Abstract
Objective:
Expansion of the G4C2 repeat tract in the C9orf72 gene is linked to frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). Here we provide comprehensive genotyping of the C9orf72 repeat region for the National Institute for Neurological Disorders and Stroke (NINDS) ALS collection (n=2095), using a novel bimodal PCR assay capable of amplifying nearly 100% GC-rich sequences.
Methods:
A single-tube 3-primer PCR assay mode, resolved using capillary electrophoresis, was used for sizing up to 145 repeats with single-repeat accuracy, for detecting expansions irrespective of their overall size, and for flagging confounding 3’ sequence variations. A modified two-primer PCR mode, resolved via agarose gel electrophoresis, provided further size information for hyper-expanded samples (>145 repeats) up to ~5.8 kb amplicons (~950 G4C2 repeats).
Results:
Within the evaluated cohort, 177 (8.4%) samples were expanded, with 175 (99%) samples being hyper-expanded. 3’ sequence variations were identified in 64 (3.1%) samples, and were most common in expanded alleles. Genotypes of all 606 (29%) homozygous samples were confirmed using an orthogonal PCR assay.
Conclusion:
This study and PCR method may improve and standardize molecular characterization of the C9orf72 locus, and have the potential to inform phenotype-genotype correlations and therapeutic development in ALS/FTD.
Keywords: C9orf72, GGGGCC hexanucleotide repeat, microsatellite, repeat-primed PCR, frontotemporal dementia, amyotrophic lateral sclerosis
INTRODUCTION
Microsatellite repeat expansions are associated with neuromuscular and neurodegenerative disorders, including frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS) (1). Both FTD and ALS are considered part of the same clinical continuum and have been linked by a hexanucleotide repeat element (G4C2)n in intron 1 of the Chromosome 9 open reading frame 72 (C9orf72) gene (1). Expansions in the C9orf72 repeat region have been reported in approximately 0.14% (~1:700) of the population (2), and are enriched in both familial FTD and ALS patients (~25% and 20–67%, respectively). The expansion also appears in up to 7% of sporadic FTD and ALS patients, making it the most prevalent genetic mutation in both disorders (3–5).
Although a validated threshold for repeat expansion pathogenicity has yet to be established, normal alleles have fewer than 20 repeats whereas disease-associated C9orf72 expansions have more than 30, and more typically, hundreds to thousands of repeats. Expanded alleles also manifest extensive somatic size mosaicism (1, 6). The impact of repeat length and associated DNA methylation status on a range of clinical phenotypes requires further elucidation (3, 7). Moreover, C9orf72 expansions may influence other neurodegenerative disorders, including Alzheimer’s disease (2).
Molecular characterization of the hexanucleotide repeat can be challenging, as the large size and high GC content of pathogenic C9orf72 expansions impede polymerization during PCR and limit the utility of conventional PCR-based fragment sizing methods. To this point, Akimoto et al. (8) evaluated C9orf72 genetic testing paradigms across labs, revealing a high incidence of false positives and negatives using laboratory-developed PCR assays. They concluded that more informative and consistent analysis methods are needed and recommended a combination of gene-specific (GS) and repeat-primed (RP) PCR as a minimum for research use, with Southern blot techniques advised for clinical diagnostics (8). In addition to PCR challenges imposed by GC-rich repetitive DNA, repeat size analysis is also confounded by the presence of sequence variations (SVs) at the 3’ end of the repeat region (5, 9). The presence of such 3’-SVs within PCR priming sites or target regions can attenuate amplification efficiency, distort allele sizing, and/or cause allele dropouts. Testing of 6891 clinical samples identified 3’-SVs in ~3% of the population (10). This report extended a previously established 3’-variable region to include an additional 50 bp downstream of the repeat tract (10).
In this study, we demonstrate the analytical capabilities and performance of a novel two-mode, multiplexed PCR chemistry for the genotyping of the C9orf72 repeat tract. This assay balances co-amplification of gene-specific (GS) and repeat-primed (RP) PCR products tailored for capillary electrophoresis (CE)-based, accurate sizing of C9orf72 repeats from genomic DNA (gDNA) samples. We show the utility of this GS/RP-PCR assay by genotyping the National Institute of Neurological Disorders and Stroke (NINDS) repository’s ALS collection. With over 2000 samples, this collection is the largest publically-available annotated set of patient-derived cell lines and matched genetic materials and is a valuable and readily accessible resource for researchers studying ALS and other neurodegenerative disorders. We used GS/RP-PCR/CE to quantify the number of hexanucleotide repeats in this collection at single-repeat resolution while also detecting 3’-SVs and low-level size-mosaicism. All expanded samples were further sized using a complementary GS-PCR/AGE workflow that resolves amplicons whose size exceeded the resolution window of CE; this companion analysis generated reproducible results for up to ~5.8 kb PCR products consistent with the amplification of ~950 repeats.
