Abstract
Soy isoflavones have been suggested as epigenetic modulating agents with effects that could be important in carcinogenesis. Hypomethylation of LINE-1 has been associated with head and neck squamous cell carcinoma (HNSCC) development from oral premalignant lesions and with poor prognosis. To determine if neoadjuvant soy isoflavone supplementation could modulate LINE-1 methylation in HNSCC, we undertook a clinical trial.
Methods:
Thirty-nine patients received 2–3 weeks of soy isoflavone supplements (300 mg/day) orally prior to surgery. Methylation of LINE-1, and 6 other genes was measured by pyrosequencing in biopsy, resection and whole blood (WB) specimens. Changes in methylation were tested using paired t-tests and ANOVA. Median follow up was 45 months.
Results:
LINE-1 methylation increased significantly after soy isoflavone (p<0.005). Amount of change correlated positively with days of isoflavone taken (p= .04). Similar changes were not seen in corresponding WB samples. No significant changes in tumor or blood methylation levels were seen in the other candidate genes.
Conclusion:
This is the first demonstration of in vivo increases in tissue-specific global methylation associated with soy isoflavone intake in patients with HNSCC. Prior associations of LINE-1 hypomethylation with genetic instability, carcinogenesis and prognosis suggest that soy isoflavones maybe potential chemopreventive agents in HNSCC.
Keywords: Isoflavones/soy, Methylation, acetylation, Cancer Prevention
Introduction
DNA methylation in head and neck squamous cell carcinoma (HNSCC) is an attractive target for cancer prevention as it is modifiable, not genotoxic and strongly associated with survival from HNSCC (1–4). Global hypomethylation has been identified as an important epigenetic feature of HNSCC (1, 5–7) and is often represented as a surrogate by hypomethylation of the highly repetitive long interspersed nucleotide element-1 (LINE-1) retrotransposon. When it is unmethylated, LINE-1 can integrate into genomic DNA for target site primed reverse transcription or create DNA breaks leading to genomic instability, insertional mutagenesis or deletion of genomic sequences (8, 9). Silencing of LINE-1 expression by promoter methylation is thought to be of evolutionary importance in preventing cancer development(9). Hypomethylation of LINE-1 has been associated with increased risk of carcinogenesis and poor prognosis for many types of cancer including colon, breast, gastric, ovarian and cervical cancers (10–14) and with development of HNSCC from oral premalignant lesions(7). Some studies report that dietary patterns and healthy life style can modulate peripheral blood leukocyte LINE-1 methylation in cervical intraepithelial neoplasia and in normal subjects(15, 16) while others report no changes in tissue levels(17).
Epidemiologic studies in patients with HNSCC have consistently indicated that diet and dietary components may be associated with a prognostic benefit in HNSCC (18–20). Patients with HNSCC are generally nutritionally deprived, with diets low in fruits, vegetables, beans or other foods that contain isoflavones (21–24). Consumption of leguminous groups high in isoflavones has been associated with decreased risk of head and neck cancer (24, 25). The efficacy of soy nutritional supplementation in patients with HNSCC or its ability to modulate biomarkers in vivo is unknown, however soy isoflavones have been associated with a prognostic benefit in many other cancers (26–28) and thus represent a potential chemotherapeutic agent with a favorable therapeutic index. In particular, there is strong evidence that genistein, a soy derived isoflavone, has specific anticancer activities such as inhibition of MMP-9 and IL-6, downregulation of VEGF, modulation of immunity and inactivation of NF-kB (29–32). Multiple in vivo and vitro anti-cancer effects of genistein have been postulated, including modulation of gene methylation (33). Genistein and related soy isoflavones were shown to reactivate methylation-silenced genes in esophageal (33), breast (34) and prostate (35) cell lines, possibly through direct inhibition of DNA methyltransferases (33).
