Abstract
tRNAHis guanylyltransferase (Thg1) specifies eukaryotic tRNAHis identity by catalyzing a 3’ to 5’ non-Watson Crick (WC) addition of guanosine to the 5’-end of tRNAHis. Thg1 family enzymes in Archaea and Bacteria, called Thg1-like-proteins (TLPs), catalyze a similar but distinct 3’ to 5’ addition in an exclusively WC-dependent manner. Here, a genetic system in Saccharomyces cerevisiae was employed to further assess biochemical differences between Thg1 and TLPs. Utilizing a novel 5’-end sequencing pipeline, we find that a Bacillus thuringiensis TLP sustains growth of a thg1Δ strain by maintaining a WC-dependent addition of U−1 across from A73. Additionally, we observe 5’-end heterogeneity in S. cerevisiae small nucleolar RNAs (snoRNAs), an observation that may inform methods of annotation and mechanisms of snoRNA processing.
Keywords: 3’ to 5’ polymerization, snoRNA, tRNAHis, RNA-Seq, Saccharomyces cerevisiae, Bacillus thuringiensis
INTRODUCTION
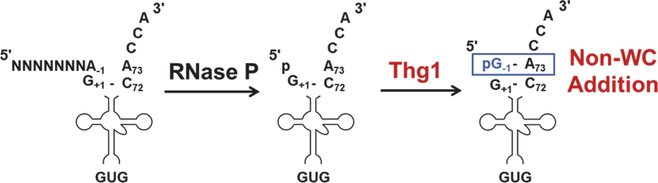
While canonical nucleic acid polymerases catalyze 5’-3’ nucleotide addition, enzymes of the tRNAHis guanylyltransferase (Thg1) family utilize similar reaction chemistry, but in the reverse (3’-5’) direction, to extend RNA substrates at their 5’-ends1,2. The founding member of this enzyme family, Thg1 from Saccharomyces cerevisiae (ScThg1), uses this unusual 3’-5’ addition activity to add a single required guanosine to the −1 position at the 5’-end of tRNAHis in a non-Watson-Crick (WC)-dependent manner (Figure 1) 1,3. This added G−1 residue serves as an essential identity element for efficient aminoacylation of tRNAHis by histidyl-tRNA synthetase (HisRS) 4–7.
Figure 1. Thg1 specifies tRNAHis identity through a non-Watson Crick 3’-5’ addition of G−1 across from A73.
Pre-tRNA 5’-leader sequences are removed by the 5’-endoribonuclease activity of Ribonuclease P (RNase P), generating the 5’-monophosphorylated tRNAHis that is the substrate for Thg1 to add G−1.
Thg1 homologs known as Thg1-like proteins (TLPs) are found in all three domains of life and are biochemically distinct from Thg1 1,8–10. Although Thg1 and TLPs share the same overall structure and mechanism of 3’-5’ nucleotide addition, TLPs exhibit a strong preference for adding WC-base paired nucleotides to RNA substrates 8–13. Moreover, in many of the archaeal and bacterial species that encode TLPs, G−1 can be obtained through an alternative post-transcriptional mechanism. Here, G−1 is genomically encoded, incorporated into the precursor tRNA during transcription, and then retained in the mature tRNAHis following atypical processing by ribonuclease P 8,14,15. These observations suggest the possibility of alternative non-tRNAHis-related functions for these enzymes. Along these lines, the bacterial TLP from Bacillus thuringiensis, (BtTLP) repairs 5’-end truncated tRNAs in vitro, adding missing 5’-nucleotides using nucleotides in the 3’-half of the aminoacyl-acceptor stem as a template to restore WC base pairing 8. Subsequently, the eukaryotic slime mold Dictyostelium discoideum was revealed to require a similar tRNA 5’-end repair reaction to repair mitochondrial tRNAs during a process known as tRNA 5’-editing1,16–18. This reaction is catalyzed by one of the D. discoideum TLPs (DdiTLP3) and represents the second established biological function for the 3’-5’ polymerase activity of Thg1/TLP enzymes 2,15,17,19. Nonetheless, biological functions for most TLPs, and especially for any bacterial or archaeal enzyme where roles in tRNAHis maturation or tRNA 5’-editing appear not to be needed, remain to be established.
An S. cerevisiae genetic system has been an important tool for investigating biological activities of Thg1/TLP enzymes3,9,15,20,21. Here, because of the essential nature of ScThg1, a thg1Δ strain is only viable in the presence of a functional version of the G−1 addition enzyme supplied on a covering plasmid. Interestingly, using this system, each of four different archaeal TLPs was not able to support growth of the thg1Δ strain, and only strains that also simultaneously expressed a mutant form of tRNAHis (C73-tRNAHis), enabling WC-dependent addition of G−1, were viable9. This observation was consistent with the in vitro preference of all TLPs to incorporate WC base pairs into their substrates. However, in similar studies the bacterial BtTLP was an exception to this rule, and supported growth of S. cerevisiae in the absence of THG1, albeit with a small but observable growth defect, since cells plated at the same OD did not grow to the same extent as wild-type after three serial dilutions in a yeast complementation assay 8,21. Thus, BtTLP was able to carry out the essential activity of ScThg1 in vivo in S. cerevisiae, but this apparent function contradicted the strong preference for WC base-paired nucleotide addition exhibited by BtTLP in vitro 8. Thus, the molecular basis for the phenotype was unclear. We aimed to determine whether the ability of BtTLP to complement the yeast thg1Δ phenotype stems from an unexpected ability to catalyze non WC-dependent G−1 addition to the A73-tRNA (like ScThg1) in vivo, or whether BtTLP catalyzes its biochemically preferred WC-dependent reaction, possibly adding U−1 to this A73-containing tRNA.