MATERIALS AND METHODS
Clinical Genomic DNA Samples
The NINDS Repository MND collection (Coriell Institute) offers open-access to de-identified gDNA samples extracted from patient-derived B lymphocyte cell lines. ALS gDNA samples (n=2095) from this repository were used (https://www.coriell.org/0/Sections/Support/Global/DNA.aspx?PgId=689). Sample reference numbers and corresponding batch records are listed in the supplementary tables. Detailed clinical information (67 variables) is provided by Coriell upon request, and available for all but one sample.
PCR/CE Assays
Samples were analyzed using the AmplideX® PCR/CE C9orf72 Kit (Cat. # 49581; Research use only, Asuragen, Inc.). A general schematic of the 2- and 3-primer assay in context of the C9orf72 gene region is shown in Figure 1. Briefly, gDNA was PCR amplified using three primers: two GS primers that flank the repeat region (GS forward primer 5’-CGCAGCCTGTAGCAAGCTCTGGAACTCAGGAGTCG-’3; GS reverse primer 56-FAM/TGCGCCTCCGCCGCCGCGGGCGCAGGCACCGCAACCGCA-’3), and one forward RP primer designed to be complementary to three G4C2 repeats. Thermal cycling was performed on Applied Biosystems’ Veriti and 9700 Thermal Cyclers (Thermo Fisher) using 98°C for 5 min, 37 cycles of 97°C for 35 sec, 62°C for 35 sec, 72°C for 3 min, and 72°C for 10 min. PCR product (2 µL) was mixed with Hi-Di™ Formamide (Thermo Fisher) and ROX 1000 Size Ladder (Asuragen, Inc.) for analysis by CE (3500xL Genetic Analyzer, Thermo Fisher). The FAM-labeled amplicons were detected using a fragment analysis protocol (50 cm capillary; 2.5 kV, 20 s injection, 19.5 kV run for 2400 s). By adjusting CE run parameters, POP7 sizing range can be extended beyond 145 repeats. Lower run voltages (<12kV) allowed for hyper-expanded allele sizing up to ~180 repeats, with an increase in run time and loss in sensitivity due to the spreading of the signal within a ‘pile-up peak’ (>145 repeats) that comprises aggregated amplicons too long to be adequately resolved by CE (Supplementary Figure 2). Electropherograms were processed as .fsa files using GeneMapper v4.1 for analysis, and manually annotated for repeat profile initiation and size of full length product. A PCR/CE control admixture sample comprised of C9orf72 alleles with 2, 5, 8, and 10 repeats and a no template control were used in all experiments. Repeat size was determined using a linear fit adjustment of the ROX ladder size peaks to the PCR/CE control sample alleles. Homozygous and discordant samples were further analyzed using an alternative 3’-distal (alternative GS reverse) priming site with the following sequence (6): 5’-ATGCAGGCAATTCCACCAGTCGCTAGAGGCGAAAGC-’3.
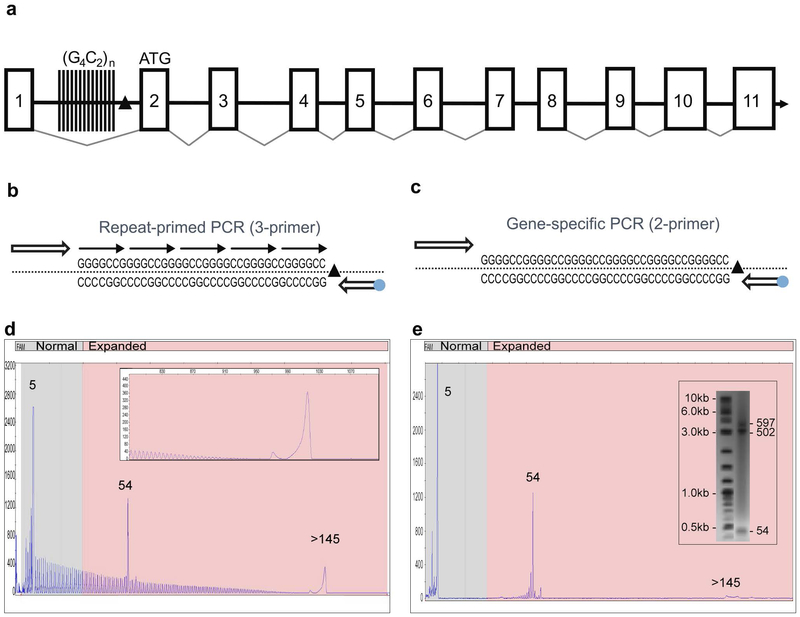
Figure 1: A novel PCR technology for the amplification of the hexanucleotide G4C2 repeat element in the C9orf72 gene.
