Abstract
Motivation
Live cell imaging plays a pivotal role in understanding cell growth. Yet, there is a lack of visualization alternatives for quick qualitative characterization of colonies.
Results
SeeVis is a Python workflow for automated and qualitative visualization of time-lapse microscopy data. It automatically pre-processes the movie frames, finds particles, traces their trajectories and visualizes them in a space-time cube offering three different color mappings to highlight different features. It supports the user in developing a mental model for the data. SeeVis completes these steps in 1.15 s/frame and creates a visualization with a selected color mapping.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Microfluidics miniaturizes macroscopic systems and enables parallel processing. Microfluidic systems integrates assay operations such as detection, sample treatment and preparation at the microscale. Moreover, they provide controlled and high-throughput environments for biological experimentation. Typically for the growth of one colony, a movie is recorded over the course of thirty to fifty hours. An initial visual inspection of the data would help in order to prepare for technical and biological replicates. When did the colony reach a certain biomass? How was the growth affected by the introduction of a certain stress in the medium (e.g. antibiotic)? The outcome of this visual inspection may help in improving experimental protocols and the quality of the result data.
After each experiment, this visual investigation occurs at a frame-by-frame basis or by employing the general computational paradigm of microfluidics data analysis (i.e. segmentation, tracking and lineage construction). While the former representation of the data highlights temporal evolution without necessarily displaying spatial changes, the latter fails at the segmentation step in cases of low temporal resolution, low object-to-background contrast, irregular cell shapes and/or high cell density. Most well-known tools deal with relatively scattered cells and an acceptable Signal-to-Noise Ratio in sample images (Bray and Carpenter, 2015; Klein et al., 2012; Li et al., 2008). However, only a handful of tools successfully handled movies where cells are amassed in a single field of view (Vallotton et al., 2009). Since a visualization is of great value for visual search and comparison of datasets, we propose to bypass the segmentation problem by using ‘virtual’ particles as previously demonstrated (Hattab et al., 2018). As found in this previous work, the particle approach faithfully represents the reported bacterial exponential growth and is on par with previous manual analyses.
SeeVis employs space-time cubes, which are one of the six classes of visualization methods for live cell imaging data (Pretorius et al., 2017). They can reveal temporal and spatial characteristics in colony development. However, to the best of our knowledge a practical application and discussion of this approach to microfluidics movies in particular has not yet been reported. SeeVis objective is to provide a qualitative visualization to perceive differences in two or more colonies.
2 Materials and methods
SeeVis––(S)egmentation-fr(ee) (Vis)ualization consists of three steps: (i) pre-processing steps to enhance the signal-to-noise and spatially aligning the images (Hattab et al., 2017), (ii) localizing cell particles with feature detection (Crocker and Grier, 1996) and (iii) visualizing the colony as a three-dimension space-time cube. The three color mapping methods are implemented to emphasize different features of colony growth. This refers to perceiving the extent of a colony in space and/or time, according to different data attributes (e.g. temporal trajectory, descent, etc.). The three visualization encodings map each particle trajectory Jk to a triplet: spot size, spot color and spot index or (s, c, f), respectively. Provided Jk, the mapping function: . With spot size s = 3, the spot color c in the RGBA color model and spot index f. The size s was chosen by trial in the local coordinate system or scene coordinates. The alpha channel a of the RGBA spot color varies in [0, 1]. By default, spots scale with the view and are opaque a = 1.
Nominal mapping (NM) highlights each trajectory to support pairwise differentiation and contrast of neighboring trajectories over space and time. It could help users identify relationships between cell pedigrees. To this end, the particle index p of a particle coordinate is treated as a nominal variable. Since the human perceptual system dictates a strong limit on the amount of categorical colors that can be distinguished (Munzner, 2014), we employ a set of unique colors ϒ = 10. The integer indices are mapped to the Tableau10 color palette (Setlur and Stone, 2016). Each color c is chosen randomly for each particle index p.
Time mapping (TM) visually promotes the extent of the population growth over time. To map each particle at a time point t to one spot color c, we use the viridis color palette (Van der Walt and Smith, 2015). It is perceptually uniform and with monotonically increasing luminance in multiple hues. TM adapts to the time span of each dataset by setting its lightest color to the data value, so the perceived brightness encodes the time on a trajectory.
Progeny mapping (PM) supports the process of tracing back single trajectories to their parents. be the set of all trajectories, sub-divided into with trajectories observed in the last frame and the remaining trajectories. The subset is defined as . That is to denote all ‘visible’ trajectories in the last frame of the image data. is displayed using NM. The of is reduced by setting the values of and .
