localized with TET8, suggesting that TET8-associated vesicles are likely derived from MVBs. Electron microscopy studies revealed in Arabidopsis that MVBs fuse with the plasma membrane and release extracellular vesicles at infection sites. Furthermore, TET8-associated vesicles have been visualized withTET8-GFP, but not ARA6, because ARA6 localizes to MVB outer membranes that fused to and remained on plasma membranes.71 Thus, TET8-labeled extracellular vesicles can be considered as a specific marker for plant exosomes. The observations that TET8-GFP-labeled exosomes can be taken up by fungal cells under in vitro conditions71 and that exosome-carried sRNAs can be translocated to fungal cells supports the involvement of a shuttle mechanism in inter-kingdom communication.
Animal tetraspanin proteins can induce the formation of specific TEMs; therefore, the finding that TET8-CFP and TET9-YFP are co-localized at fungal infection sites suggests that a particular TEM exists in those regions where hetero-tetraspanin organization is required. Such a TEM could play a key role in exosome biogenesis and translocation since the tet8 T-DNA knock-out mutant and two independent tet9 CRISPR frame-shift lines resulted in a weak, but consistently enhanced susceptibility to B. cinerea, as compared to the wild type.71 Moreover, tet8tet9 double mutants showed pronounced enhanced susceptibility, suggesting that TET8 and TET9 have partially redundant functions, and supporting the idea that TET8- and TET9-associated exosomes contribute to plant immunity against fungal infection by transferring host sRNAs into fungal cells.71 The transferred sRNAs are thought to suppress fungal pathogenicity by targeting fungal virulence genes. Most Arabidopsis sRNAs transferred to B. cinerea have predicted target genes, such as those involved in trafficking pathways, that are downregulated after infection. When such target genes were mutated in B. cinerea, the resulting strains showed reduced virulence on Arabidopsis, supporting the idea that vesicle trafficking pathways are important for fungal virulence.71,86 Furthermore, transgenic Arabidopsis lines overexpressing those sRNAs displayed a reduced susceptibility to B. cinerea and the pathogen showed reduced expression of the target genes.71 Therefore, exosomes play an essential role in cross-kingdom sRNA trafficking between Arabidopsis and the fungal pathogen B. cinerea.
An interesting parallel has been reported in humans: cross-kingdom trafficking of human miRNAs into the parasite Plasmodium falciparum inhibits expression of its pathogenicity genes, which explains why sickle cell anemia patients, who have elevated levels of these transferred miRNAs, are more resistant to malaria.71,96 It is unclear whether human exosomes are also responsible for sRNA delivery, and how ubiquitous such sRNA trafficking-mediated defense mechanisms are in animals and plants. Functional studies of host-transferred sRNAs may reveal important virulence genes in pathogens and pests. Knowledge of tetraspanin activity during exosome biogenesis, cargo loading, and cross-kingdom RNAi may suggest a strategy to develop effective delivery methods of pathogen-targeting artificial RNAs with the goal of controlling plant diseases during pre- and post-harvesting of crops.
MVBs proliferate in susceptible and resistant barley (Hordeum vulgare) leaves in response to infection by the biotrophic powdery mildew fungus Blumeria graminis f. sp. hordei, and this response is associated with the cell wall defense response.97,98 This response is also exacerbated in the cytoplasm of haustorium-containing epidermal cells, where MVBs proliferate in the vicinity of the cell wall-associated oxidative microburst.99 Since MVBs are related to exosome biogenesis, this could explain the exosome production observed at infection sites.