A) Schematic representation of the C9orf72 gene structure showing the predicted 11 exons (boxes), the location of the intronic hexanucleotide repeat expansion (vertical lines) and the 3’SV region (black triangle). B) Schematics of the 3-primer GS/RP-PCR design (blue dot – FAM fluorophore) and C) 2-primer GS-PCR design. D) A representative 3-primer GS/RP-PCR/CE profile for an expanded sample (ND10206) that overlays GS peaks (a normal allele at 5 repeats, an expanded allele at 54 repeats) with a hyper-expanded RP-PCR profile and corresponding pile-up peak (inset, >145 repeats). E) Matching 2-primer GS-PCR products for Coriell sample ND10206 resolved on CE and AGE (inset) shows the two alleles (sized at 5 and 55 repeats). The hyper-expanded peak is resolved in the gel image as a 502 and 597 repeats.
Agarose Gel Electrophoresis (AGE)
To assess expanded alleles with >145 repeats, the AmplideX® PCR/CE C9orf72 Kit was used with modifications: 1) 80 ng gDNA input, 2) Omission of RP primer (GS primers only), and 3) An alternative thermal cycling protocol (Supplementary materials). A total of 177 expanded samples were reflexed to this 2-primer modified assay and resolved on CE and AGE. A positive PCR/AGE control, comprised of samples ND06751 and ND09492 mixed in equal parts, was used to evaluate assay processivity, sensitivity and sizing accuracy. PCR products (13 µL) and 2-Log DNA Ladder (NEB) were loaded onto a 12-well Reliant Precast 1% Seakem Gold DNA MiniGel (Lonza Walkersville, Inc.) and visualized with ethidium bromide. AGE data were analyzed using the GelAnalyzer software (2010a, Dr. Istvan Lazar). To enhance the accuracy of software-assisted sizing and address non-linear electrophoretic mobility, fragment sizing was partitioned into three partially-overlapping AGE size ranges using the reference ladder. Target DNA bands were assigned visual intensity levels as high (H), medium (M), or low (L). Intensities were assigned by visual comparison to the 2-log DNA ladder bands, where 4.0–10.0 kb ladder bands were medium intensity.
DNA Sequence Analysis
Following PCR amplification and size analysis, samples with abnormal repeat profiles were cloned and sequenced to resolve allele-specific changes. C9orf72 repeat amplicons were cloned into the pGEM-T Easy Vector System II (Promega). Positive colonies were expanded and sequenced (Sanger) on an ABI Prism 3730xl DNA analyzer (Thermo Fisher).
RESULTS
PCR Assay Design and Performance Evaluation
We designed two sets of single-tube PCR reagents configured with overlapping components to amplify the ALS/FTD-associated (G4C2)n repeats in the C9orf72 gene (Figure 1A). A 3-primer PCR design operates in two modes by combining degenerate repeat priming with repeat-flanking GS priming (GS/RP-PCR; Figure 1B). Omitting the RP results in a 2-primer GS-PCR and generates large amplicons free from the background of the RP profile to further clarify the detection of full-length products (Figure 1C). GS/RP-PCR amplicons produce an overlapping electrophoretic repeat profile with a 6 bp peak frequency and create two distinct yet complementary data signatures that are ideally resolved using high-resolution CE (Figure 1D). When hyper-expanded alleles (>145 repeats) are present, the high molecular weight amplicons can exceed the sizing range of CE (~950 bp; Figure 1D inset) and produce a ‘pile-up peak’. GS-PCR products generated from the 2-primer assay design can be resolved by CE or by AGE for hyper-repeat expansions up to ~950 repeats (Figure 1E).
The 3-primer GS/RP-PCR configuration enables CE-based sizing up to 145 repeats (Figure 2). Allele sizing can be assessed from the mobility of GS amplicons or by directly counting RP amplicon peaks. Importantly, the RP peak profile independently identifies hyper-expanded alleles and thus resolves uncertainty in allele zygosity (Figure 2A). Analytical verification experiments showed that the assay detected expanded alleles over a 2-log gDNA input range using as little as 1 ng, and revealed low-level mosaic alleles down to a 5% mass fraction of an expanded allele spiked into a 95% normal background (Figure 2B-C). Similarly, reproducible low-level mosaic amplicon profiles were observed in 67% of expanded samples, absent in the normal samples, and could be consistently resolved on CE and AGE (Figure 2D and Supplementary Figure 3). Finally, the assay was evaluated against 569 previously characterized gDNA samples from the NINDS ALS repository (11), demonstrating full categorical concordance and 99.3% sizing agreement within ±1 repeat (Supplementary Table 1). Interestingly, genotypes for the four size-discordant samples (ND11513, ND11685, ND120467, ND12382), previously reported as homozygous normal (11), were reconciled using an alternative 3’-distal primer configuration and were determined to be heterozygous (two normal alleles in each case), consistent with results obtained using the primary PCR assay.