High-throughput RNA-sequencing techniques can generate libraries with single-nucleotide resolution of RNA ends 22–24. 5’-end addition catalyzed by BtTLP in S. cerevisiae was investigated using RNA-Seq by quantifying differential 5’-end reads between RNA derived from strains expressing ScThg1 vs. BtTLP. We found an over-representation of U−1 nucleotides at the 5’-ends of the reads aligned to tRNAHis genes in the BtTLP-expressing strain. However, the altered 5’-end of tRNAHis was the only major change, suggesting that BtTLP is not catalyzing detectable activity on other RNAs in this system. Additionally, these data also revealed that a subset of 14 small nucleolar RNAs (snoRNAs) exhibit 5’-end peaks that are shifted upstream or downstream of the annotated 5’-end, in contrast to the majority of well-represented non-coding RNA (ncRNA) genes for which reads supported the annotated 5’-start site. This unexpected variability in 5’-ends of snoRNAs was validated by primer extension and raises questions about mechanisms of 5’-end processing for these ncRNAs.
MATERIALS AND METHODS
Preparation of yeast strains
A previously described plasmid for galactose-inducible expression of BtTLP [CEN LEU2 PGAL–BtTLP] along with analogous ScThg1-expressing and empty vector control plasmids, were transformed into yeast thg1Δ strain JJY20 containing a [CEN URA3 PTHG1-THG1] covering plasmid 8. A plasmid shuffle assay was used to select strains expressing either BtTLP or ScThg1 as the only source of Thg1 activity (see Supplemental Methods).
Yeast low molecular weight RNA isolation
Cultures from independent single colonies were grown at 30 °C to an OD600 of 1 in rich media with 2% galactose (YPGal) and RNA was extracted from pellets using phenol and ethanol precipitation, as previously described 3. RNAs were treated with DNAse and quantified by NanoDrop by assuming 1 unit of A260=40 μg/mL total RNA.
Library preparation for RNA-Seq
Isolated RNAs were size selected to deplete RNA larger than ~200 nucleotides using the mirVana Kit (Ambion), then libraries were prepared using the NEBNext Small RNA Library Prep Set for Illumina (NEB) and sequenced (see Supplemental Methods). Initial sequencing results indicated few differences between BtTLP- and ScThg1-expressing strains, therefore no further biological replicates were sequenced and expression statistics are not reported.
Determining differences in ncRNA 5’-ends
To assess changes in 5’-end status, alignments around the annotated 5’-ends of ncRNAs were compared between ScThg1-expressing and BtTLP-expressing samples. For this analysis, let Nx(b) be the number of read alignments that start at location x (with respect to the annotated start of an RNA, defined as +1 location) and are sequenced with base b at that location (see Supplemental Methods for additional details). We quantified upstream distributions of 5’-ends defined by ρup = [N-3, N-2, N−1(A), N−1(U), N−1(C), N−1(G), N+1] and downstream distributions defined by: ρdown = [N+1, N+2, N+3, N+4]. (Figure 2B) for reads derived from each strain.
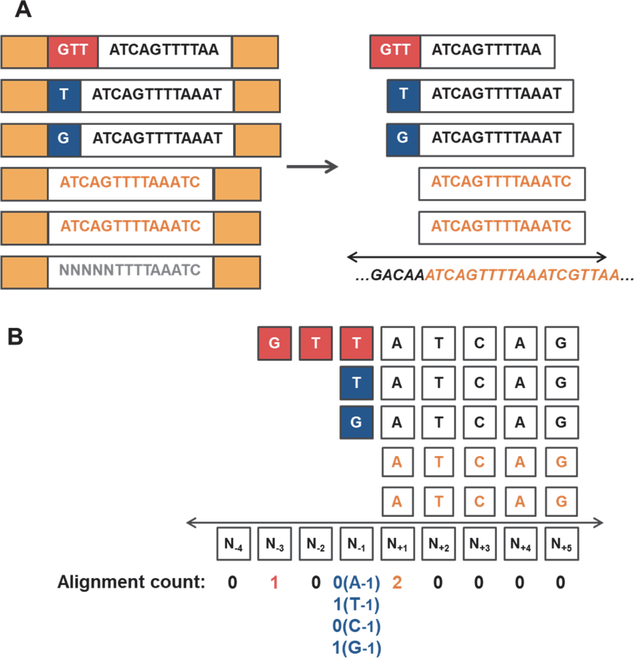
Figure 2. 5’-end sequence analysis pipeline to detect activities of Thg1/TLPs.
5’-addition activity could generate 5’-sequences that do not match genome-encoded nucleotides expected from incorporation by transcription. A) The pipeline developed here distinguishes between read sequences (shown in white boxes) that precisely match annotated mature RNA 5’-ends (shown in orange text) vs. read sequences that contain different nucleotides at the 5’-end, possibly added by Thg1/TLP enzymes (red/blue boxes). After removal of adapter sequences (solid yellow boxes), trimming, and quality filtering of reads (poor quality read indicated by gray text), 5’-end nucleotides that map upstream of the annotated mature RNA 5’-end are preserved by soft-clipping, which allows up to 10 nucleotides at the 5’-end that do not match the genomically-encoded nucleotide(s) at this location (genomic sequence is indicated under the double-arrow). B) Quantification of alignment count values for reads corresponding to a representative non-coding RNA. After reads are aligned, the number of reads with a 5’-end that corresponds to each of 10 nucleotides surrounding the annotated mature RNA 5’-end (N+1) are quantified. For position N−1, the reads were further analyzed according to the base identity (A, T, C, or G) at this location.
2,476 annotated ncRNA were sourced from the sgdOther database. For each ncRNA, the difference in the upstream and downstream distributions of 5’-ends between the ScThg1- and BtTLP-expressing strains were assessed by Fisher’s Exact Test -- when the read counts from the entire 2×7 or 2×4 table totaled < 10,000 -- or from the χ2 Test otherwise. Distribution changes were considered of statistical significance corresponding for p-values < 0.05 after adjusting for multiple hypothesis testing using the Benjamini-Hochberg method.