Our group and others have characterized epigenetic changes in HNSCC associated with diet (36), smoking (2, 37, 38) and survival (1, 3, 39, 40). We have previously shown the stability of epigenetic methylation markers between pretreatment biopsies and subsequent tumor resection samples(41). We also recently assessed associations of gene methylation with patient prognosis using a panel of 6 candidate genes (CCNA1, NDN, CD1a, DCC, GADD45a, and p16) that are frequently methylated in HNSCC. In a large cohort study of 346 patients, we found that pretreatment DNA hypermethylation of two of the genes (CD1A and NDN) was associated with significant survival benefit (1). Here we further extend these studies to test the hypothesis that LINE-1 hypomethylation levels and DNA hypermethylation of these specific candidate genes could be favorably modulated by dietary soy supplementation and potentially improve survival. To determine if high dose soy isoflavone treatment can modulate such epigenetic changes in tumor tissue, we conducted a short term, neoadjuvant soy isoflavone supplementation clinical trial in HNSCC patients undergoing definitive surgical management. We determined genomic and gene-specific methylation changes in tissue and whole blood samples collected before and after soy administration in these patients that suggest potential therapeutic benefit of soy isoflavone supplementation through increasing LINE-1 methylation in tumor tissue.
Methods
Patient Population
This Phase II clinical trial was conducted as an inter-institutional translational component of a National Cancer Institute sponsored University of Michigan Head and Neck Specialized Program of Research Excellence (SPORE). Thirty-nine patients participated in this clinical trial. Patients were recruited from the University of Michigan (n = 31), Winship Cancer Institute of Emory University (n = 5) and Karmanos Cancer Center of Wayne State University (n = 3). To be eligible, patients had to be potentially curable with definitive surgery for either primary or locally recurrent head and neck squamous carcinoma. Of 74 eligible subjects approached at the University of Michigan, there were 10 refusals, 19 screen failures, and four patients with inadequate tissue samples. There were no screen failures reported for Winship or Karmanos institutions, and data on patient refusal to participate at those sites were not available. This trial was approved by the Institutional Review Board for human subjects at all of the treating centers. All patients signed informed consent. Median follow up was 45 months.
A total of 23 patients (59%) were previously untreated for their primary cancer and 16 (41%) were treated either for a second primary cancer (8 patients) or locally recurrent cancer (8 patients). Sites (n) of primary tumor for previously untreated patients included oral cavity (21), larynx (1) and oropharynx cancer (1), while patients with recurrent disease included oral cavity (5) and laryngeal (3) cancer patients. The 8 patients with second primary tumors included 5 oral cavity and 3 oropharynx cancers. The average age of the patients in this trial was 60.1 years (range 26.0 – 78.4 years). A total of 12 participants were female (31%). The tumor site for the majority of the cancers was oral cavity (80%) and nearly half of the patients (44%) were diagnosed with stage IVA cancer, with the rest equally distributed between Stages I, II and III. Only 8 participants were never smokers and two were never drinkers. A total of 45% of subjects were considered normal weight by BMI and 55% were considered overweight or obese (Table 1).
Table 1.
Descriptive statistics of study participants
| Age: mean (sd) | 60.1 (12.5) |
| % Female | 31% |
| % ever/former smoker | 79%/42% |
| Pack years: mean (sd) | 32.3 (26.0) |
| Underweight/overweight/obese | 14/8/9 |
| BMI: mean (sd) | 27.2 (6.1) |
| Tumor Stage I/II, III/IV (%) | 12 (31%), 27 (69%) |
| Disease Site: larynx/oral cavity/oropharynx | 4/31/4 |
| Events: death/recurrence | 10/9 |
Patients were scheduled to receive a minimum of 2 weeks of daily soy supplementation, corresponding to the time between biopsy and surgical resection (range 7–39 days, median = 15.2). Soy isoflavone supplementation consisted of 300 mg/day of G-2535, a purified soy extract (NCI-DCP Repository MRI Global, Kansas City, MO., NCI, NIH) administered orally prior to surgery. The dose selected was based on prior clinical studies that established a maximum tolerated dose. The entire dose was given as two capsules once a day at breakfast or at a subsequent meal if breakfast was missed. Capsules could be opened and contents dissolved and administered via gastrostomy tube if necessary. The duration of the neoadjuvant treatment interval was deemed acceptable and was based on the average time from biopsy to surgical resection in our historical cohort of surgically treated patients in our SPORE database which averages 28.5 days (range 5–115). The active pharmaceutical ingredient, G-2535, is a purified soy extract containing ≥97% total unconjugated isoflavones, composed primarily of genistein and daidzein in a 2:1 ratio (genistein to daidzein). Compliance was determined by pill counts and patient recall at patient follow up visits.