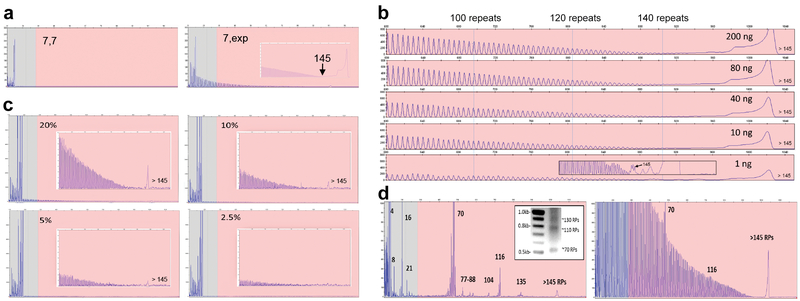
Figure 2: Performance characteristics of the C9orf72 PCR/CE assay.
A) Accurate sizing of up to 145 repeats with a distinct RP-PCR profile for expanded samples is achieved using the 3-primer PCR design. The RP profile enables zygosity resolution, i.e. distinguishing a homozygous sample with two alleles with 7 repeats (ND09188; left) and an expanded heterozygous sample (ND12102; right) with one allele with 7 repeats and a hyper-expanded allele. B) The 3-primer PCR demonstrates robust performance and detection of expanded alleles across a 200-fold gDNA range down to 1 ng/reaction (ND12754). Long-range RP peak counting is denoted at the 100, 120 and 140 repeat peaks. C) The presence of expanded allele RP profiles are visible down to a 5% mass fraction (admixture of expanded ND10966 in the background of unexpanded ND09188). D) Low-level minor alleles are readily detected in the 2-primer assay configuration on both CE and AGE (ND10689, inset). Numbers represent allele sizes (repeats – RPs).
Genotyping of the NINDS ALS Sample Collection
Following verification of the assay’s analytical performance, the GS/RP-PCR was utilized to genotype cell-line gDNA samples from the NINDS ALS Repository (n=2095). C9orf72 genotype information for the sample set is in Supplementary Table 1 and summarized in Figure 3A.
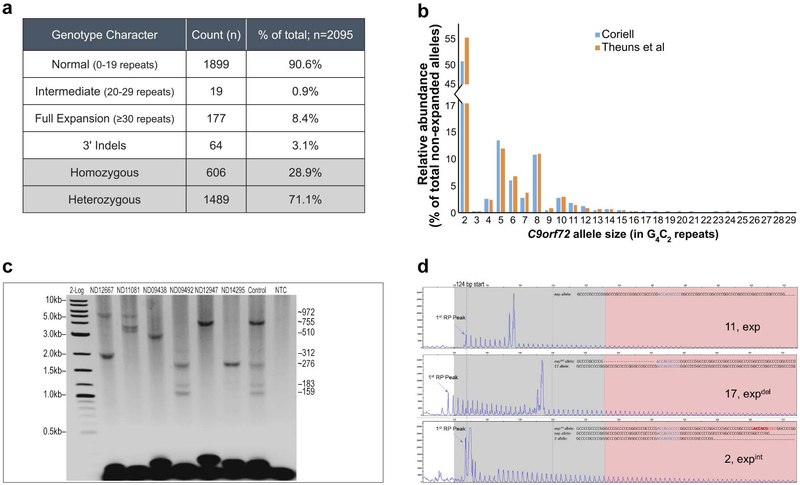
Figure 3: Comprehensive C9orf72 genotyping of 2095 NINDS ALS samples from the Coriell cell repository.
A) Genotype summary table. B) The normal and intermediate C9orf72 repeat size distribution (by allele; <30 repeats) within the tested Coriell NINDS ALS sample set was consistent with previous reports (12). C) Representative GS-PCR results, repeat sizes are listed (right). Detection of hyper-expanded ~5.8kb (950 repeat) amplicons and low-level size mosaicism. An admixture of samples ND06751 and ND09492 in equal parts was used to assess processivity, sensitivity, and AGE sizing accuracy. D) A noticeable offset in the RP single start site (~124 bp), or signal dips in the RP profile are evident when 3’ deletions or insertions are present. Representative samples and their confirmed underlying sequence are shown. Top pane (no 3’-SV), ND09438; middle pane (deletion), ND12947; bottom pane (insertion), ND09492.