The successful establishment of the arbuscular mycorrhizal (AM) symbiosis relies on the formation of arbuscules in the inner cortex of the root, which mediates a nutrient exchange between the plant and fungus (Figure 2). A variety of transporter proteins on the peri-arbuscular membrane facilitate nutrient exchange and communication. Arbuscule formation depends on fungal recognition by the host plant, which requires the tight coordination of several cellular processes ranging from the formation of the pre-penetration apparatus to initial fungal colonization and development of arbuscules. During the early stages of infection, mycorrhizal associations also require cross-talk between the host and the beneficial fungus, and this signaling can occur before the hyphae have come into contact with the host. Plants secrete strigolactones, which induce hyphal branching, and stimulate the production and secretion of Myc factors, which allow the colonization process.100,101 It would be interesting to determine if MVBs proliferate at the arbuscule region and to determine at which stages of the mutualistic interaction exosomes are produced. So far, there is clear evidence that the secretory pathway of the plant host plays a critical role in the arbuscule formation, but it is unclear how this pathway is redirected during arbuscule development. It is attractive to speculate that exosomes coordinate the exchange of proteins, lipids, and sRNA during the establishment of AM symbiosis. Transcription factors are known to function in the transcriptional reprogramming of cortical cells during arbuscule development, and it is possible that they are modulated by exosomes released from fungi and the plant host. Studies involving the silencing and overexpression of key proteins involved in exosome biogenesis and function, such as the tetraspanins, will provide further insight into exosome biogenesis and the possible reprogramming of hyphal polarity in the hyphopodium structure which requires a further re-initiation of polarity to infect the host cells and generates the arbuscule formation.
Proteomic analyses of exosome cargo components formed during the AM symbiosis will be instrumental in determining their composition relative to those reported in pathogenic interactions.71,102 Recently, H2O2 has been described as an important component of axonal regeneration after acute injury.103 However, the injured neurons do not express the NADPH oxidase required for ROS generation. Instead, macrophages recruited to the vicinity secrete exosomes carrying NOX enzymes, which are taken up by the injured neuron via endocytosis and promote axonal growth in a ROS-dependent manner.103–105 This is the first report describing the role of exosomes in NADPH oxidase translocation in injured neurons. This is an interesting finding since NOX enzymes also function in the injury response of the fungus Trichoderma atroviride.106 It remains to be determined whether an exosome shuttle system carrying ROS generating enzymes contributes to localized ROS production in mutualistic and pathogenic interactions, at the infection thread or during arbuscule formation. Since the arbuscules and the infected cells require an intense vesicular trafficking, it will be important to decipher the role of tetraspanins and the exosomes in the exchange of key proteins, miRNA and mRNA, required for the mutualistic interaction (Figure 2). It could be involved in hyphal colonization during mycorrhizal association and arbuscular formation. During the legume–rhizobia interaction, the infection thread allows migration of the rhizobium through the root hairs until they reach the cortical cells. It is not clear how root hairs are inverted during infection thread formation, but this process clearly involves a complete change in root hair polarity. Given the involvement of tetraspanins in membrane fusion and endocytosis, we anticipate that tetraspanins might be key players in the regulation of root hair or fungal hypha polarity and in the exocytosis and endocytosis occurring at the tip of migrating infection threads. It is clear that the exosome role is just emerging in the plant biology field, but the parallelism between the pathogenesis and mutualism, and the different strategies used by the host, pathogen or symbiont to modulate the interaction, point out to the exosomes as a key player for tuning the interaction by exchanging important components required for a successful interaction. In this scenery, specific tetraspanin such as AtTET8, which are specific markers for exosomes, will open the door for the manipulating the number of exosomes and understand the role of exosomes in pathogenic and mutualistic interaction.
Funding Statement
We acknowledge the funding support from Conacyt for grants 240595, 132155, 253247 and Investigación en Fronteras de la Ciencia 2015-I-319, and DGAPA-UNAM for grants PAPIIT IV200519 and IN209118. Saúl Jiménez Jiménez was supported by Conacyt PhD Scholarship number 314168.
Acknowledgments
We greatly appreciate the technical support from Juan E. Olivares Grajales and the National Advanced Microscopy Laboratory (LNMA).
