Abstract
Summary
We report VCPA, our SNP/Indel Variant Calling Pipeline and data management tool used for the analysis of whole genome and exome sequencing (WGS/WES) for the Alzheimer’s Disease Sequencing Project. VCPA consists of two independent but linkable components: pipeline and tracking database. The pipeline, implemented using the Workflow Description Language and fully optimized for the Amazon elastic compute cloud environment, includes steps from aligning raw sequence reads to variant calling using GATK. The tracking database allows users to view job running status in real time and visualize >100 quality metrics per genome. VCPA is functionally equivalent to the CCDG/TOPMed pipeline. Users can use the pipeline and the dockerized database to process large WGS/WES datasets on Amazon cloud with minimal configuration.
Availability and implementation
VCPA is released under the MIT license and is available for academic and nonprofit use for free. The pipeline source code and step-by-step instructions are available from the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (http://www.niagads.org/VCPA).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The Alzheimer’s Disease Sequencing Project (ADSP) is an integral component of the National Alzheimer’s Project Act (NAPA) towards a cure of Alzheimer’s Disease (AD). ADSP will eventually analyze whole-genome sequencing (WGS) and whole-exome sequencing (WES) data from more than 20 000 late-onset AD patients and cognitively normal elderly to find new genetic variants associated with disease risk. To ensure all sequencing data are processed consistently and efficiently according to best practices, a common workflow called ‘Variant Calling Pipeline and data management tool’ (VCPA) was developed by the Genome Center for Alzheimer’s Disease (GCAD) in collaboration with ADSP. VCPA is capable to process any kind of germline DNA sequencing data and available for general use. VCPA (i) is optimized for large-scale production of WGS and WES data, (ii) includes a tracking database with web frontend for users to track production process and review quality metrics, (iii) is implemented using the Workflow Description Language (WDL) for better deployment and maintenance and (iv) is designed for the latest human reference genome build (GRCh38/hg38, version GRCh38DH) and follows best practices for WGS analysis with input from TOPMed (Trans-Omics for Precision Medicine) and CCDG (Centers for Common Disease Genomics).
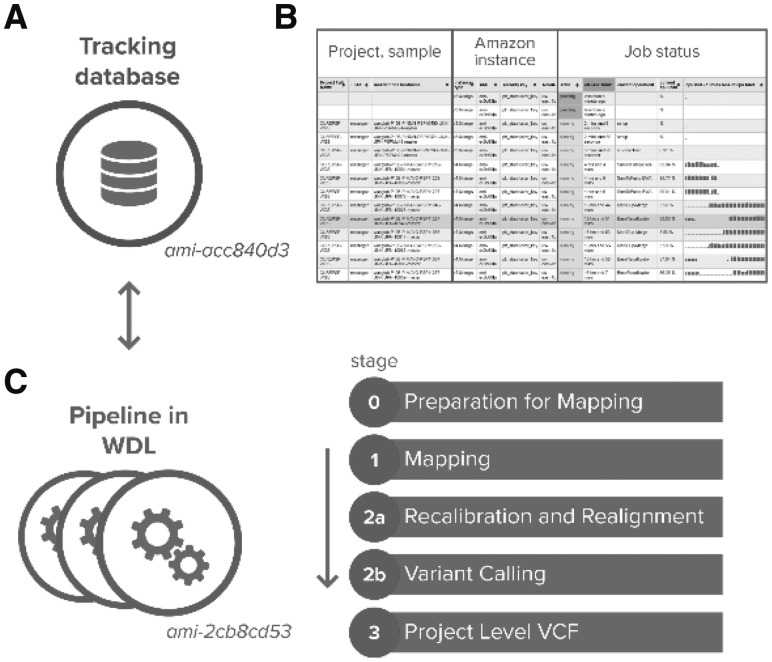
VCPA consists of two independent but interoperable components: a tracking database (Fig. 1A) with a web frontend (Fig. 1B) and a SNP/indel calling pipeline (Fig. 1C). The pipeline was optimized for automatic processing WGS/WES data in various file formats, from mapping sequence reads to the latest human reference genome (GRCh38/hg38) and variant calling. The tracking database (available as another AMI) was designed for monitoring the job status and recording quality metrics for each processed sample (Fig. 1B). With a dynamic web interface of the database, researchers can easily compare, share and visualize all these individual level quality metrics.
Fig. 1.
(A) VCPA tracking database; (B) dynamic view of job status; (C) VCPA Pipeline overview
2 SNP/indel calling pipeline
The variant calling pipeline for the WGS (stages 1 and 2a) was developed with input from CCDG/TOPMed for functional equivalence (Regier et al., 2018) and follows best practices of Germline Single Nucleotide Polymorphisms (SNPs) & Insertion/deletion (indel) Discovery for Genomic Analysis Toolkit (GATK) v3.7 (DePristo et al., 2011). A uniqueness of VCPA is that it accepts either WGS or WES pair-end reads in FASTQ, BAM (binary sequence alignment map format), or CRAM (compressed BAM) formats with flow cell information and genomic regions for exome sequencing enrichment/capture kits. The workflow is modularized and consists of four stages (Fig. 1C). Users can configure the workflow to skip individual stages to reduce the time and cost.