Consistent with the established prevalence of C9orf72 expansions in ALS-positive populations (10), 177 (~8.4%) of the 2095 samples harbored expanded C9orf72 alleles, while only two of these samples had primary expanded alleles between 30 and 145 repeats (ND10554 and ND12780 with 56 and 70 repeats, respectively). Furthermore, the size distribution of normal C9orf72 alleles agreed with earlier analyses in European populations (12), underscoring the relatively high frequency of 2-, 5- and 8-repeat alleles (78% of all normal alleles; Figure 3B) and the resulting high prevalence of homozygous samples (28.9%).
All 177 expanded samples were analyzed using the GS-PCR assay and resolved by CE and AGE. Amplicons as large as ~5.8 kb were produced corresponding to ~950 repeats (Figure 3C, ND12667, ND11081). The results also demonstrated the mosaic nature of C9orf72 cell-line gDNA (Figure 2D and3C). AGE images for the 177 samples, and their corresponding table of expanded genotypes, are in Supplementary Figure 2 and Table 2, respectively.
Sequence variations have been identified at the 3’ end (3’-SVs) of the repeat region (5, 9, 10). While 3’-SVs have unknown clinical significance, they can confound interpretations by causing allele dropouts or by distorting amplicon mobility and skewing C9orf72 allele size conversions. To circumvent these effects, primers were positioned outside regions with reported variability (Figure 1A, and Materials and Methods). Furthermore, the complementary repeat primer orientation included this variable region, and thus was able to identify and indirectly characterize 3’-SVs through their impact on sample repeat profiles (Figure 3D).
Sixty-four of the 2095 ALS samples (3.1%) had indels downstream of the repeat tract. These indels were primarily found in expanded samples, and represented 30% of expanded cell lines. By comparison, indels were marginally present (0.6%) in non-expanded samples. Insertions were 10.6-fold less common than deletions and manifested as either RP signal dips, or offsets in the otherwise fixed 124 bp RP start site (Figure 3D). A subset of these samples (n=6) were further investigated using Sanger sequencing. Sequencing confirmed the RP-based analysis and identified previously reported 3’-SVs (10). Furthermore, an insertion event of a common 6 bp element within the repeat stretch (Figure 3D) was found in two unrelated samples. Interestingly, this common 6 bp insertion also maps directly downstream of the repeat stretch and was previously identified as a frequent 3’-deletion site (5, 9).
To help ensure that no alleles dropped out from the primary analysis, all 606 samples (~29%) initially identified as homozygous were verified using PCR with an alternative distal priming site that was 194 bp downstream of the standard primer. The orthogonal results confirmed primary findings using the primary GS/RP-PCR assay, despite significant presence of 3’-SVs in this sample set.
DISCUSSION
PCR innovations have created reliable systems for the amplification of long tracts of repetitive GC-rich DNA, including those with >95% GC content and >1000 CGG repeats in the 5’-UTR of the FMR1 gene (13, 14). In this study, we leveraged these advances to design, verify, and apply a novel PCR assay to characterize the C9orf72 repeat region across the complete NINDS ALS sample collection (n=2095). The results demonstrated broad agreement with earlier population studies (5, 12) across distributions of allele sizes and categorical sample genotypes, as well as the frequency and location of 3’-SVs. Additionally, we observed genotypic concordance for an available subset of previously annotated NINDS samples (11) (n=569). These findings underscore the analytical validity of the C9orf72 PCR assay, as well as the relevance of the NINDS cell lines for ALS research.
The PCR technology offers several benefits compared to conventional C9orf72 PCR or recently published long-read assays (15, 16). First, the PCR can navigate extreme GC content (>98% GC character) and accurately size <145 repeats on CE, with an extended range of ~950 repeats by AGE. Second, the sensitivity of the assays enables detection of expanded alleles using 1 ng of gDNA, and the identification of expanded major and minor alleles down to a 5% mass fraction. Third, the single-tube, 3-primer (GS/RP-PCR) design overlays the RP profile onto prominent GS peaks, thereby generating a multiplex, information-rich readout which expands analytical value in several ways: 1) enhances sizing accuracy through direct RP peak counting that can confirm sizing from GS peaks; 2) confirms sample zygosity through the sensitive and size-agnostic detection of expanded repeat profiles and pile-up peaks; and 3) flags 3’-SVs through repeat profile irregularities such as signal dips or peak offsets. Finally, the assay may be used with gDNA isolated from whole blood and other biosamples with comparable results to cell-line DNA.
