Abstract
Motivation
Coalescent- and reconciliation-based methods are now widely used to infer species phylogenies from genomic data. They typically use per-gene phylogenies as input, which requires conducting multiple individual tree inferences on a large set of multiple sequence alignments (MSAs). At present, no easy-to-use parallel tool for this task exists. Ad hoc scripts for this purpose do not only induce additional implementation overhead, but can also lead to poor resource utilization and long times-to-solution. We present ParGenes, a tool for simultaneously determining the best-fit model and inferring maximum likelihood (ML) phylogenies on thousands of independent MSAs using supercomputers.
Results
ParGenes executes common phylogenetic pipeline steps such as model-testing, ML inference(s), bootstrapping and computation of branch support values via a single parallel program invocation. We evaluated ParGenes by inferring > 20 000 phylogenetic gene trees with bootstrap support values from Ensembl Compara and VectorBase alignments in 28 h on a cluster with 1024 nodes.
Availability and implementation
GNU GPL at https://github.com/BenoitMorel/ParGenes.
Supplementary information
Supplementary material is available at Bioinformatics online.
1 Introduction
The availability of genomic data for an increasing number of organisms allows to use thousands of genomic loci (henceforth: genes) to delineate evolutionary relationships between species. Species tree inference methods can be divided into supermatrix and supertree approaches. The former infer the species tree directly from a large concatenated MSA (supermatrix), whereas the latter infer individual per-gene trees which are then reconciled into a species phylogeny. Supermatrix methods are widely used due their simplicity and availability of efficient implementations (Kozlov et al., 2015; Nguyen et al., 2015). However, supertree inference methods gain popularity as they can model events such as incomplete lineage sorting (Mirarab and Warnow, 2015), gene duplication and loss (Arvestad et al., 2003), as well as horizontal gene transfer (Linz et al., 2007).
As input, supertree methods typically require a set of per-gene trees (potentially also including bootstrap trees) that shall be reconciled (Boussau et al., 2012). Inferring this set of per-gene trees using maximum likelihood (ML) methods is computationally intensive and requires the use of cluster computing resources.
While popular parallel tools for ML tree inference [e.g. RAxML (Stamatakis, 2014), IQ-TREE (Nguyen et al., 2015)] can efficiently process large supermatrices, no dedicated parallel tool exists for inferring per-MSA trees on a large set of MSAs. In current studies users deploy ad hoc, and thus potentially error-prone, scripts for submitting each individual gene tree inference to a cluster as a single job. As cluster systems typically limit the number of sequential jobs a single user can execute in parallel, this can substantially increase the time-to-solution.
To this end, we have developed and made available a novel tool called ParGenes. It offers a simple command-line interface that allows to select the best-fit model, infer ML trees and compute bootstrap support values on thousands of gene MSAs in a single MPI run. ParGenes relies on ModelTest-NG (https://github.com/ddarriba/modeltest) and RAxML-NG (Kozlov, 2018) to perform model selection and tree inference, respectively.
2 Features
ParGenes encapsulates all per-gene calculations into one single MPI invocation. To improve load balance and decrease time-to-solution, ParGenes schedules per-gene inferences and allocates a variable number of cores to these inferences within its MPI runtime environment. In the following, we describe some of the key features.
2.1 Simultaneous processing of MSAs
Unlike standard tools for ML inference, ParGenes analyzes multiple MSAs. Thus, the user needs to provide a directory containing all MSAs in PHYLIP or FASTA format. One can either specify global or MSA-specific options for both, RAxML-NG and ModelTest-NG. We pre-process each MSA, to check that the file is valid, compress it, save it in a binary file, and read its number of taxa and unique patterns.
2.2 Model selection
ParGenes employs ModelTest-NG, a re-designed, substantially more efficient version of the widely used Modeltest tool (Posada and Crandall, 1998), to select the best-fit model of evolution for a given MSA. If model testing is enabled in ParGenes, it will first execute ModelTest-NG on each MSA, and then use the best-fit model for subsequent ML inferences.
