Abstract
Summary
A new version (version 2) of the genomic dose-response analysis software, BMDExpress, has been created. The software addresses the increasing use of transcriptomic dose-response data in toxicology, drug design, risk assessment and translational research. In this new version, we have implemented additional statistical filtering options (e.g. Williams’ trend test), curve fitting models, Linux and Macintosh compatibility and support for additional transcriptomic platforms with up-to-date gene annotations. Furthermore, we have implemented extensive data visualizations, on-the-fly data filtering, and a batch-wise analysis workflow. We have also significantly re-engineered the code base to reflect contemporary software engineering practices and streamline future development. The first version of BMDExpress was developed in 2007 to meet an unmet demand for easy-to-use transcriptomic dose-response analysis software. Since its original release, however, transcriptomic platforms, technologies, pathway annotations and quantitative methods for data analysis have undergone a large change necessitating a significant re-development of BMDExpress. To that end, as of 2016, the National Toxicology Program assumed stewardship of BMDExpress. The result is a modernized and updated BMDExpress 2 that addresses the needs of the growing toxicogenomics user community.
Availability and implementation
BMDExpress 2 is available at https://github.com/auerbachs/BMDExpress-2/releases.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Transcriptomic dose-response modeling seeks to establish a relationship between doses of test substances and the corresponding changes in gene expression at both a gene and gene set level. The potency estimates that come from these studies can provide a quick assessment of the mechanistic processes by which test substances modify biological systems, and more importantly, the doses [benchmark doses (BMD)] at which these changes occur (Moffat et al., 2015). Work has shown that BMDs identified from in vivo transcriptomic dose-response studies provide an estimate of a test article’s toxicological potency (Thomas et al., 2013a, b). The speed at which these studies can be performed and reported has led to significant interest in the use of genomic dose-response studies to support screening level risk assessments for environmental agents.
BMDExpress is a biologist-friendly, graphical user interface (GUI) software package that facilitates transcriptomic dose-response analysis (Yang et al., 2007). It employs validated continuous parametric models from the US Environmental Protection Agency (EPA) BMDS software (https://www.epa.gov/bmds) to compute curve fits for each gene expression feature, and then estimates BMDs. The individual probe/gene-level modeling process is largely consistent with recommendations by the US EPA for toxicological assessments (US EPA, 2012). One of the unique aspects of the software is the functional classification analysis module that sorts transcriptomic features into pre-defined gene categories [e.g. Gene Ontologies (GOs)] and determines gene set level potency estimates for each of the populated gene sets (i.e. a gene set BMD).
The original BMDExpress software has seen wide use across government, industry and academia, as it supports transcriptomic analysis that is much less resource intensive than guideline toxicity tests, and provides an approximation of toxicological potency. There is a growing consensus that these types of studies analyzed by the approach implemented in BMDExpress can increase efficiencies and expand the biological space that is considered in risk assessment (Thomas et al., 2013a, b). In this work, we re-engineered and improved BMDExpress. For example, we have updated the analytical model versions, added exponential models, provided extensive data visualizations similar to those implemented in BMDExpress Viewer (Kuo et al., 2016), incorporated on-the-fly data filtering, updated gene set annotations, added gene expression platforms and increased the number of operating systems that are supported.
2 BMDExpress 2 software application
2.1 Software design
BMDExpress 2 is a desktop application, designed and coded using the Model-View-Presenter (MVP) software architecture pattern, and implemented in Java using JavaFX libraries for user interface design. The MVP design pattern specifies decoupling of various aspects of the software project, enabling ease of maintenance and testing.
2.2 Download and install
The full-featured application is available for installation on Windows, Macintosh and Linux (https://github.com/auerbachs/BMDExpress-2/releases). A detailed user manual is also available (https://github.com/auerbachs/BMDExpress-2/wiki). A command line version of the software is included with all installations.
2.3 Data analysis
A set of tutorials including a quick start guide quick start describing the work flow is available (https://tinyurl.com/bmde-tut). The current BMDExpress workflow has four steps. First, a gene expression data file (tab-delimited .txt file) is imported, and gene annotations are assigned according to the data file’s gene identifiers. Second, the number of features to be analyzed are (optionally) reduced by employing an ANOVA or a trend test (parametric or non-parametric options) in combination with a fold change filter. Third, curve fits are computed for a subset of the models from the US EPA BMDS software (the models included in BMDExpress 2 are shown in Supplementary Fig. S1) and BMDs for best fit models for each of the features are computed along with their upper (BMDU) and lower bound (BMDL) estimates. Fourth, features demonstrating a user defined acceptable model fit (e.g. global goodness of fit P-value >0.1, BMDU/BMDL ratio <40) are parse into pre-defined gene sets (e.g. GO Biological Processes) and gene set level BMD values are determined. Gene set BMD/BMDL/BMDU are calculated multiple ways including by median, mean and percentile. The resultant output of the analysis is a collection of gene set BMD/BMDL/BMDU values that can be used to estimate biological potency, along with descriptive statistics and visualizations. A detailed workflow diagram can be found in Supplementary Figure S1. In addition, a parallel workflow diagram of an example analysis using previously published genomic dose-response data (Dunnick et al., 2017) can be found in Supplementary Figure S2.
2.4 Filtering and visualization
A set of user interface panels for real-time filtering of the displayed data is provided. In Figure 1, functional classification data and visualizations are displayed. By selecting limits on various statistical parameters using the filter panel on the right, the graphical views and data table are updated accordingly. The integration of filtering with visualization allows the user to rapidly interact with the results of their analysis.