References
- 1.Garcia-Espana A, Chung P-J, Sarkar IN, Stiner E, Sun T-T, Desalle R.. Appearance of new tetraspanin genes during vertebrate evolution. Genom. 2008;91(4):326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Hassuna N, Monk PN, Moseley GW, Partridge LJ.. Strategies for targeting tetraspanin proteins. BioDrugs. 2009;23(6):341–359. doi: 10.2165/11315650-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Muto A, Van de Velde J, Neyt P, Himanen K, Vandepoele K. Van Lijsebettens M, Molecular cloning, expression pattern, and phylogenetic analysis of a tetraspanin CD82-like molecule in lamprey Lampetra japonica. Dev Genes Evol. 2016;226(2):87–98. doi: 10.1007/s00427-016-0530-y. [DOI] [PubMed] [Google Scholar]
- 4.Wang F, Vandepoele K, Van Lijsebettens M. Tetraspanin genes in plants. Plant Sci. 2012;190:9–15. doi: 10.1016/j.plantsci.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Espana A, Chung PJ, Zhao X, Lee A, Pellicer A, Yu J, Sun TT, DeSalle R. Origin of the tetraspanin uroplakins and their co-evolution with associated proteins: implications for uroplakin structure and function. Mol Phylogenet Evol. 2006;41(2):355–367. doi: 10.1016/j.ympev.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 7.Levy S, Shoham T. Protein-protein interactions in the tetraspanin web. Physiol (Bethesda). 2005;20:218–224. doi: 10.1152/physiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 8.Rubinstein E, Ziyyat A, Wolf J-P, Le Naour F, Boucheix C. The molecular players of sperm-egg fusion in mammals. Semin Cell Dev Biol. 2006;17(2):254–263. doi: 10.1016/j.semcdb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Harada Y, Yoshida K, Kawano N, Miyado K. Critical role of exosomes in sperm-egg fusion and virus-induced cell-cell fusion. Reprod Med Biol. 2013;12(4):117–126. doi: 10.1007/s12522-013-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jankovicova J, Simon M, Antalíková J, Cupperová P, Michalková K. Role of tetraspanin CD9 molecule in fertilization of mammals. Physiol Res. 2015;64(3):279–293. [DOI] [PubMed] [Google Scholar]
- 11.Jin L, Wang J, Guan F, Zhang J, Yu S, Liu S, Xue Y, Li L, Wu S, Wang X, Yang Y. Dominant point mutation in a tetraspanin gene associated with field-evolved resistance of cotton bollworm to transgenic Bt cotton. Proc Natl Acad Sci USA. 2018;115(46):11760–11765. doi: 10.1073/pnas.1812138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zöller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9(1):40. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Muto A, Van de Velde J, Neyt P, Himanen K, Vandepoele K, Van Lijsebettens M. Functional analysis of the Arabidopsis tetraspanin gene family in plant growth and development. Plant Physiol. 2015;169(3):2200–2214. doi: 10.1104/pp.15.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reimann R, Kost B, Dettmer J. Tetraspanins in plants. Front Plant Sci. 2017;8:545. doi: 10.3389/fpls.2017.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boavida LC, Qin P., Broz M., Becker JD, McCormick S. Arabidopsis tetraspanins are confined to discrete expression domains and cell types in reproductive tissues and form homo- and heterodimers when expressed in yeast. Plant Physiol. 2013;163(2):696–712. doi: 10.1104/pp.113.216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berditchevski F, Odintsova E. Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J Cell Biol. 1999;146:477–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8(2):89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 18.Kovalenko OV, Metcalf DG, DeGrado WF, Hemler ME. Structural organization and interactions of transmembrane domains in tetraspanin proteins. BMC Struct Biol. 2005;5:11. doi: 10.1186/1472-6807-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci. 2003;28(2):106–112. doi: 10.1016/S0968-0004(02)00014-2. [DOI] [PubMed] [Google Scholar]