2.3 ML searches and bootstrapping
ParGenes schedules the per-MSA inference jobs that are executed using RAxML-NG (Kozlov, 2018). ParGenes allows to run multiple RAxML-NG tree searches per MSA from independent starting trees, which is recommended to better explore the tree search space. Then, it identifies the best-scoring ML tree for each gene. To increase job granularity and thereby improve load balance, each independent tree search is separately scheduled. ParGenes can also conduct a user-specified number of bootstrap (BS) inferences. It schedules independent tree inferences of BS replicates (10 BS replicates per job), and subsequently concatenates the resulting trees into one per-MSA BS tree file. Then, it runs RAxML-NG again to compute support values.
2.4 Checkpointing
Since ParGenes performs massively parallel and compute-intensive runs, it also offers a checkpointing feature that allows to resume calculations (e.g. if program execution was interrupted due to typical cluster run-time limitations of 24 or 48 h).
ParGenes keeps track of all jobs that have finished so far, and skips them upon restart from a checkpoint. A job is typically an individual per-gene ML search, a batch of 10 bootstrap replicate searches, or a ModelTest-NG run.
Furthermore, RAxML-NG and ModelTest-NG also have their own intrinsic checkpointing mechanisms: RAxML-NG writes a checkpoint after each inference step (e.g. model optimization, topological optimization cycle, etc.) of the tree search, and ModelTest-NG after each model it tests. ParGenes uses these checkpointing mechanisms as well, thereby allowing for a very fine-grained checkpointing.
2.5 Estimating the optimal number of cores
Given the input MSAs, ParGenes can calculate an a priori estimate of the number of overall cores that will yield ‘good’ parallel efficiency. This is important, as it is difficult for users to set this value prior to running the analysis.
3 Job scheduling
ParGenes implements a scheduler that simultaneously executes independent jobs with a varying number of cores per job. A job is either a per-MSA RAxML-NG or ModelTest-NG run. We first outline the parallelization scheme and then the scheduling strategy.
3.1 Parallelization scheme
For the typical use case, the input data will contain thousands of independent (per-gene) MSAs with hundreds to a few thousand sites each. While standard tools like RAxML parallelize likelihood computations over MSA sites, ParGenes parallelizes the computations over the MSAs. Note that, the parallel efficiency of the RAxML parallelization is limited by MSA length (rule-of-thumb: 1000 MSA sites per core). While most of input MSAs are small, their size exhibits substantial variance with respect to both, the number of taxa, and sites (Supplementary Fig. S1). Therefore, inferring trees on large per-gene MSAs on a single core has two drawbacks. First, the MSA size might exceed the available main memory per core. Second, this can decrease parallel efficiency as a large job might take longer to complete than all other jobs (Supplementary Fig. S2a). To this end, ParGenes allocates several cores for the largest jobs (MSAs) by invoking the respective multi-threaded RAxML-NG executable (Fig. 1). For each MSA, ParGenes first calls RAxML-NG in parsing mode to obtain the recommended number of cores for optimal parallel efficiency via the fine-grained parallelization of the likelihood function in RAxML-NG (Stamatakis, 2015). The actual number of cores assigned to a job is then rounded down to the next power of two to simplify scheduling. We also assign twice the number of cores to the 5% MSAs with the largest number of taxa (Supplementary Material, Section 2).
Fig. 1.