Stage 0 includes preparation steps for read mapping. For samples already mapped previously, PICARD (http://broadinstitute.github.io/picard) is used to roll back BAM files to uBAM (unaligned BAM) files (https://gatkforums.broadinstitute.org/gatk/discussions/tagged/ubam).
Stage 1 generates BAM files. First, reads are mapped to GRCh38/hg38 using BWA-MEM (Li H., 2013) and duplicate reads are marked by BamUtil (https://genome.sph.umich.edu/wiki/BamUtil). Next, BAM files are processed by Samblaster (adding MC and MQ tags to pair-end reads) (Faust and Hall, 2014) and sorted by genomic coordinates using SAMtools (Li et al., 2009). Finally, coverage statistics are computed using Sambamba (Tarasov et al., 2015).
Stage 2A performs local realignment near known indel sites (1000 Genome indels) and recalibration of base call quality scores using GATK v3.7 (McKenna et al., 2010).
Stage 2B implements the GATK best practice steps for variant calling and annotation on SNPs and indels, and generates genotype call files in genomic Variant Call Format (gVCF) for each sample individually. Quality metrics of called variants are computed using GATK (DePristo et al., 2011).
Stage 3 combines gVCF files from multiple samples and performs joint genotype calling using GATK best practices. A project-level VCF file containing genotype information for all polymorphic sites across all samples is generated.
Details for job submission are described in Supplementary Methods. The resulting directory architecture and important VCPA outputs are described in Supplementary Figure S1.
3 Tracking database
The tracking database enables the user to monitor production status (Fig. 1B) and review sequencing quality such as mapping percentage, depth coverage and quality of called variants. All 113 quality metrics are collected during the pipeline execution and imported into the database, and are organized by workflow stages and projects and viewable through an interactive web user interface.
The tracking database is built on a LAMP (Linux, Apache Httpd, MySQL and PHP) application stack using the SLIM-PHP framework. The application has a small memory and storage footprint, provides a RESTful API interface to the MySQL back-end, and supports password protection to restrict access. The tracking database is dockerized and can be installed on-site (off the cloud) if preferred.
4 Using VCPA on Amazon EC2 or local Linux environment
We evaluated our pipeline using the NA12878 sample from the Genome in a Bottle project using the hg38 high confidence set (Supplementary Methods). Sensitivity/precision of VCPA calls were 0.999/0.994 for SNPs and 0.985/0.987 for indels respectively and comparable to TOPMed/CCDG workflows (Regier et al., 2018). We also ran VCPA on two replicates of NA19238 (Yoruban) in CRAM and FASTQ format and the pairwise variant discordance rate is 1.000 for both SNVs and indels (Supplementary Methods), comparable to TOPMed/CCDG workflows (Regier et al., 2018). Benchmark of cost and time on Amazon Elastic Compute Cloud (EC2) for running VCPA on these samples can be found in Supplementary Table S1. VCPA is available as Amazon Machine Images (AMI), ami-acc840d3. A dockerized version is also available for deployment on other linux-based environments.
To conclude, VCPA is an efficient, high quality and scalable pipeline for processing WGS/WES data on the Amazon EC2 environment. VCPA is used for the ADSP production and can track information from >1000 genome analysis runs simultaneously. Future plans include incorporating other variant calling pipelines such as xAtlas (Farek et al., 2018) and GATK4.
Supplementary Material
Acknowledgement
We thank members from the Genome Center for Alzheimer's Disease (GCAD) and ADSP for their input. For full acknowledgement session, refer to https://www.niagads.org/adsp/content/acknowledgement-statement/.
Funding
This work is supported by the National Institute on Aging (U54-AG052427, U01-AG032984, U24-AG041689).
Conflict of Interest: none declared.
References
- Chandran S., et al. (2018) uBAM-unmapped BAM format, https://software.broadinstitute.org/gatk/documentation/article?id=11008. [Google Scholar]
- DePristo M.A., et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farek J., et al. (2018) xAtlas: scalable small variant calling across heterogeneous next-generation sequencing experiments. bioRxiv, doi: 10.1101/295071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust G.G., Hall I.M. (2014) SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics, 30, 2503–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint, 1303, 3997. [Google Scholar]
- Li H., et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov A., et al. (2015) Sambamba: fast processing of NGS alignment formats. Bioinformatics, 31, 2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss K., et al. (2017) Full-stack genomics pipelining with GATK4 + WDL + Cromwell. F1000Res., 6, 1379. [Google Scholar]
- Regier A.A., et al. (2018) Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. bioRxiv, doi: 10.1101/269316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.