- 20.Termini CM, Gillette JM. Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol. 2017;5:34. doi: 10.3389/fcell.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knepper C, Savory EA, Day B. Arabidopsis NDR1 is an integrin-like protein with a role in fluid loss and plasma membrane-cell wall adhesion. Plant Physiol. 2011;156(1):286–300. doi: 10.1104/pp.110.169656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2009;315(9):1584–1592. doi: 10.1016/j.yexcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 23.van Deventer SJ, Dunlock VE, van Spriel AB. Molecular interactions shaping the tetraspanin web. Biochem Soc Trans. 2017;45(3):741–750. doi: 10.1042/BST20160284. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman B., Kelly B., McMillan BJ, Seegar TC, Dror RO, Kruse AC, Blacklow SC. Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell. 2016;167(4):1041–1051. e11. doi: 10.1016/j.cell.2016.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cnops G, Neyt P, Raes J, Petrarulo M, Nelissen H, Malenica N, Luschnig C, Tietz O, Ditengou F, Palme K, Azmi A. The TORNADO1 and TORNADO2 genes function in several patterning processes during early leaf development in Arabidopsis thaliana. Plant Cell. 2006;18(4):852–866. doi: 10.1105/tpc.105.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cunha ES, Sfriso P, Rojas AL, Roversi P, Hospital A, Orozco M, Abrescia NG. Mechanism of structural tuning of the Hepatitis C Virus Human Cellular Receptor CD81 large extracellular loop. Structure. 2018;26(1):181. doi: 10.1016/j.str.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Lambou K, Tharreau D, Kohler A, Sirven C, Marguerettaz M, Barbisan C, Sexton AC, Kellner EM, Martin F, Howlett BJ and Orbach MJ. Fungi have three tetraspanin families with distinct functions. BMC Genomics. 2008;9:63. doi: 10.1186/1471-2164-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rous BA, Reaves BJ, Ihrke G, Briggs JAG, Gray SR, Stephens DJ, Banting G, Luzio JP. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13(3):1071–1082. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latysheva N, Muratov G, Rajesh S, Padgett M, Hotchin NA, Overduin M, Berditchevski F. Syntenin-1 is a new component of tetraspanin-enriched microdomains: mechanisms and consequences of the interaction of syntenin-1 with CD63. Mol Cell Biol. 2006;26(20):7707–7718. doi: 10.1128/MCB.00849-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sánchez-Madrid F, Yáñez-Mó M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J Biol Chem. 2006;281(28):19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- 31.Olmos E, Reiss B, Dekker K. The ekeko mutant demonstrates a role for tetraspanin-like protein in plant development. Biochem Biophys Res Commun. 2003;310:1054–1061. [DOI] [PubMed] [Google Scholar]
- 32.Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, Tharreau D, Notteghem JL, Lebrun MH. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc Natl Acad Sci USA. 2001;98(12):6963–6968. doi: 10.1073/pnas.111132998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moribe H, Mekada E. Co-occurrence of tetraspanin and ROS generators: conservation in protein cross-linking and other developmental processes. Worm. 2013;2(2):e23415. doi: 10.4161/worm.23415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moribe H, Konakawa R, Koga D, Ushiki T, Nakamura K, Mekada E, Chisholm AD. Tetraspanin is required for generation of reactive oxygen species by the dual oxidase system in Caenorhabditis elegans. PLoS Genet. 2012;8(9):e1002957. doi: 10.1371/journal.pgen.1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charrin S, Manié S, Billard M, Ashman L, Gerlier D, Boucheix C, Rubinstein E. Multiple levels of interactions within the tetraspanin web. Biochem Biophys Res Commun. 2003;304(1):107–112. [DOI] [PubMed] [Google Scholar]