Validation of 5’-ends by primer extension
Primer extension was performed with RNA isolated from two different yeast strains, BY4743, the wild type parental strain for the strains that were sequenced, (MATa/α, his3Δ1/ his3Δ1, leu2Δ0/ leu2Δ0, LYS2/ lys2Δ0, met15Δ0/ MET15, ura3Δ0/ ura3Δ0) and SC126, a unrelated yeast strain, (MATa ade2–1°, met1, can1–100°, ura3–52, leu2–3,112, SUP4°).
Primers were designed to hybridize ~8–10 nucleotides away from the annotated 5’-end of snoRNAs, labeled, and used in primer extension as previously described 3,25 (Supplemental Table 1). Annealed primer was extended for 5 min at room temperature with 400, 150, and 50 μM dNTP and 0.9 U/μL AMV-reverse transcriptase (USB) in AMV-RT reaction buffer (USB) followed by 1 hour at 37 °C. dNTP titration was performed to determine the optimal concentration to terminate cDNA synthesis at the precise 5’-end of the RNA (150 μM), since RT is known to add additional nucleotides at higher concentrations. Sequencing reactions contained 400 μM of each corresponding ddNTP and 100 μM dNTP. Reactions were stopped by addition of equal volume of RNA loading buffer (80% formamide, 1 mM EDTA). Products were resolved by 10% PAGE with 4M urea, and visualized by PhosphorImager.
RESULTS
BtTLP adds a U−1 to tRNAHis in vivo in S. cerevisiae.
We applied an unbiased RNA-Seq approach to targeting small ncRNA species isolated from the BtTLP-complemented thg1Δ strain, and compared this to ncRNA from an isogenic control strain overexpressing ScThg1. As with previous tRNA 5’-end sequencing protocols testing D. discoideum TLPs, purified RNAs were treated with pyrophosphatase to remove ligation-inhibiting 5’-triphosphates that could be generated by the action of Thg1/TLPs 17. Although modifications on some RNA substrates, such as tRNA, are known to interfere with reverse transcription, no additional steps were taken to minimize polymerase fall-off during library preparation. Calculations of median distance of the reads from tRNA 3’-ends revealed an average alignment position of 54 nucleotides into the tRNA sequence across 49 tRNA species with at least 50 alignment counts, indicative of significant polymerase fall-off occurring as expected. However, since we did not expect tRNA modification status to differ significantly between the BtTLP- and ScThg1-expressing strains, the limited number of 5’-end containing reads derived from tRNA were sufficient for further analysis as described below.
The resulting paired-end reads were aligned to the S. cerevisiae reference genome using STAR, a transcript aligner that considers alignments with soft-clipped bases at the 5’ and 3’ ends (Figure 2A). This feature is essential to obtain the type of read signatures we were interested in, since TLP enzymes have so far all been demonstrated to add nucleotides to the 5’-end using another part of the substrate RNA as a template, and therefore the added nucleotides could differ from those encoded in the genome. Also, since activities of Thg1/TLP enzymes characterized so far involve limited addition of nucleotides to tRNA substrates (up to 3 nucleotides so far) 1,8,19, we restricted our analysis to quantifying the number of reads aligned such that the 5’-end is positioned within 3 nucleotides either upstream (numbered −1 to −3) or downstream (numbered +2 to +4) of the annotated 5’-end (+1) of the mature RNA (Figure 2B).
With these sequence patterns in mind, a computational screen was developed to compare 5’-end alignments of reads derived from BtTLP- vs. ScThg1-expressing strains for 2,476 S. cerevisiae ncRNA. Distributions of 5’-end alignments in the vicinity of annotated start sites were calculated for the ScThg1 and BtTLP-expressing samples and compared, resulting in a p-value quantifying statistical significance of the difference between the two conditions for each distribution at each annotated ncRNA (see Methods).
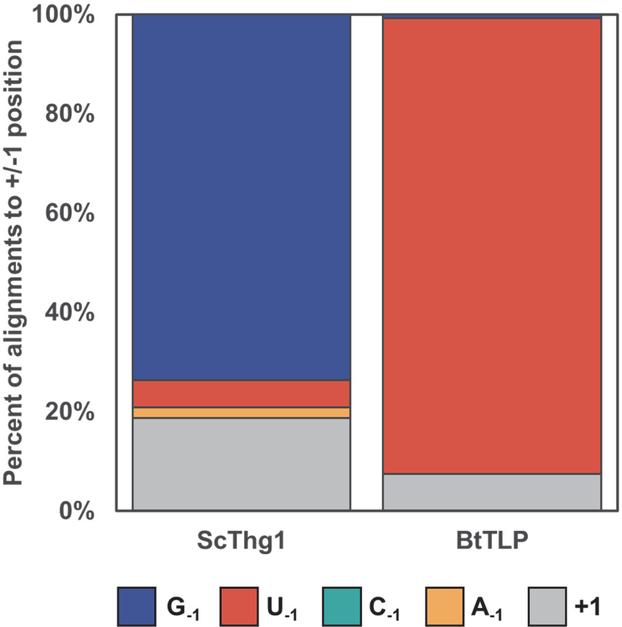
Annotated ncRNAs were ranked by the associated p-value separately for upstream and downstream distributions. The only genes that showed a shift in 5’-end composition between the ScThg1 vs. BtTLP strains that was at the same time highly statistically significant (adjusted p-value < 0.001) and featured an absolute change in 5’-end location and identity of more than 10% were the tRNAHis genes in their upstream distribution, and specifically these corresponded to differences in the −1 base identity (Figure 3, Table 1). Across seven tRNAHis loci, the overwhelming majority of reads from both strains contain a −1 nucleotide, with less than 20% of reads in each case corresponding to the RNaseP-processed intermediate species prior to G−1 addition (aligned to N+1). However, the identity of the nucleotide at −1 differs dramatically between the two strains. The percentage of reads that end in G−1 is 74% in the ScThg1 strain, but <1% in the BtTLP-expressing strain. Correspondingly, U−1-ending reads increase from 5% in the ScThg1 strain to 92% in the BtTLP-expressing strain (Table 1). Thus, BtTLP prefers to incorporate the WC base paired U−1 nucleotide onto the 5’-end of tRNAHis, consistent with its in vitro preferences 8.