Fig. 1.
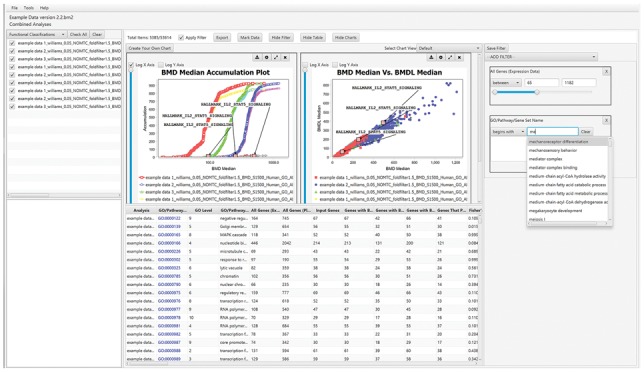
Layout of functional classification view, showing tabular and graphical data visualizations (center panel), real-time data filters (right panel), experiment workflow tree (upper left panel) and selected experiment metadata (lower left panel)
2.5 Platforms and annotations
We have added support for several new transcriptomic platforms. BMDExpress now supports 20 Affymetrix platforms, 8 Agilent platforms, 4 Illumina platforms, 3 BioSpyder platforms and multiple species RNA-Seq annotations (both RefSeq and Ensembl gene identifiers). The application downloads updated platform annotation files from a remote server. We actively maintain the latest GO annotations (Gene Ontology Consortium, 2008) that are offered to the user when available. Pathway classification is defined via REACTOME (Fabregat et al., 2017). Custom defined annotations may also be employed via the Defined Category Analysis feature.
3 On-going development efforts
BMD modeling of toxicogenomic data is an active field of research. Hence, we anticipate future releases of BMDExpress with new features and improved performance. The following notable features are planned for future release: model averaging, non-parametric dose-response modeling, automated report generation, in vitro to in vivo extrapolation functionality for extrapolation of in vitro results, updates to the data exploration views, wAUC (weighted Area Under the Curve) metrics to allow for more robust connectivity mapping (Sedykh, 2016), and improved GUI performance, among others. As improved curve fitting methods (e.g. model averaging), statistical, and bioinformatics methods are developed, they will be introduced into the workflow.
4 Conclusion
BMDExpress 2 offers a structured workflow for analyzing transcriptomic dose-response data with improved software performance, re-engineered code base, expanded bioinformatics methods, increased support for varied transcriptomic platforms and updated pathway annotations. The updated version of the software (version 2) is already in use in academia, government, non-profit research institutions and private industry for performing BMD analysis using toxicogenomic data, and it is the designated analysis platform for the National Toxicology Program in its approach to transcriptomic dose-response modeling (National Toxicology Program, 2017).
Supplementary Material
Acknowledgements
We would like to acknowledge, Joshua Harrill, Stephen Ferguson and Julia Rager for their feedback on early versions of the software and Shyamal Peddada and Sean Harris provided guidance on the implementation of the ORIOGEN trend test. Finally, we would like to acknowledge Andy Shapiro, Pierre Bushel, Nikolai Chepelev, Andrew Williams, Matthew Wheeler and Ken Shao for helpful commentary on the manuscript.
Funding
Financial support for this project was provided by the National Toxicology Program at the National Institute of Environmental Health Sciences (ZIA ES103318-01); and the National Institute of Environmental Health Sciences Bioinformatics Support contract with Sciome LLC (HHSN273201700001C).
Conflict of Interest: none declared.
References
- Dunnick J.K., et al. (2017) Hepatic transcriptomic alterations for N, N-dimethyl-p-toluidine (DMPT) and p-toluidine after 5-day exposure in rats. Arch. Toxicol., 91, 1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., et al. (2017) The Reactome Pathway Knowledgebase. Nucleic Acids Res., 46, D649–D655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. (2008) The Gene Ontology project in 2008. Nucleic Acids Res., 36, D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo B., et al. (2016) BMDExpress Data Viewer - a visualization tool to analyze BMDExpress datasets. J. Appl. Toxicol., 36, 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat I., et al. (2015) Comparison of toxicogenomics and traditional approaches to inform mode of action and points of departure in human health risk assessment of benzo a pyrene in drinking water. Crit. Rev. Toxicol., 45, 1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Toxicology Program. (2018) NTP Research Report on National Toxicology Program Approach to Genomic Dose-Response Modeling: Research Report 5. NTP Research Reports, Durham, NC. [PubMed] [Google Scholar]
- Sedykh A. (2016) CurveP method for rendering high-throughput screening dose-response data into digital fingerprints. Methods Mol. Biol., 1473, 135–141. [DOI] [PubMed] [Google Scholar]
- Thomas R.S., et al. (2013a) Incorporating new technologies into toxicity testing and risk assessment: moving from 21st century vision to a data-driven framework. Toxicol. Sci., 136, 4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.S., et al. (2013b) Temporal concordance between apical and transcriptional points of departure for chemical risk assessment. Toxicol. Sci., 134, 180–194. [DOI] [PubMed] [Google Scholar]
- US EPA. (2012) Benchmark dose technical guidance. In:Risk Assessment Forum. U.S. Environmental Protection Agency. Washington, DC [Google Scholar]
- Yang L., et al. (2007) BMDExpress: a software tool for the benchmark dose analyses of genomic data. BMC Genomics, 8, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