3 Results
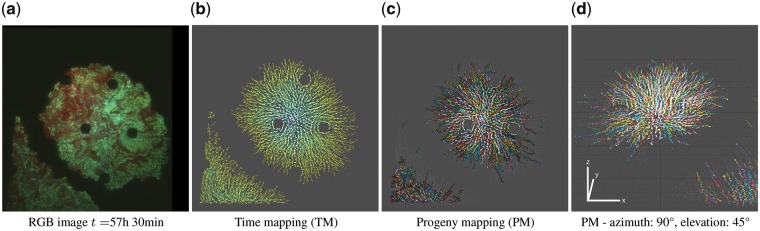
We reference two sets of image data (McIntosh and Bettenworth, 2017; Schlüter et al., 2015) and five simulated movies (Wiesmann et al., 2017). We compared the computation time of all datasets for both pre-processing and particle steps based on 100 runs; see Supplementary Figures S12 and S13 in (Hattab et al., 2018). SeeVis ran on four datasets averaging a speed of 1.15 s/frame. Once the pre-processing and tracking has been done, a CSV output file was created. In this scenario, the average speed increased to 45 ms/frame displaying the visualization of a 44 frames movie in 2 s (see Supplementary Material for further details). SeeVis worked successfully for heterogeneous colonies in dynamic and low fluorescence image content. A cell region is detected as a particle before the cell expands, until it expands and long after the cell has separated into two bacteria. Computationally, results were robust and reproducible (Hattab et al., 2018). An annotated and worked example is provided in the Supplementary Material. In Figure 1a, we observed that another colony invaded the initial field of view. The color mappings are showcased in Figure 1b. TM laid clear emphasis on growth by weighing the factor of time. PM colored trajectories that trace back particles present in the last frame of the image data. It provided temporal emphasis and proved to reduce any possible clutter by decreasing the number of displayed trajectories.
Fig. 1.
Color mappings demonstrated for dataset D1 (Schlüter et al., 2015). (a) RGB image (exposure: +60%). (b, c) The space-time cube is displayed with azimuth = 0∘ and elevation = 90° for two different color mappings and a 30% greyscale background: (b) TM, (c) PM, respectively. (d) PM shown onto a grid
Funding
This work was funded by the German–Canadian DFG International Research Training Group GRK 1906/1.
Conflict of Interest: none declared.
Supplementary Material
References
- Bray M.A., Carpenter A.E. (2015) CellProfiler Tracer: exploring and validating high-throughput, time-lapse microscopy image data. BMC Bioinform., 16, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J., Grier D. (1996) Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci., 179, 298–310. [Google Scholar]
- Hattab G., et al. (2017) Vicar: an adaptive and landmark-free registration of time lapse image data from microfluidics experiments. Front. Genet., 8, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattab G., et al. (2018) A novel methodology for characterizing cell sub-populations in automated time-lapse microscopy. Front. Bioeng. Biotechnol., 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J., et al. (2012) TLM-Tracker: software for cell segmentation, tracking and lineage analysis in time-lapse microscopy movies. Bioinformatics, 28, 2276–2277. [DOI] [PubMed] [Google Scholar]
- Li K., et al. (2008) Cell population tracking and lineage construction with spatiotemporal context. Med. Image Anal., 12, 546–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh M., Bettenworth V. (2017) Onset of Quorum Sensing and Exopolysaccharide Production in Single Cells within Growing Microcolonies. Philipps University of Marburg; https://pub.uni-bielefeld.de/record/2913120. [Google Scholar]
- Munzner T. (2014) Visualization Analysis and Design. A.K. Peters Visualization Series. CRC Press. [Google Scholar]
- Pretorius A.J., et al. (2017) A survey of visualization for live cell imaging. Computer Graph. Forum, 36, 1–18. [Google Scholar]
- Schlüter J.-P., et al. (2015) Phase Contrast and Fluorescence Bacterial Time-Lapse Microscopy Image Data. Bielefeld University, https://pub.uni-bielefeld.de/record/2777409. [Google Scholar]
- Setlur V., Stone M. (2016) A linguistic approach to categorical color assignment for data visualization. IEEE Trans. Vis. Computer Graph., 22, 698–707. [DOI] [PubMed] [Google Scholar]
- Vallotton P., et al. (2009) Segmentation and tracking individual pseudomonas aeruginosa bacteria in dense populations of motile cells. In: 24th International Conference on Image and Vision Computing New Zealand, 2009. IVCNZ ‘09. IEEE, Wellington, New Zealand, pp. 221–225. [Google Scholar]
- Van der Walt S., Smith N. (2015) A Better Default Colormap for Matplotlib. [Google Scholar]
- Wiesmann V., et al. (2017) Simulated Movies of Fluorescently Stained Bacteria. Fraunhofer Institute for Integrated Circuits, Erlangen, Germany. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.