Deviations in RP-PCR profiles reflect repeat sequence variations, which are associated with phenotypic or heritable genetic features in several other repeat disorders (10, 17–19). In this study, we identified 3’SV in the C9orf72 repeat tract in 3.1% of the NINDS samples, predominantly in expanded samples. Furthermore, sequencing data confirmed these observations and identified an insertion of a reported 6 bp deletion ‘hot spot’ element (5’-CGTGGT-’3; (5, 9, 10)) in the repeat tract of two unrelated samples. These results reiterate the highly variable nature of this region and the need to interrogate the hexanucleotide sequence space for insertions and repeat interruptions. To this end, the ‘dips’ in the GS/RP-PCR repeat profile are ideally suited for sensitive and rapid identification of such interruptions deep into the repeat region, and can be employed in future screening studies to help elucidate their prevalence and possible impact on C9orf72 biology.
From an analytical standpoint, 3’-SVs may attenuate PCR efficiency and/or cause allele dropouts if they impact PCR priming sites. This effect, in turn, could lead to misclassification of samples as homozygous (only single allele detected). Indeed, this may well be the underlying cause of discordance for four size-discrepant samples identified in our study that were previously classified as homozygous (11) (Supplementary Table 1).
3’-SVs have been shown to extend 80 bp beyond the repeat element (10). If 3’-SVs are captured within the GS-PCR amplicons, they can cause allele-sizing inaccuracies and even categorical miscalls (20, 21). This is a particular concern for ‘at-risk’ alleles that straddle the expanded categorical cutoff of 30 repeats. We estimate from our data that such at-risk alleles are rare and occur at a frequency of approximately 1 in 10,000 alleles; this is based on a 0.6% indel rate in alleles with <145 repeats and a 1.8% frequency of at-risk alleles with 17–42 repeats (Figure 3B). Furthermore, the GS/RP-PCR assay design allows for inspection of RP peak profile irregularities which can help identify 3’-SVs and at-risk samples. When a skewed RP start site is observed and individual allele repeat profiles are out of phase, meticulous repeat-peak tracing can accurately resolve skewed size and even inform of the underlying 3’-SV allele association (Figure 3D).
The analytical risks associated with the detection of rare variants is shared across all sizing assays, including Southern blotting (e.g. when variants occur at restriction enzyme cut sites, within a probe hybridization area, or as indels that alter the repeat region size). These risks can be mitigated by running orthogonal or multiplexed/multi-modal confirmatory tests. Our results highlight the robust performance of the described PCR primer designs, as sample genotypes were in accord with previous genotyping results, and with results obtained using an alternate PCR design.
The majority of C9orf72 expanded samples have hyper-expanded alleles yet several reports, including this study, have identified patients or patient-derived samples with primary expanded alleles shorter than 100 repeats (22–25). Interestingly, some of these patients remain asymptomatic at ages in which full penetrance is normally observed suggesting a more complex mechanism of pathogenicity and inheritance for shorter expansions which may be influenced by genetic modifiers, sequence variations, environmental conditions, and somatic mosaicism (22, 24, 25). Albeit rare, these observations warrant investigation into factors influencing pathology and the categorical cutoffs for expansions, which can be addressed using the sizing capabilities of the PCR assays described.
In conclusion, we described a multi-purpose, streamlined PCR assay that can quantify molecular features of the C9orf72 repeat region. The assay reports repeat sizing from 2 to ~950 repeats, detects expansions of >950 repeats in agreement with other assays, and flags sequence variations around the repeat tract. Importantly, this reagent system provides a rapid time-to-result in a single-tube format, requiring ~4.5 hours for PCR/CE and ~7 hours PCR/AGE, enables inputs of 10- to 100-fold less than published assays (8), and is configured and verified to help standardize results across laboratories. These capabilities can address well-documented inconsistencies in performance in current assays (8) and offer potentially more reliable results in screening and genotype-phenotype studies. Finally, the technology may also be helpful in accelerating new therapeutic approaches such as antisense oligonucleotides (26–29) or Cas9-based agents (30, 31) for repeat expansion disorders such as ALS/FTD.
Supplementary Material
Supplementary Figure 1: Lower CE run voltages enabled hyper-expanded allele sizing up to ~180 repeats. Lower run voltages (<12kV; bottom pane) allowed for hyper-expanded allele sizing up to ~180 repeats on CE, albeit with an increase in run time and a noticeable loss in sensitivity due to the size-separation of the ‘pile-up peak’ (see insets and asterisks).
Supplementary Figure 2: Composite gel image of GS-PCR band patterns for all 177 expanded samples. Modified thermal cycling conditions were used with a 2-primer GS-PCR configuration for all 177 samples previously identified as expanded via the GS/RP-PCR assay. GS-PCR products were resolved using AGE. For each gel, side-by-side direct and inverted images are provided, to allow better sensitivity and resolution. Running lane numbers correspond to sample annotations in the supplementary genotype table (PCR_AGE_Expanded_177 Sample).