Figure 3. BtTLP adds U−1 to tRNAHis when Thg1 is deleted.
Counts and percentages of reads aligned to the −1 position, by base, or to the +1 position of tRNAHis. This RNA exhibits a significant change in the composition of 5’-ends between the ScThg1 and BtTLP samples as evident from the statistical significance (adjusted p-value <0.001) and large effect size (% change in read count). Most reads aligning near the 5’-end of tRNAHis contain the −1 nucleotide (reads that start with +1 indicated in gray), with almost all of them beginning with base G−1 (blue) in the ScThg1-expressing strain and base U−1 (red) in the BtTLP-complemented sample.
Table 1:
Changes in distribution of reads corresponding to 5’-ends of selected ncRNA
| ncRNA | Straina | −3 | −2 | A −1 | U −1 | C −1 | G −1 | +1 |
|---|---|---|---|---|---|---|---|---|
| tH(GUG) | ScThg1 | <1% | <1% | 2% | 5% | <1% | 74% | 19% |
| BtTLP | <1% | <1% | <1% | 92% | <1% | <1% | 7% | |
| snR47 | ScThg1 | <1% | 5% | <1% | 49% | <1% | <1% | 46% |
| BtTLP | <1% | 4% | <1% | 59% | <1% | <1% | 36% | |
| snR51 b | ScThg1 | <1% | 5% | <1% | 43% | <1% | <1% | 51% |
| BtTLP | <1% | 4% | <1% | 50% | <1% | <1% | 45% |
thg1Δ strain expressing indicated Thg1/TLP gene.
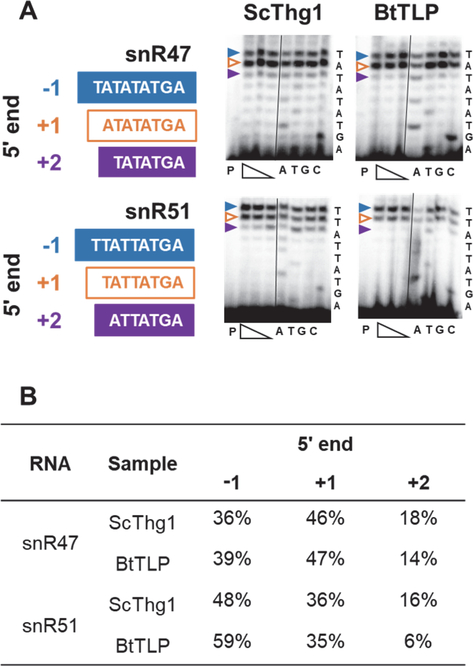
Although the change in snR51 peak patterns between the two strains (p-value ~0.07) is not statistically significant to the <0.05 level that was observed for the other two ncRNA, it follows a similar pattern (gain in representation of U−1 in going from the ScThg1- to the BtTLP-complemented sample) that was also validated by primer extension data (see Figure 4).
snoRNA snR47 also exhibited a statistically significant difference (adjusted p-value < 0.05) in the upstream distribution between the two strains (Table 1). The percentage of reads aligning to the +1 position decreased from 46% to 36% between the ScThg1- and BtTLP-expressing strains, respectively. The read distribution for snR51 showed a similar, although not robustly significant (adjusted p-value ~0.07), shift in read alignment between the two strains. For both snoRNAs, the decrease in +1 alignment count correlates with an increased percentage of aligned reads beginning with U−1 (Table 1). Primer extension experiments corroborate this modest but detectable increase in the −1 position population for both RNAs in the BtTLP-expressing strain compared to the ScThg1 strain, although the primer extension data suggest that the increase of N−1-containing RNA in the BtTLP-expressing strain primarily derives from a decreased amount of RNA with a 5’-end that aligns to N+2 (Figure 4). Nonetheless, the pattern of additional 5’-end nucleotides observed by both RNAseq and primer extension of RNA from the BtTLP strain would be consistent with the bacterial enzyme utilizing these snoRNA to some extent as substrates for 3’-5’ addition in vivo in S. cerevisiae.
Figure 4. snR47 and 51 exhibit an increase in population ending at the −1 position from the ScThg1 to the BtTLP complemented strain.
A) Primer extension assays were used to analyze RNA from the ScThg1- or BtTLP-complemented strains, as indicated. Expected 5’-end sequence of each snoRNA is listed to the right of each primer extension panel. The identity of the band corresponding to each 5’-end stop position is indicated by colored arrows, with blue corresponding to −1-terminating product, white with orange outline corresponding to +1 terminating product (annotated 5’-end), and purple corresponding to +2-terminating product. Each primer extension experiment contains primer only control (lane P), followed by three experimental lanes with the indicated target RNA extended in the presence of a range of dNTP concentration (400, 150, and 50 μM). The dNTP titration was used to ensure that observed primer extension products correspond to the actual RNA 5’-end, and not to additional nucleotide incorporation by RT (which can be observed at very high dNTP concentrations). The similar patterns of primer extension products observed here across the concentration range suggests that the length of each cDNA product represents an actual RNA 5’-end. Lanes labeled A,T,G,C correspond to sequencing lanes performed in the presence of the indicated ddNTP. B) Quantification of the representative assay shown in panel A to measure percent of total primer extension products corresponding to each 5’-end stop position for the indicated RNAs from each strain. Extension results were quantified using the reactions containing 150 μM dNTP (middle concentration of the three extension lanes).