- 36.Charrin S, Manié S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 2002;516(1–3):139–144. [DOI] [PubMed] [Google Scholar]
- 37.Zuidscherwoude M, et al. Tetraspanin microdomains control localized protein kinase C signaling in B cells. Sci Signal. 2017;10(478):1–15. [DOI] [PubMed] [Google Scholar]
- 38.Espenel C, Margeat E, Dosset P, Arduise C, Le Grimellec C, Royer CA, Boucheix C, Rubinstein E, Milhiet P-E. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J Cell Biol. 2008;182(4):765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuidscherwoude M, Göttfert F, Dunlock VME, Figdor CG, Van Den Bogaart G, Van Spriel AB. The tetraspanin web revisited by super-resolution microscopy. Sci Rep. 2015;5:12201. doi: 10.1038/srep12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cnops G, Wang X, Linstead P, Van Montagu M, Van Lijsebettens M, Dolan L. Tornado1 and tornado2 are required for the specification of radial and circumferential pattern in the Arabidopsis root. Development. 2000;127(15):3385–3394. [DOI] [PubMed] [Google Scholar]
- 41.Lieber D, Lora J, Schrempp S, Lenhard M, Laux T. Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr Biol. 2011;21(12):1009–1017. doi: 10.1016/j.cub.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Couto N, Caja S, Maia J, Strano Moraes MC, Costa-Silva B. Exosomes as emerging players in cancer biology. Biochimie. 2018. doi: 10.1016/j.biochi.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 43.Tucci M, Mannavola F, Passarelli A, Stucci LS, Cives M, Silvestris F. Exosomes in melanoma: a role in tumor progression, metastasis and impaired immune system activity. Oncotarget. 2018;9(29):20826–20837. doi: 10.18632/oncotarget.24846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang CL, Ueno M., Liu D, Masuya D, Nakano J, Yokomise H, Nakagawa T, Miyake M. MRP-1/CD9 gene transduction regulates the actin cytoskeleton through the downregulation of WAVE2. Oncogene. 2006;25(49):6480–6488. doi: 10.1038/sj.onc.1209654. [DOI] [PubMed] [Google Scholar]
- 45.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 46.Murk JL, Humbel BM, Ziese U, Griffith JM, Posthuma G, Slot JW, Koster AJ, Verkleij AJ, Geuze HJ, Kleijmeer MJ. Endosomal compartmentalization in three dimensions: implications for membrane fusion. Proc Natl Acad Sci USA. 2003;100(23):13332–13337. doi: 10.1073/pnas.2232379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Duffield A, Kamsteeg E-J, Brown AN, Pagel P, Caplan MJ. The tetraspanin CD63 enhances the internalization of the H,K-ATPase beta-subunit. Proc Natl Acad Sci USA. 2003;100(26):15560–15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spoden G, Freitag K, Husmann M, Boller K, Sapp M, Lambert C, Florin L, Sommer P. Clathrin- and caveolin-independent entry of human papillomavirus type 16–involvement of tetraspanin-enriched microdomains (TEMs). PLoS One. 2008;3(10):e3313. doi: 10.1371/journal.pone.0003313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guix FX, et al. Tetraspanin 6: a pivotal protein of the multiple vesicular body determining exosome release and lysosomal degradation of amyloid precursor protein fragments. Mol Neurodegener. 2017;12(1):25. doi: 10.1186/s13024-017-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9(6):871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin F, Roth DM, Jans DA, Pouton CW, Partridge LJ, Monk PN, Moseley GW. Tetraspanins in viral infections: a fundamental role in viral biology? J Virol. 2005;79(17):10839–10851. doi: 10.1128/JVI.79.17.10839-10851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grove J, Hu K, Farquhar MJ, Goodall M, Walker L, Jamshad M, Drummer HE, Bill RM, Balfe P, McKeating JA. A new panel of epitope mapped monoclonal antibodies recognising the prototypical tetraspanin CD81. Wellcome Open Res. 2017;2:82. doi: 10.12688/wellcomeopenres.12058.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Veneault-Fourrey C, Lambou K, Lebrun MH. Fungal Pls1 tetraspanins as key factors of penetration into host plants: a role in re-establishing polarized growth in the appressorium? FEMS Microbiol Lett. 2006;256(2):179–184. doi: 10.1111/j.1574-6968.2006.00128.x. [DOI] [PubMed] [Google Scholar]