Supplementary Figure 3: Detection and reproducibility of complex mosaic genotypes in C9orf72 expanded samples. The 2-Primer GS-PCR assay enables the reproducible detection of low-level mosaic genotypes observed in expanded samples. Reproducibility is demonstrated using two representative expanded samples (ND10312 and ND09907) in the overlapping CE trace profiles from 2 separate runs (blue and green traces; see also enlarged insets). Corresponding mosaic allele patterns are noticeable in parallel lower resolution AGE data for these samples.
ACKNOWLEDGMENTS
The project described was supported by R44NS089423 (to EB) from the NINDS. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINDS or the National Institutes of Health. This study used cell line DNA samples from the NINDS Repository, as well as clinical data. NINDS Repository sample numbers corresponding to the samples used are listed in Supplementary Table 1.
Footnotes
DISCLOSURE OF INTEREST
All authors are employed by Asuragen, Inc. Asuragen markets an RUO assay for genotyping C9orf72 hexanucleotide repeats (AmplideX® C9orf72 PCR/CE Kit).
REFERENCES
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. Epub 2011/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, et al. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. American journal of human genetics. 2013;92(3):345–53. Epub 2013/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benussi L, Rossi G, Glionna M, Tonoli E, Piccoli E, Fostinelli S, et al. C9ORF72 hexanucleotide repeat number in frontotemporal lobar degeneration: a genotype-phenotype correlation study. Journal of Alzheimer’s disease : JAD. 2015;45(1):319. Epub 2015/03/05. [DOI] [PubMed] [Google Scholar]
- 4.Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. The Lancet Neurology. 2012;11(4):323–30. Epub 2012/03/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Human mutation. 2013;34(2):363–73. Epub 2012/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. Epub 2011/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang M, Tartaglia MC, Moreno D, Sato C, McKeever P, Weichert A, et al. DNA methylation age-acceleration is associated with disease duration and age at onset in C9orf72 patients. Acta neuropathologica. 2017;134(2):271–9. Epub 2017/04/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akimoto C, Volk AE, van Blitterswijk M, Van den Broeck M, Leblond CS, Lumbroso S, et al. A blinded international study on the reliability of genetic testing for GGGGCC-repeat expansions in C9orf72 reveals marked differences in results among 14 laboratories. Journal of medical genetics. 2014;51(6):419–24. Epub 2014/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rollinson S, Bennion Callister J, Young K, Ryan SJ, Druyeh R, Rohrer JD, et al. Small deletion in C9orf72 hides a proportion of expansion carriers in FTLD. Neurobiology of aging. 2015;36(3):1601 e1–5. Epub 2015/01/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordin A, Akimoto C, Wuolikainen A, Alstermark H, Forsberg K, Baumann P, et al. Sequence variations in C9orf72 downstream of the hexanucleotide repeat region and its effect on repeat-primed PCR interpretation: a large multinational screening study. Amyotrophic lateral sclerosis & frontotemporal degeneration. 2017;18(3–4):256–64. Epub 2016/12/13. [DOI] [PubMed] [Google Scholar]
- 11.Rutherford NJ, DeJesus-Hernandez M, Baker MC, Kryston TB, Brown PE, Lomen-Hoerth C, et al. C9ORF72 hexanucleotide repeat expansions in patients with ALS from the Coriell Cell Repository. Neurology. 2012;79(5):482–3. Epub 2012/07/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theuns J, Verstraeten A, Sleegers K, Wauters E, Gijselinck I, Smolders S, et al. Global investigation and meta-analysis of the C9orf72 (G4C2)n repeat in Parkinson disease. Neurology. 2014;83(21):1906–13. Epub 2014/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipovic-Sadic S, Sah S, Chen L, Krosting J, Sekinger E, Zhang W, et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clinical chemistry. 2010;56(3):399–408. Epub 2010/01/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Hadd A, Sah S, Filipovic-Sadic S, Krosting J, Sekinger E, et al. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. The Journal of molecular diagnostics : JMD. 2010;12(5):589–600. Epub 2010/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biasiotto G, Archetti S, Di Lorenzo D, Merola F, Paiardi G, Borroni B, et al. A PCR-based protocol to accurately size C9orf72 intermediate-length alleles. Molecular and cellular probes. 2017;32:60–4. Epub 2016/10/26. [DOI] [PubMed] [Google Scholar]