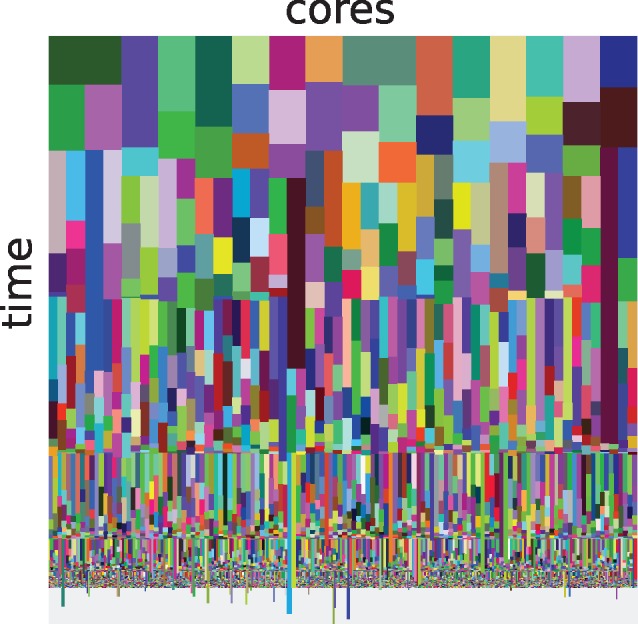
Typical ParGenes core utilization, for a run with 512 cores. Each colored block represents a per-MSA job including the number of cores allocated to the job (x-axis, block width) and the execution time of the job relative to the overall ParGenes execution time (y-axis, block height). The gray blocks depict idle time. The exact experimental setup is described in the Supplementary Material
3.2 Scheduling strategy
ParGenes first sorts all jobs by (i) decreasing number of required cores and (ii) decreasing overall number of characters per MSA. As the number of cores per job (see Section 3) is always a power of two, ParGenes can always keep all cores busy, as long as there are jobs left to process. This works because the MSAs requiring the largest number of cores are scheduled first.
4 Results
We evaluated ParGenes on two large empirical datasets obtained from Ensembl (Zerbino et al., 2018) and VectorBase (Emrich et al., 2015). They comprise 8880 and 12 000 gene families, respectively. Executing the entire ParGenes pipeline on 1024 cores (model testing, ML tree search from 20 starting trees, bootstrapping analysis with 100 replicates) took 25 h for the Ensembl dataset and 3 h for the VectorBase dataset. The VectorBase dataset required less time as its MSAs are smaller. In the Supplementary Material, we show scalability results for different core counts.
5 Conclusions and future work
We have presented an efficient parallel tool for comprehensive phylogenetic inference of gene trees on thousands of MSAs via a single MPI invocation. Apart from being flexible with respect to the inference options, ParGenes also yields ‘good’ parallel efficiency via appropriate scheduling mechanisms. We expect that ParGenes will contribute to increasing throughput times and productivity in gene-tree/species-tree reconciliation studies. Future directions entail the improvement of fault-tolerance mechanisms (e.g. core failures or single jobs failing for other reasons) and more accurate RAxML-NG runtime prediction approaches (e.g. machine learning).
Supplementary Material
Acknowledgements
This work was financially supported by the Klaus Tschira Foundation and by DFG grant STA 860/4-2. We are grateful to B. Bousseau, C. Rey and E. Tannier for providing the datasets.
Conflict of Interest: none declared.
References
- Arvestad L. et al. (2003) Bayesian gene/species tree reconciliation and orthology analysis using MCMC. Bioinformatics, 19, 1–10. [DOI] [PubMed] [Google Scholar]
- Boussau B. et al. (2012) Genome-scale coestimation of species and gene trees. Genome Res., 23, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emrich S.J. et al. (2015) VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res., 43, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov A.M. et al. (2015) Examl version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics, 31, 2577–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov O. (2018) Models, optimizations, and tools for large-scale phylogenetic inference, handling sequence uncertainty, and taxonomic validation, https://sco.h-its.org/exelixis/pubs/dissAlexey.pdf.
- Linz S. et al. (2007) A likelihood framework to measure horizontal gene transfer. Mol. Biol. Evol., 24, 1312–1319. [DOI] [PubMed] [Google Scholar]
- Mirarab S., Warnow T. (2015) Astral-ii: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics, 31, i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T. et al. (2015) Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol., 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics, 14, 817–818. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) Raxml version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2015) Using RAxML to infer phylogenies. Curr. Protoc. Bioinformatics, 51, 6.14.1–6.14.14. [DOI] [PubMed] [Google Scholar]
- Zerbino D.R. et al. (2018) Ensembl 2018. Nuclic Acids Res., 46, D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