- 54.Gourgues M, Brunet-Simon A, Lebrun M-H, Levis C. The tetraspanin BcPls1 is required for appressorium-mediated penetration of Botrytis cinerea into host plant leaves. Mol Microbiol. 2004;51(3):619–629. [DOI] [PubMed] [Google Scholar]
- 55.Bourett TM, Howard RJ. Actin in penetration pegs of the fungal rice blast pathogen, Magnaporthe grisea. Protoplasma. 1992;168:20–26. doi: 10.1007/BF01332647. [DOI] [Google Scholar]
- 56.Park G, Bruno KS, Staiger CJ, Talbot NJ, Xu J-R. Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol Microbiol. 2004;53(6):1695–1707. doi: 10.1111/j.1365-2958.2004.04220.x. [DOI] [PubMed] [Google Scholar]
- 57.Gourgues M, Clergeot PH, Veneault C, Cots J, Sibuet S, Brunet-Simon A, Levis C, Langin T, Lebrun MH. A new class of tetraspanins in fungi. Biochem Biophys Res Commun. 2002;297(5):1197–1204. [DOI] [PubMed] [Google Scholar]
- 58.Veneault-Fourrey C, Parisot D, Gourgues M, Laugé R, Lebrun M-H, Langin T. The tetraspanin gene ClPLS1 is essential for appressorium-mediated penetration of the fungal pathogen Colletotrichum lindemuthianum. Fungal Genet Biol. 2005;42(4):306–318. doi: 10.1016/j.fgb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Edens WA, et al. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J Cell Biol. 2001;154(4):879–891. doi: 10.1083/jcb.200103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moribe H, Yochem J, Yamada H, Tabuse Yo, Fujimoto T, Mekada E. Tetraspanin protein (TSP-15) is required for epidermal integrity in Caenorhabditis elegans. J Cell Sci. 2004;117(Pt 22):5209–5220. doi: 10.1242/jcs.01403. [DOI] [PubMed] [Google Scholar]
- 61.Siegmund U, Heller J, van Kann JAL, Tudzynski P, Schmidt HH. The NADPH oxidase complexes in Botrytis cinerea: evidence for a close association with the ER and the tetraspanin Pls1. PLoS One. 2013;8(2):e55879. doi: 10.1371/journal.pone.0055879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schurmann J, Buttermann D, Herrmann A, Giesbert S, Tudzynski P. Molecular characterization of the NADPH oxidase complex in the ergot fungus Claviceps purpurea: cpNox2 and CpPls1 are important for a balanced host-pathogen interaction. Mol Plant Microbe Interact. 2013;26(10):1151–1164. doi: 10.1094/MPMI-03-13-0064-R. [DOI] [PubMed] [Google Scholar]
- 63.Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci. 2014;127(Pt 17):3641–3648. doi: 10.1242/jcs.154906. [DOI] [PubMed] [Google Scholar]
- 64.Lee Y, Rubio MC, Alassimone J, Geldner N. A mechanism for localized lignin deposition in the endodermis. Cell. 2013;153(2):402–412. doi: 10.1016/j.cell.2013.02.045. [DOI] [PubMed] [Google Scholar]
- 65.Foreman J, et al. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422(6930):442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- 66.Takeda S, Gapper C, Kaya H, Bell E, Kuchitsu K, Dolan L. Local positive feedback regulation determines cell shape in root hair cells. Science. 2008;319(5867):1241–1244. doi: 10.1126/science.1152505. [DOI] [PubMed] [Google Scholar]