- 16.Cleary EM, Pal S, Azam T, Moore DJ, Swingler R, Gorrie G, et al. Improved PCR based methods for detecting C9orf72 hexanucleotide repeat expansions. Molecular and cellular probes. 2016;30(4):218–24. Epub 2016/06/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yrigollen CM, Durbin-Johnson B, Gane L, Nelson DL, Hagerman R, Hagerman PJ, et al. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genetics in medicine : official journal of the American College of Medical Genetics. 2012;14(8):729–36. Epub 2012/04/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohshima K, Sakamoto N, Labuda M, Poirier J, Moseley ML, Montermini L, et al. A nonpathogenic GAAGGA repeat in the Friedreich gene: implications for pathogenesis. Neurology. 1999;53(8):1854–7. Epub 1999/11/24. [DOI] [PubMed] [Google Scholar]
- 19.Musova Z, Mazanec R, Krepelova A, Ehler E, Vales J, Jaklova R, et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. American journal of medical genetics Part A. 2009;149A(7):1365–74. Epub 2009/06/11. [DOI] [PubMed] [Google Scholar]
- 20.Bachinski LL, Czernuszewicz T, Ramagli LS, Suominen T, Shriver MD, Udd B, et al. Premutation allele pool in myotonic dystrophy type 2. Neurology. 2009;72(6):490–7. Epub 2008/11/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bean L, Bayrak-Toydemir P. American College of Medical Genetics and Genomics Standards and Guidelines for Clinical Genetics Laboratories, 2014 edition: technical standards and guidelines for Huntington disease. Genetics in medicine : official journal of the American College of Medical Genetics. 2014;16(12):e2. Epub 2014/10/31. [DOI] [PubMed] [Google Scholar]
- 22.Xi Z, van Blitterswijk M, Zhang M, McGoldrick P, McLean JR, Yunusova Y, et al. Jump from pre-mutation to pathologic expansion in C9orf72. American journal of human genetics. 2015;96(6):962–70. Epub 2015/05/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dols-Icardo O, Garcia-Redondo A, Rojas-Garcia R, Sanchez-Valle R, Noguera A, Gomez-Tortosa E, et al. Characterization of the repeat expansion size in C9orf72 in amyotrophic lateral sclerosis and frontotemporal dementia. Human molecular genetics. 2014;23(3):749–54. Epub 2013/09/24. [DOI] [PubMed] [Google Scholar]
- 24.Dobson-Stone C, Hallupp M, Loy CT, Thompson EM, Haan E, Sue CM, et al. C9ORF72 repeat expansion in Australian and Spanish frontotemporal dementia patients. PloS one. 2013;8(2):e56899. Epub 2013/02/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xi Z, Zinman L, Grinberg Y, Moreno D, Sato C, Bilbao JM, et al. Investigation of c9orf72 in 4 neurodegenerative disorders. Archives of neurology. 2012;69(12):1583–90. Epub 2012/09/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–28. Epub 2013/10/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. Epub 2015/08/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Science translational medicine. 2013;5(208):208ra149. Epub 2013/10/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron. 2016;90(3):535–50. Epub 2016/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo EH, Goh WL, Chow SC. The aminopeptidase inhibitor, z-L-CMK, is toxic and induces cell death in Jurkat T cells through oxidative stress. Toxicology mechanisms and methods. 2017:1–10. Epub 2017/08/30. [DOI] [PubMed] [Google Scholar]
- 31.Pinto BS, Saxena T, Oliveira R, Mendez-Gomez HR, Cleary JD, Denes LT, et al. Impeding Transcription of Expanded Microsatellite Repeats by Deactivated Cas9. Molecular cell. 2017;68(3):479–90 e5. Epub 2017/10/24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Lower CE run voltages enabled hyper-expanded allele sizing up to ~180 repeats. Lower run voltages (<12kV; bottom pane) allowed for hyper-expanded allele sizing up to ~180 repeats on CE, albeit with an increase in run time and a noticeable loss in sensitivity due to the size-separation of the ‘pile-up peak’ (see insets and asterisks).
Supplementary Figure 2: Composite gel image of GS-PCR band patterns for all 177 expanded samples. Modified thermal cycling conditions were used with a 2-primer GS-PCR configuration for all 177 samples previously identified as expanded via the GS/RP-PCR assay. GS-PCR products were resolved using AGE. For each gel, side-by-side direct and inverted images are provided, to allow better sensitivity and resolution. Running lane numbers correspond to sample annotations in the supplementary genotype table (PCR_AGE_Expanded_177 Sample).
Supplementary Figure 3: Detection and reproducibility of complex mosaic genotypes in C9orf72 expanded samples. The 2-Primer GS-PCR assay enables the reproducible detection of low-level mosaic genotypes observed in expanded samples. Reproducibility is demonstrated using two representative expanded samples (ND10312 and ND09907) in the overlapping CE trace profiles from 2 separate runs (blue and green traces; see also enlarged insets). Corresponding mosaic allele patterns are noticeable in parallel lower resolution AGE data for these samples.