Tissue Samples and Methylation
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were collected from biopsy and surgical resection specimens, and an expert head and neck pathologist (JM) confirmed tumor histology and screened representative blocks for areas of >70 % cellularity and minimal necrosis. Only subjects with sufficient tissue and DNA to yield methylation results were included in the analyses. Designated areas of FFPE tissue were microdissected from unstained slides and DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. DNA concentration and purity was measured with a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). Sodium bisulfite treatment was performed on 250 ng of DNA using the EpiTect Bisulfite Kit (Qiagen, Valencia, CA) according to the manufacturer’s recommended protocol. Peripheral whole blood samples were collected at time of initiation of soy isoflavone treatment and at time of tumor resection. DNA was extracted using the QIAsymphony Automated DNA extraction system (Qiagen, Valencia CA). Two hundred ng of DNA were bisulfite converted using the EpiTect Bisulfite Kit (Qiagen, Valencia, CA) for methylation analyses.
Methylation assays for promoter regions of candidate DCC, CD1A, CCNA1, GADD45α, NDN and p16 genes were performed as described previously (1). LINE-1 methylation was measured using a previously published assay (42). Bisulfite singleplex PCR amplification was performed using FastStart Taq Polymerase (Roche Diagnostics, Indiana, USA) with primer concentrations of 0.2 mM and 10ng/μL of bisulfite-converted DNA. Fifteen microliters of PCR product was combined with sequencing primer and methylation analysis was conducted using the PyroMark™ MD System (Biotage) according to manufacturer’s protocol (PyroGold reagents). Four bisulfite and four pyrosequencing controls were generated by mixing unmethylated and methylated control DNA (genomic: EpigenDX; bisulfite-converted: EpiTect) to obtain controls with 0%, 30%, 60% and 100% methylation. Each sample plate was run with all controls. If methylation values of controls were incorrect, all samples on plate were re-run. Measurement of all samples for every methylation marker was not possible due to insufficient quantity of extracted DNA.
Statistical Methods
Changes in methylation levels were assessed using paired t-tests of pretreatment and post-treatment methylation levels in tissue specimens and whole blood. Associations of pretreatment levels and crude changes in methylation levels with clinical tumor and patient characteristics were explored with ANOVA. Multivariable linear regression models were used to test for associations between methylation levels and clinical characteristics controlling for previous treatment. Associations between methylation change and survival or recurrence free time were tested with Cox proportional hazards models. The association between change in LINE-1 and duration of soy isoflavone administration was tested with linear regression and ANOVA. All analyses were conducted in SAS (Cary, NC).