- 67.Coakley G, Maizels RM, Buck AH. Exosomes and other extracellular vesicles: the new communicators in parasite infections. Trends Parasitol. 2015;31(10):477–489. doi: 10.1016/j.pt.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, Liu Y, Xiu F, Wang J, Cong H, He S, Shi Y, Wang X, Li X, Zhou H. Characterization of exosomes derived from Toxoplasma gondii and their functions in modulating immune responses. Int J Nanomedicine. 2018;13:467–477. doi: 10.2147/IJN.S151110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weidle UH, Birzele F, Kollmorgen G, Rueger R. The multiple roles of exosomes in metastasis. Cancer Genom Proteom. 2017;14(1):1–15. doi: 10.21873/cgp.20015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W, Jiang X, Bao J, Wang Y, Liu H, Tang L. Exosomes in pathogen infections: A bridge to deliver molecules and link functions. Front Immunol. 2018;9:90. doi: 10.3389/fimmu.2018.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360(6393):1126–1129. doi: 10.1126/science.aar4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Y, Xiu F, Mou Z, Xue Z, Du H, Zhou C, Li Y, Shi Y, He S, Zhou H. Exosomes derived from Toxoplasma gondii stimulate an inflammatory response through JNK signaling pathway. Nanomedicine (Lond). 2018;13(10):1157–1168. doi: 10.2217/nnm-2018-0035. [DOI] [PubMed] [Google Scholar]
- 73.Valenzuela MM, Bennit HRF, Gonda A, Osterman CJD, Hibma A, Khan S, Wall NR. Exosomes secreted from human cancer cell lines contain Inhibitors of Apoptosis (IAP). Cancer Microenviron. 2015;8(2):65–73. doi: 10.1007/s12307-015-0167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stahl PD, Raposo G. Exosomes and extracellular vesicles: the path forward. Essays Biochem. 2018;62(2):119–124. doi: 10.1042/EBC20170088. [DOI] [PubMed] [Google Scholar]
- 75.Cai ZY, Xiao M, Quazi SH, Ke ZY. Exosomes: a novel therapeutic target for Alzheimer’s disease? Neural Regen Res. 2018;13(5):930–935. doi: 10.4103/1673-5374.232490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buck AH, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 78.Gonzalez JF, Venturi V. A novel widespread interkingdom signaling circuit. Trends Plant Sci. 2013;18(3):167–174. doi: 10.1016/j.tplants.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 79.Yoon YJ, Kim OY, Gho YS. Extracellular vesicles as emerging intercellular communicasomes. BMB Rep. 2014;47(10):531. doi: 10.5483/BMBRep.2014.47.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lakkaraju A, Rodriguez-Boulan E. Itinerant exosomes: emerging roles in cell and tissue polarity. Trends Cell Biol. 2008;18(5):199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hassuna N, Monk PN, Moseley GW, Partridge LJ. Strategies for targeting tetraspanin proteins: potential therapeutic applications in microbial infections. BioDrugs. 2009;23(6):341–359. doi: 10.2165/11315650-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellavia D, Raimondi L, Costa V, De Luca A, Carina V, Maglio M, Fini M, Alessandro R, Giavaresi G. Engineered exosomes: A new promise for the management of musculoskeletal diseases. Biochim Biophys Acta. 2018;1862:1893–1901. [DOI] [PubMed] [Google Scholar]
- 83.Wang M, Weiberg A, Lin FM, Thomma BP, Huang HD, Jin H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nature Plants. 2016;2(10):16151. doi: 10.1038/nplants.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342(6154):118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weiberg A, Jin HL. Small RNAs - the secret agents in the plant-pathogen interactions. Curr Opin Plant Biol. 2015;26:87–94. doi: 10.1016/j.pbi.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang M, Weiberg A, Dellota E Jr, Yamane D, Jin H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017;14(4):421–428. doi: 10.1080/15476286.2017.1291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gangoda L, Boukouris S, Liem M, Kalra H, Mathivanan S. Extracellular vesicles including exosomes are mediators of signal transduction: are they protective or pathogenic? Proteomics. 2015;15(2–3):260–271. doi: 10.1002/pmic.201400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, de la Canal L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J Exp Bot. 2017;68(20):5485–5495. doi: 10.1093/jxb/erx355. [DOI] [PubMed] [Google Scholar]