For the remaining RNAs for which comparison of alignment counts yielded a p-value <0.05 (see Supplemental Table 2) none have a significant enough effect size to warrant further investigation, and many of these can likely be attributed to low numbers of reads mapping near the 5’-end. Notably, the abundance of reads that aligned to 5.8S rRNA (more than 80% from both strains) may have exacerbated this problem by preventing deeper coverage of other RNA species. However, no additional steps had been taken to remove this rRNA, since it was also in principle a potential BtTLP target. Also, only 20 tRNA families exhibited alignment accumulation of more than 50 reads at any one site within 10 nt of the annotated 5’-end of the mature tRNA, most likely due to interference of RT-blocking modifications, which is a known issue for obtaining reads from tRNA 5’-ends. Thus, no reliable conclusions can be drawn about the effects of BtTLP on the 5’-end status of these RNAs.
Subset of snoRNAs show 5’-end alignment peaks that vary from annotated ends.
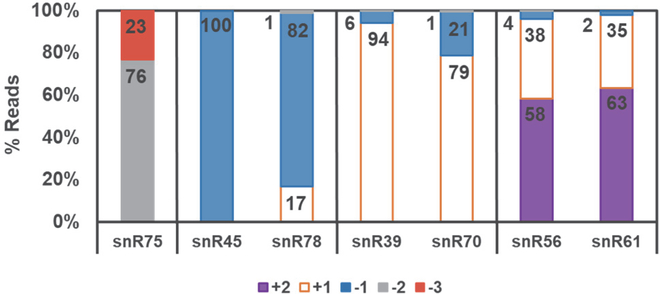
The above analysis assumes that the annotated start site of the RNA (also referred to as the +1 position) should represent the most common 5’-end alignment observed among the population of reads for any given RNA. Indeed, of the top 200 gene loci (by alignment peak count) that were analyzed in the ScThg1-expressing (control) strain, only 42 have an alignment peak that does not occur precisely at the annotated 5’-end. Of these 42, 20 are from six tRNAs, which include the tRNAHis loci with peaks at the −1 position and tRNAs with alignment peaks 5–10 nt from the annotated 5’-end and one corresponds to 21S rRNA with an alignment peak at the −1 position (Supplementary Table 3). Interestingly, the remaining 21 are from snoRNA.
To examine these off-annotation reads, we further classified snoRNAs as having a “distinct” or an “ambiguous” 5’-end alignment peak depending on whether the position with the maximum number of 5’-end read alignments was neighbored by any position with at most 2/3 as many alignments. For comparison, 38 snoRNA were also included in this analysis that exhibited alignment peaks at the annotated 5’-end. Considering this total sample, most of these (49) exhibited distinct alignment peaks. However, for 14 of these snoRNA, the distinct 5’-end read peak is 1–5 nt away from the annotated +1 nucleotide (Table 2). As most of the distinct peaks occur at the expected annotated 5’-end, these off-annotation peaks are not likely due to systematic errors in alignment of snoRNA reads. Examples from several different highly expressed snoRNAs show a clear shift in the location of alignment starts relative to their respective annotation (Figure 5). Similar classification of alignment peaks from the BtTLP-expressing strain revealed no significant differences overall between the two strains (data not shown), indicating that the observed 5’-end heterogeneity is a not associated with expression of the BtTLP or ScThg1 enzyme. Instead, this is likely an intrinsic property arising from differences in 5’-end processing of the precursor snoRNA species that have been transcribed with genomically-encoded nucleotides upstream of the +1 position.
Table 2.
Distribution of peak locations in snoRNA alignments
| Peak location | No. of snoRNAs with distinct peaks |
|---|---|
| −5 | 1 |
| −4 | 0 |
| −3 | 1 |
| −2 | 1 |
| −1 | 3 |
| +1 | 35 |
| +2 | 6 |
| +3 | 1 |
| +4 | 0 |
| +5 | 1 |
Peak locations denoted with respect to the annotated 5’-end. Of the 49 snoRNAs sequenced with “distinct” peaks, 14 align off of the annotated 5’-end position.
Figure 5. 5’-end heterogeneity is predicted by RNA-Seq.
For each indicated snoRNA analyzed in the ScThg1-expressing (control) strain described in methods, the fraction of 5’-end containing reads that correspond to the specified 5’-end nucleotides was quantified and shown as a percent of total 5’-end-containing reads. The annotated 5’-end for each RNA (+1 position) is indicated by white bars with orange outline. Other observed 5’-ends are indicated by red (−3), gray (−2) or blue (−1) for reads corresponding to sequences with additional 5’-nucleotides relative to the annotated RNA start, or purple (+2) corresponding to sequences with shorter 5’-ends relative to the annotated RNA start site.
Primer extension corroborates 5’-ends predicted from RNA-Seq and reveal that in vivo, snoRNA 5’-ends are heterogeneous.
To validate the snoRNA reads, primer extension was used to measure the length of the 5’-end of five representative RNAs with RNA-seq read patterns that varied from the annotated 5’-ends (snR75, snR78, snR56, snR61, snR45) (Figure 6). Two snoRNAs whose distinct 5’-end read peak agreed with the annotated RNA 5’-end (snR39 and snR70) were used as controls.