Results
Changes in LINE-1 Methylation after Neoadjuvant Soy Isoflavone Supplementation
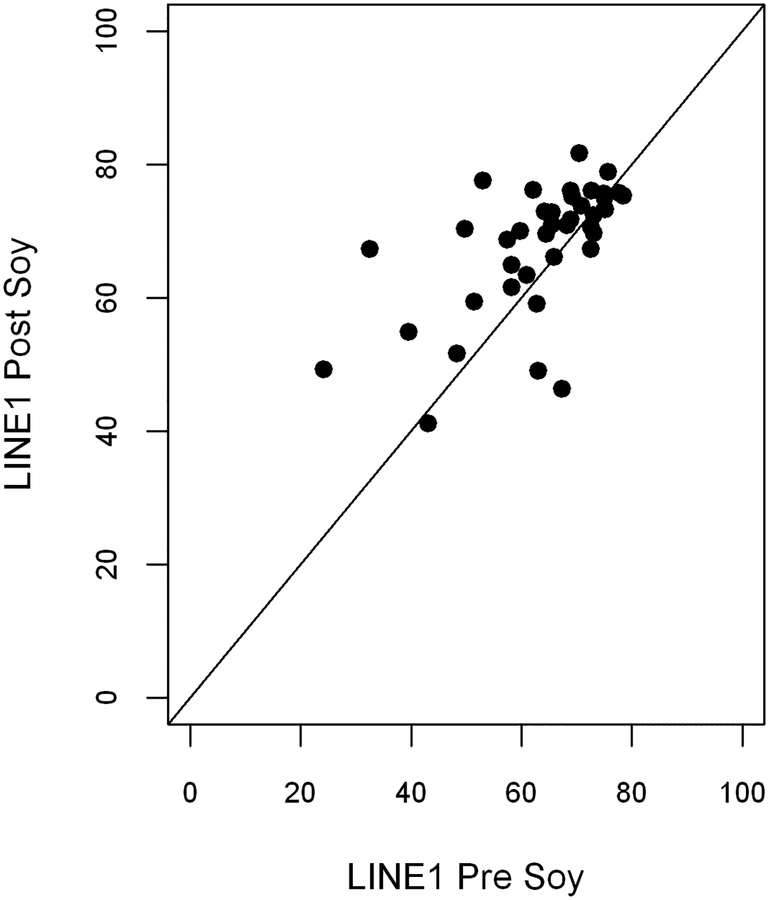
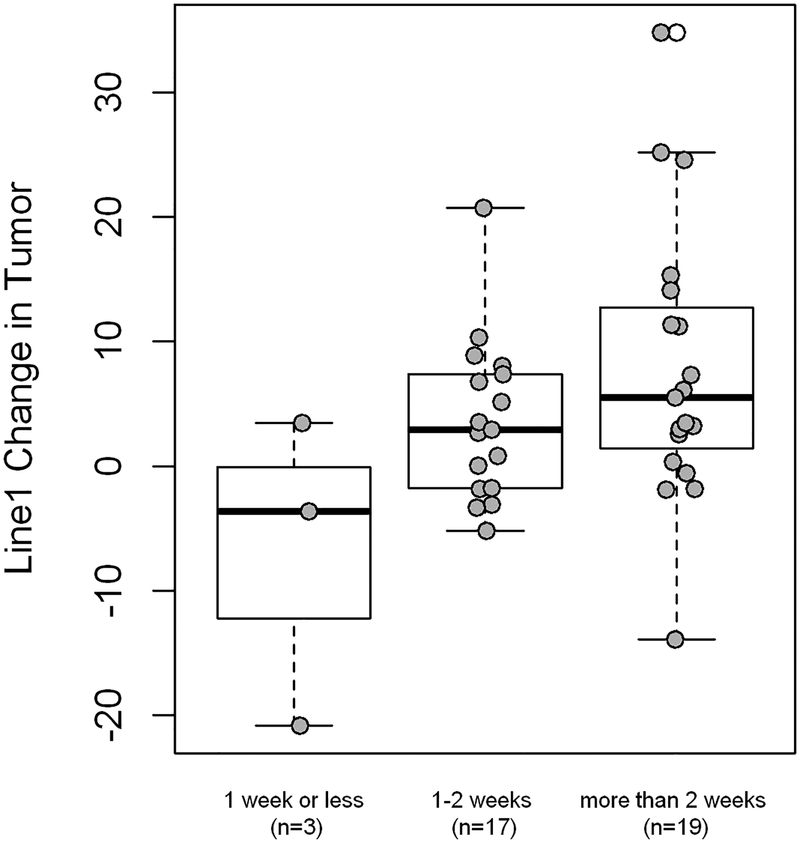
The most significant findings in this study were mean increases in LINE-1 methylation in tumor resection specimens after short term oral soy supplementation. LINE-1 methylation was significantly increased in tumor specimens after soy isoflavone (mean increase 4.9%; range −34.8% to +20.9%; p<0.005, Table 2 and Figure 1). A dose response effect was suggested in that the amount of methylation change correlated positively with duration of isoflavone supplementation in days (p= .04, Figure 2). The increases in LINE-1 also tended to differ by patient BMI and prior treatment, with greatest increases in underweight individuals (p=0.04) and in the patients who were previously untreated (p=0.19). These changes were noted in the microenvironment tumor specimens while similar changes for LINE-1 methylation were not seen in DNA analysis from corresponding whole blood samples. Levels of pretreatment LINE-1 methylation and CD1A methylation were significantly lower in tumor tissue compared to whole blood levels (p<0.0001 each). This contrasted the levels of the other methylation markers (CCNA, Gadd45alpha, NDN, p16, DCC) which were all significantly higher in tumor tissue compared to whole blood.