- 89.Rutter BD, Innes RW. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol. 2017;173(1):728–741. doi: 10.1104/pp.16.01253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–335. doi: 10.1038/nrm3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blanc C, et al. The cuticle mutant eca2 modifies plant defense responses to biotrophic and necrotrophic pathogens and herbivory insects. Mol Plant Microbe Interact. 2018;31(3):344–355. doi: 10.1094/MPMI-07-17-0181-R. [DOI] [PubMed] [Google Scholar]
- 92.Serrano M, Coluccia F, Torres M, L’Haridon F, Métraux JP. The cuticle and plant defense to pathogens. Front Plant Sci. 2014;5:274. doi: 10.3389/fpls.2014.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.L’Haridon F, Besson-Bard A, Binda M, Serrano M, Abou-Mansour E, Balet F, Schoonbeek HJ, Hess S, Mir R, Léon J, Lamotte O. A permeable cuticle is associated with the release of reactive oxygen species and induction of innate immunity. PLoS Pathog. 2011;7(7):e1002148. doi: 10.1371/journal.ppat.1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol. 2007;144(1):367–379. doi: 10.1104/pp.107.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ebine K, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13(7):853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 96.LaMonte G, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12(2):187–199. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.An Q, Ehlers K, Kogel KH, Van Bel AJ, Hückelhoven R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006;172(3):563–576. doi: 10.1111/j.1469-8137.2006.01844.x. [DOI] [PubMed] [Google Scholar]
- 98.An Q, Hückelhoven R, Kogel KH, Van Bel AJ. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 2006;8(6):1009–1019. doi: 10.1111/j.1462-5822.2006.00683.x. [DOI] [PubMed] [Google Scholar]
- 99.An Q, van Bel AJ, Huckelhoven R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal Behav. 2007;2:4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luginbuehl LH, Oldroyd GED. Understanding the Arbuscule at the Heart of endomycorrhizal symbioses in plants. Curr Biol. 2017;27(17):R952–R963. doi: 10.1016/j.cub.2017.06.042. [DOI] [PubMed] [Google Scholar]
- 101.Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science. 2017;356(6343):1175–1178. doi: 10.1126/science.aan0081. [DOI] [PubMed] [Google Scholar]
- 102.Choi D-S, Yang JS, Choi EJ, Jang SC, Park S., Kim OY, Hwang D., Kim KP, Kim YK, Kim S., Gho YS. The protein interaction network of extracellular vesicles derived from human colorectal cancer cells. J Proteome Res. 2012;11(2):1144–1151. doi: 10.1021/pr200842h. [DOI] [PubMed] [Google Scholar]
- 103.Hervera A, Hervera A, De Virgiliis F, Palmisano I, Zhou L, Tantardini E, Kong G, Hutson T, Danzi MC., Perry RBT, Santos CX, Kapustin AN. Reactive oxygen species regulate axonal regeneration through the release of exosomal NADPH oxidase 2 complexes into injured axons. Nat Cell Biol. 2018;20(3):307–319. doi: 10.1038/s41556-018-0039-x. [DOI] [PubMed] [Google Scholar]
- 104.Krishnamoorthy L, Chang CJ. Exosomal NADPH Oxidase: delivering Redox Signaling for Healing. Biochemistry. 2018. doi: 10.1021/acs.biochem.8b00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, Zahorchak AF, Logar AJ, Wang Z, Watkins SC, Falo LD. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104(10):3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 106.Hernandez-Onate MA, Esquivel-Naranjo EU, Mendoza-Mendoza A, Stewart A, Herrera-Estrella AH. An injury-response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proc Natl Acad Sci USA. 2012;109(37):14918–14923. doi: 10.1073/pnas.1209396109. [DOI] [PMC free article] [PubMed] [Google Scholar]