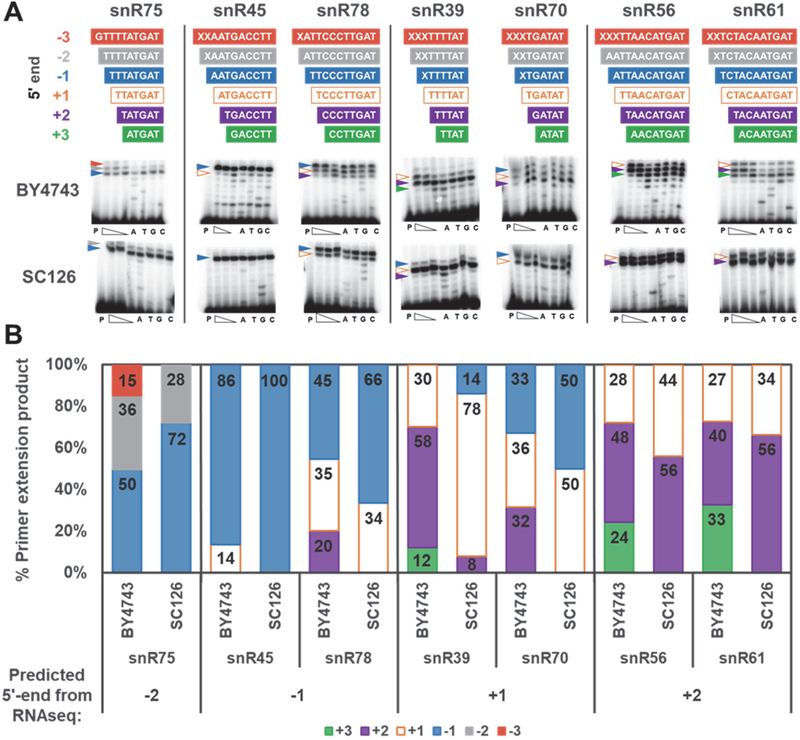
Figure 6. Validation of alternative 5’-end sequences associated with some snoRNAs.
A) Primer extension of RNA derived from either of two different wild-type S. cerevisiae strains (BY4743 or SC126) to corroborate 5’-end heterogeneity in yeast snoRNAs suggested by RNAseq reads. The colored bars shown above each panel correspond to possible 5’-end sequences of each snoRNA, with the annotated 5’-end (+1) indicated in white boxes with orange outline. Sequences corresponding to additional nucleotides present at the 5’-end (−3, −2, −1) are indicated in red, gray, and blue respectively. Sequences corresponding to shorter RNA 5’-ends (+2, +3) are indicated by purple and green, respectively. For longer RNA 5’-ends, if no reads with this 5’-end sequence were observed by RNAseq, the identity of the nucleotide is indicated by X, since the primer extension experiment applied here in the presence of all four dNTPs only indicates the length of the RNA, and not sequence. The first lane (P) in each panel corresponds to the primer only control reaction, and the three subsequent lanes represent extension of the RNA with varied dNTP (400, 150, 50 μM) to rule out additional nucleotides added by RT at high concentration of dNTP. Lanes A,T,G,C correspond to ddNTP sequencing lanes. Colored arrows are used to mark the positions of abundant primer extension products corresponding to each RNA 5’-end, as indicated above the panel. B) Quantification of percent of total primer extension products corresponding to each 5’-end stop position for the indicated RNAs from each strain. Extension results were quantified using the reactions containing 150 μM dNTP (middle concentration of the three extension lanes) and bars are colored to indicate the 5’-end that is represented using the same colors as in panel A. The most abundant 5’-end that was observed in the RNA-seq read data is indicated below each RNA for comparison to the primer extension results.
Primer extension experiments were used to assess location of 5’-ends for all seven snoRNAs in wild-type yeast strains, since expression of the TLP did not affect the locations of 5’-ends in the RNA-seq reads. To test the possibility of natural strain-specific variation, RNA was derived from two different wild-type yeast strains (BY4743, the parental strain for the yeast thg1Δ deletion, and SC126, an unrelated lab strain). Interestingly, while the general pattern of 5’-end location agreed generally well with the RNA-seq read distributions (described further below), in 13 out of 14 primer extension experiments, the snoRNA population exhibited a larger degree of 5’-end heterogeneity than was indicated by the RNA-seq data, with a distribution of 5’-end lengths centered around the predominant RNA-seq stop position (Figure 6). In one case, (snR45) the overwhelming majority (86–100%) of the population observed by primer extension was found to end precisely at the −1 position that was also predicted by RNA-Seq.
Primer extensions were quantified according to the proportion of bands corresponding to each 5’-end length, analogous to the alignment count data plotted in Figure 5 (Figure 6B). In snR45 and snR78, RNAs that both exhibited distinct −1 stop peaks in the RNAseq data, the largest fraction of primer extension products also corresponds to this position, with a smaller population corresponding to the annotated +1 position in snR78. Likewise, for two RNA that were predicted by RNAseq to have shorter 5’-ends than the annotated sequence (snR56 and snR61), primer extension also validated this result, with the largest fraction of products corresponding to the shorter 5’-end (+2 stop). Interestingly, the control snoRNAs snR39 and snR70 exhibited greater heterogeneity using the primer extension approach, but in most cases the products corresponded to a distribution of 5’-ends in the RNA centered on the annotated +1 start site. Only in the case of snR75 does the majority population at the −1 stop observed by primer extension not match the 5’-end that is one nucleotide longer (−2 stop) predicted by RNA-Seq. However even in this example the analysis revealed a distinct distribution of 5’-end products corresponding to generally longer RNAs (terminating at −1, −2, and −3 positions), thus corroborating the observations that this RNA exists in a slightly 5’-extended state relative to its original annotation, due to the presence of one or more genomically-encoded nucleotides that have not been removed during processing as was previously thought.