Table 2.
Change in pre- and post-treatment methylation levels in tumor (t) and blood (b)
| Marker | Mean pre-treatment methylation level (sd) | Mean post-treatment methylation level (sd) | Mean change (pre-post) (sd) | p value change in methylation |
|---|---|---|---|---|
| p16 (t) | 16.71 (23.55) | 13.35 (19.03) | 3.36 (24.81) | 0.40 |
| p16 (b) | 1.14 (1.74) | 0.94 (0.64) | 0.33 (2.02) | 0.41 |
| DCC (t) | 32.63 (20.11) | 30.91 (24.98) | 1.07 (23.30) | 0.78 |
| DCC (b) | 7.43 (2.26) | 7.66 (2.42) | −0.05 (1.83) | 0.89 |
| CCNA1 (t) | 30.53 (17.90) | 32.32 (18.82) | −2.14 (22.95) | 0.61 |
| CCNA1 (b) | 15.95 (4.55) | 16.33 (3.21) | −0.17 (2.82) | 0.77 |
| NDN (t) | 43.36 (16.32) | 43.60 (12.99) | −0.24 (21.0) | 0.94 |
| NDN (b) | 36.94 (4.90) | 37.59 (5.26) | 0.15 (2.46) | 0.76 |
| GADD45 (t) | 1.76 (1.33) | 1.53 (0.67) | 0.24 (1.56) | 0.35 |
| GADD45 (b) | 0.76 (0.53) | 1.06 (0.51) | −0.33 (0.93) | 0.08 |
| LINE-1 (t) | 62.88 (12.48) | 67.78 (9.75) | −4.90 (10.15) | 0.005 |
| LINE-1 (b) | 72.43 (1.16) | 72.62 (1.17) | −0.18 (1.60) | 0.57 |
| CD1A (t) | 65.85 (12.99) | 66.41 (13.91) | −0.56 (14.98) | 0.82 |
| CD1A (b) | 87.56 (3.87) | 88.23 (3.38) | −0.75 (2.80) | 0.18 |
Figure 1:
Change in LINE-1 Methylation Levels in pre- and post-supplementation tumor tissue samples. LINE-1 methylation increased significantly in tumor specimens after soy isoflavone (p<0.005).
Figure 2:
LINE-1 methylation level changes by days of soy administration. Methylation changes differed significantly with longer duration of supplementation (ANOVA, p=0.04) and amount of change correlated positively with longer duration by linear regression analysis (p=0.009).
Changes in Candidate Gene Methylation
Comparing changes in methylation levels between pre- and post-soy treatment samples, no significant changes in methylation for most of the candidate genes were noted including p16, DCC, CCNA1, CD1A, GADD45α, and NDN (Table 2). Although CCNA1 methylation in tumor samples did not differ significantly after soy isoflavone treatment, levels of methylation of this gene in peripheral blood samples tended to increase in underweight individuals (p=0.03). Interestingly, levels of CD1A methylation which we previously demonstrated as associated with overall survival, tended to increase in peripheral blood samples after soy isoflavone supplementation (p=0.18) and correlated with the total dose of soy administration (p=0.08). Despite lack of statistically significant changes in CD1A methylation in the tumor specimens, the peripheral blood changes were most notable in the previously untreated patients (p=0.10).
Pretreatment Gene Methylation and Clinical Characteristics and Prior Treatment
Pretreatment levels of methylation of the 6 candidate genes in tumor specimens and peripheral blood were explored with respect to differences in tumor site, stage, pretreatment BMI, drinking and smoking habit and prior treatment. There was very little evidence of any significant associations of clinical characteristics with methylation of p16, DCC, CCNA1, CD1A, GADD45α, NDN or LINE-1 genes. LINE-1 methylation in tumor tended to be higher in obese subjects (p=0.17). In tumors of never smokers, CCNA1 methylation was higher (p<0.04) while DCC methylation tended to be lower (p=0.18). In pretreatment peripheral blood samples, NDN methylation was higher in obese subjects (p=0.04). CD1A methylation was lower in current drinkers (p=0.04). Comparing previously untreated patients to patients with recurrent disease, the levels of NDN methylation (p=.003) and CCNA1 methylation (p=.05) were lower while LINE-1 methylation (p=0.10) was lower in patients with recurrent disease. Interestingly, methylation of CD1A, a marker that was previously associated with increased survival in our prior epigenetic cohort study, tended to be marginally lower in the blood of patients with recurrent disease (p=0.07). The majority of clinical trial patients had oral cavity cancer and pretreatment NDN methylation was lower (p<.008) and LINE-1 higher (p<0.0001) in oral cavity cancers compared to oropharynx or larynx cancers.
Clinically, neoadjuvant soy isoflavone supplementation was found to be very feasible and well tolerated without any serious adverse events. Patient compliance was excellent. The average duration of genistein intake was 15.3 days, ranging from 7 to 26 days of administration. The primary reasons for any patient noncompliance were forgetfulness on the part of the patients and mild nausea. No serious adverse events were otherwise reported that would preclude patients from taking genistein. No clinical tumor regressions or progressions were noted during the brief neoadjuvant treatment period.
Epigenetic Change and Survival
Among the 39 evaluable patients in the trial, we observed 9 recurrences and 10 deaths during follow-up. The median follow-up was 45 months (Kaplan Meier estimate). There did not appear to be any overall survival differences between patients with previously untreated or recurrent cancers (log rank p=0.43) or for time to progression (log rank p=0.20) but such a small study is likely underpowered to detect expected differences. In this small prospective cohort study, LINE-1 methylation was not associated with objective survival or recurrence benefit [HR 0.99, 95% C.I. 0.93 – 1.06 and HR 1.05, 95% C.I. 1.00 – 1.11, respectively (data not shown)]. The total dose of soy isoflavone administered similarly was not associated with survival or recurrence during this short follow up period. The lack of a significant association of the changes in LINE-1 methylation with clinical outcome did not differ when patients were separately grouped and analyzed as previously untreated or recurrent cancer.
Discussion
The search for effective and tolerable chemopreventive agents for HNSCC is ongoing. Plant derived phytochemicals, such as soy isoflavones, have been suggested as active agents with pleiotropic in vitro and in vivo effects on gene methylation, angiogenesis, prostaglandin mediated inflammation, and apoptosis that could be potentially important in carcinogenesis (26, 29, 30, 33). Global LINE-1 hypomethylation (5, 43–45) and specific gene hypermethylation patterns (1, 46) have been implicated in HNSCC and in carcinogenesis and prognosis in a variety of other cancers such as colon, breast, and prostate. In HNSCC, pretreatment intake of soy products (tofu) was associated with significantly improved prognosis in a prospective University of Michigan epidemiologic dietary study in 374 patients (unpublished data) but only one study to our knowledge (prostate cancer) has evaluated the effects of purified soy isoflavone supplementation on epigenetic biomarkers (47). Of the many proposed mechanisms of action of soy isoflavones, epigenetic alteration of gene methylation is an attractive target for chemoprevention since cigarette smoking is a major predisposing factor for cancer development and has been associated with oncogene silencing of important tumor suppressor genes such as p16 (33, 48). Thus, we undertook a proof of principle window of opportunity clinical trial to determine the clinical feasibility and potential epigenetic effects of short term soy isoflavone supplementation in patients with head and neck cancer.