DISCUSSION
In this work we used RNA-Seq to analyze the effects of overexpression of a bacterial 3’-5’ polymerase enzyme, BtTLP, in S. cerevisiae in the absence of the endogenous ScThg1 enzyme. While the ScThg1-expressing strain contains nearly 100% G−1-tRNAHis, the same tRNA isolated from the BtTLP-expressing strain is nearly completely modified with U−1. Thus, BtTLP adds U−1 to tRNAHis in vivo in S. cerevisiae, consistent with its known in vitro preference for WC addition 8. As our analysis examined the 5’-ends of ncRNAs, we also discovered discrepancies between the annotated 5’-ends of snoRNAs and the most frequently-observed 5’-ends in the RNA-Seq data, which we validated for two different wild-type yeast strains.
Growth of S. cerevisiae depends upon relatively efficient aminoacylation of tRNAHis 4,5. Thus, the most straightforward interpretation of the ability of BtTLP to complement deletion of THG1 would be that tRNAHis contains the important G−1 identity element that is essential for aminoacylation. Yet, here we demonstrated that tRNAHis in the BtTLP-complemented strain instead contains a U−1 nucleotide, raising questions about the impact of this non-standard residue on HisRS activity. Interestingly, kinetic studies of S. cerevisiae HisRS demonstrated that replacement of G−1 with either A−1 or C−1 only modestly affected in vitro aminoacylation efficiency, as opposed to the severe (102-103-fold) defects observed for the complete absence of a −1 nucleotide in tRNAHis transcripts 26. While the U−1:A73 tRNAHis analogous to the tRNA that is produced in the BtTLP strains was not tested in vitro, the results described here suggest that the U-1-containing tRNA would be similarly well-tolerated by HisRS and have a minimal impact on overall aminoacylation. Previously, several tested archaeal TLPs were demonstrated not to complement the thg1Δ strain in the presence of the A73-tRNA, thus implying that they are not capable of catalyzing a similar U−1 addition activity in S. cerevisiae 9. Although the bona fide physiological function of TLPs in Archaea and Bacteria remain unknown, these differences may imply distinct functions and origins for enzymes from the different domains of life1,9,27.
Although the massive shift in N−1 nucleotide identity clearly correlates with the in vitro properties of ScThg1 vs. BtTLP, we cannot rule out the possibility that some fraction of the U−1-containing reads result from mis-processing by the 5’-end maturation enzyme RNase P, since four of the eight tRNAHis gene loci contain an encoded U at the −1 position that would be transcribed as part of the pre-tRNA 5’-leader sequence. Several lines of evidence suggest that any background levels of U−1 derived from the pre-tRNA instead of post-transcriptional addition are quite low. First, RNase P is known to exhibit generally high fidelity for 5’-end cleavage, consistent with precise tRNA structural requirements necessary for efficient translation 28. Second, in a strain conditionally lacking Thg1, the +1-terminating tRNAHis clearly accumulates, suggesting that 5’-end processing occurs correctly for the majority of tRNAHis transcripts 3. Finally, in our own RNA-Seq data from the ScThg1-expressing strain, only 5% of the observed reads (where presumably 5’-end maturation occurs normally) contain U−1. Moreover, the 5% U−1 containing reads likely represent an overestimate of the amount of misprocessing to retain genomically-encoded U−1 by RNase P, since Thg1 enzymes (including ScThg1) can also incorporate U−1 into tRNAHis at low levels, as recently demonstrated for the human Thg1 homolog 2,29. Taken together, it seems likely that addition of U−1 observed at the 5’-end of tRNAHis was primarily catalyzed by BtTLP.
An unexpected feature of our analysis of ncRNA 5’-ends was a previously undescribed deviation from annotated 5’-ends for several snoRNA species in S. cerevisiae. snoRNAs are typically annotated via homology with existing snoRNAs 30 or by a fit to a probabilistic model of the predicted snoRNA structure 31–33. One feature integrated into the model of C/D box snoRNAs is a terminal stem of expected length between 4–8 bp33. These base-paired stems are often shortened during pre-snoRNA processing by 5’-3’ exonucleases 34–36 and variable stem lengths have been observed in other eukaryotes37. For the majority of snoRNAs observed here, the sequenced 5’-ends accumulated on the annotated site (Table 2), but a small though significant number (14) showed deviations from this in the range of 1–5 nt, consistent with the range of predicted terminal stems for Box C/D snoRNAs, though this range is not as clear for the processing of H/ACA snoRNAs ends 31. Notably, for nine snoRNAs, the nature of the 5’-end remains ambiguous due to no single position where the majority of reads accumulate. Validation by primer extension assays revealed that the observed 5’-end deviations occur reproducibly in a strain-independent manner for certain snoRNA species, suggesting that this is not the result of stochastic differences in processing, but rather represent reliable and consistent differences from the 5’-end sequences that are commonly annotated in the genome databases. The 5’-end changes of the C/D box snoRNAs were evaluated by snoscan33 showing only one (snR78) with a newly predicted target. It would be interesting to determine whether the observed heterogeneity may affect other properties such as the lifetime of the snoRNA, or its association with proteins in the cell. Moreover, this work underscores the need to experimentally determine 5’-end status for structured ncRNAs as a complement to powerful computational approaches for predicting these sequences.
Supplementary Material
ACKNOWLEDGEMENTS
Funding for this work was provided by NIH GM087543 to JEJ, NSF DMR-1719316 to RB, and Center for RNA Biology Fellowships to BM and SD.
ABBREVIATIONS
- Thg1
tRNAHis guanylyltransferase
- TLP
Thg1-like protein
- WC
Watson–Crick
- HisRS
histidyl-tRNA synthetase
REFERENCES
- 1.Jackman JE, Gott JM & Gray MW Doing it in reverse: 3’-to-5’ polymerization by the Thg1 superfamily. RNA 18, 886–99 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackman JE & Phizicky EM tRNAHis guanylyltransferase catalyzes a 3’-5’ polymerization reaction that is distinct from G-1 addition. Proc. Natl. Acad. Sci. U. S. A 103, 8640–5 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu W, Jackman JE, Lohan AJ, Gray MW & Phizicky EM tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5’ end of tRNAHis. Genes Dev. 17, 2889–901 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu W, Hurto RL, Hopper AK, Grayhack EJ & Phizicky EM Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol. Cell. Biol 25, 8191–201 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preston MA & Phizicky EM The requirement for the highly conserved G-1 residue of Saccharomyces cerevisiae tRNAHis can be circumvented by overexpression of tRNAHis and its synthetase. RNA 16, 1068–77 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooley L, Appel B & Söll D Post-transcriptional nucleotide addition is responsible for the formation of the 5’ terminus of histidine tRNA. Proc. Natl. Acad. Sci. U. S. A 79, 6475–6479 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nameki N, Asahara H, Shimizu M, Okada N & Himeno H Identity elements of Saccharomyces cerevisiae tRNA(His). Nucleic Acids Res. 23, 389–94 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao BS, Maris EL & Jackman JE tRNA 5’-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucleic Acids Res. 39, 1833–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abad MG, Rao BS & Jackman JE Template-dependent 3’-5’ nucleotide addition is a shared feature of tRNAHis guanylyltransferase enzymes from multiple domains of life. Proc. Natl. Acad. Sci. U. S. A 107, 674–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura A et al. Structural basis of reverse nucleotide polymerization. Proc. Natl. Acad. Sci. U. S. A 110, 20970–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyde SJ et al. tRNA(His) guanylyltransferase (THG1), a unique 3’-5’ nucleotidyl transferase, shares unexpected structural homology with canonical 5’-3’ DNA polymerases. Proc. Natl. Acad. Sci. U. S. A 107, 20305–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hyde SJ, Rao BS, Eckenroth BE, Jackman JE & Doublié S Structural studies of a bacterial tRNA(HIS) guanylyltransferase (Thg1)-like protein, with nucleotide in the activation and nucleotidyl transfer sites. PLoS One 8, e67465 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura S et al. Template-dependent nucleotide addition in the reverse (3’-5’) direction by Thg1-like protein. Sci. Adv 2, e1501397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orellana O, Cooley L & Söll D The additional guanylate at the 5’ terminus of Escherichia coli tRNAHis is the result of unusual processing by RNase P. Mol. Cell. Biol 6, 525–9 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abad MG et al. A role for tRNA(His) guanylyltransferase (Thg1)-like proteins from Dictyostelium discoideum in mitochondrial 5’-tRNA editing. RNA 17, 613–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohan AJ & Gray MW Analysis of 5’- or 3’-Terminal tRNA Editing: Mitochondrial 5’ tRNA Editing in Acanthamoeba castellanii as the Exemplar. Methods Enzymol. 424, 223–242 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Abad MG et al. Mitochondrial tRNA 5′-editing in Dictyostelium discoideum and Polysphondylium pallidum. J. Biol. Chem 289, 15155–15165 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betat H, Long Y, Jackman JE & Mörl M From end to end: tRNA editing at 5’- and 3’-terminal positions. Int. J. Mol. Sci 15, 23975–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long Y, Abad MG, Olson ED, Carrillo EY & Jackman JE Identification of distinct biological functions for four 3’-5’ RNA polymerases. Nucleic Acids Res. 44, 8395–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackman JE & Phizicky EM Identification of critical residues for G-1 addition and substrate recognition by tRNA(His) guanylyltransferase. Biochemistry 47, 4817–25 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Heinemann IU, Randau L, Tomko RJ & Söll D 3’-5’ tRNAHis guanylyltransferase in bacteria. FEBS Lett. 584, 3567–72 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park D, Morris AR, Battenhouse A & Iyer VR Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 42, 3736–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosinski-Chupin I et al. Single nucleotide resolution RNA-seq uncovers new regulatory mechanisms in the opportunistic pathogen Streptococcus agalactiae. BMC Genomics 16, 419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shell SS, Chase MR, Ioerger TR & Fortune SM in Methods in Molecular Biology 1285, 31–45 (Humana Press, New York, NY, 2015). [DOI] [PubMed] [Google Scholar]
- 25.Swinehart WE, Henderson JC & Jackman JE Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA 19, 1137–46 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudinger J, Florentz C & Giegé R Histidylation by yeast HisRS of tRNA or tRNA-like structure relies on residues −1 and 73 but is dependent on the RNA context. Nucleic Acids Res. 22, 5031–7 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinemann IU, Nakamura A, O’Donoghue P, Eiler D & Söll D tRNAHis-guanylyltransferase establishes tRNAHis identity. Nucleic Acids Res. 40, 333–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirsebom LA RNase P RNA mediated cleavage: Substrate recognition and catalysis. Biochimie 89, 1183–1194 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Shigematsu M & Kirino Y 5’-Terminal nucleotide variations in human cytoplasmic tRNAHisGUG and its 5’-halves. RNA 23, 161–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartschat S, Kehr S, Tafer H, Stadler PF & Hertel J snoStrip: a snoRNA annotation pipeline. Bioinformatics 30, 115–6 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Schattner P et al. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 32, 4281–96 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J-H et al. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 34, 5112–23 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe TM A Computational Screen for Methylation Guide snoRNAs in Yeast. Science (80-. ). 283, 1168–1171 (1999). [DOI] [PubMed] [Google Scholar]
- 34.Watkins NJ, Leverette RD, Xia L, Andrews MT & Maxwell ES Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2, 118–33 (1996). [PMC free article] [PubMed] [Google Scholar]
- 35.Vincenti S, De Chiara V, Bozzoni I & Presutti C The position of yeast snoRNA-coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre-mRNA. RNA 13, 138–50 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darzacq X & Kiss T Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5’,3’-terminal stem structure. Mol. Cell. Biol 20, 4522–31 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishore S et al. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 14, R45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.