The major finding in this study was a significant and dose related increase in LINE-1 methylation in tumor tissue after neoadjuvant soy isoflavone supplementation. This could have important implications as it demonstrates a potentially beneficial epigenetic change in vivo with a non-toxic dietary supplement in patients with HNSCC. Genome-wide hypomethylation and localized, gene-specific hypermethylation are hallmarks of cancers (49). Hypomethylation of LINE-1 elements is an early event in carcinogenesis (50) and is indicative of overall methylation in the genome (42). LINE-1 elements comprise approximately 17% of the genome (11). Thus, even small changes in LINE-1 methylation can be associated with larger changes at the genomic level. These elements are environmentally responsive, with changes seen in subjects exposed to benzene (51, 52), persistent organic pollutants (53, 54), lead (55), poor diet (56), and smoking (57). LINE-1 hypomethylation, as a marker of global hypomethylation, is an important clinical biomarker for epigenetic change. LINE-1 hypomethylation has been associated with nutrition in patients with cancer (15, 58), early cancer diagnosis(10, 53), tumor aggressiveness (14) and prognosis (13, 59) suggesting potential usefulness as a surrogate marker in interventional studies. Combinational therapy with nutritional epigenetic modulation and gene targeted therapies has been suggested as an important new frontier in cancer prevention and treatment(60).
There are few interventional studies that have used LINE-1 methylation as a biomarker of response. In HNSCC, hypomethylation of LINE-1 has also been associated with genomic centromere instability and loss of heterozygosity (43, 44). In one of the larger studies, Smith et al measured LINE-1 hypomethylation in 119 HNSCC tumors and found hypomethylation in 67% of cases with average genomic methylation of 46.8% compared to 54% in normal tissue (5). Hsiung et al suggested that global hypomethylation in whole blood could be a useful biomarker for cancer. They found a 1.6-fold increase in relative risk of HNSCC in subjects in the lowest tertile of LINE-1 methylation in whole blood samples comparing cases to controls (45). Delgado-Cruzata et al (61) described significant LINE-1 methylation increases in peripheral blood leukocyte DNA after weight loss intervention at six and twelve months in breast cancer survivors. The magnitude of the LINE-1 differences were similar to those seen here (3% and 2.2% change at six and twelve months respectively). Likewise, our data comparing whole blood and tumor are consistent with basal levels and differences in percent hypomethylation of LINE-1 between tumor and normal tissue reported by Chalitchagorn et al in HNSCC (50). Although we didn’t see significant changes in whole blood LINE-1 methylation after our short soy isoflavone intervention, the fact that changes were seen in methylation in the unstable tumor microenvironment could be meaningful and may indicate a possible, beneficial mechanism of action on DNA stability in the tumor microenvironment (62) that was not reflected in the whole blood at these dose levels and duration. From a prevention standpoint, Foy et al found that patients with oral pre-malignant lesions and with low levels of LINE-1 methylation had a significantly shorter time to progression to cancer (7). Although we did not observe an overall survival benefit from the soy isoflavone intervention in this study, larger sufficiently powered studies with longer follow up would be needed to determine any potential prognostic and clinical benefit of longer term soy supplementation in HNSCC patients.
The findings show differing results of pretreatment soy isoflavone supplementation on epigenetic changes in tumor and peripheral blood samples from patients undergoing potentially curative surgery for head and neck squamous cell carcinoma. Although significant changes in tumor LINE-1 methylation suggest that soy treatment is associated with increased genomic stability that could have prognostic importance, the results also suggest that changes in tumor do not parallel changes seen in peripheral blood gene methylation. We saw very little variation between patient levels for pretreatment whole blood methylation for each gene in our cohort, whereas differences between respective blood and tissue methylation levels for each gene were large. “It is unknown why significant changes were only seen in LINE-1 methylation in tumor or if longer treatment duration or higher doses of soy isoflavone might have been associated with other epigenetic changes. Clinical characteristics typically associated with poor outcomes (e.g. underweight, recurrent cancer) tended to show the greatest changes in LINE-1 methylation. Although not statistically significant, the direction of change for whole blood methylation after the soy intervention was for increased methylation for all genes tested except p16. We also observed suggestive methylation changes in the gene CD1A. CD1A, which modulates Langerhans and dendritic cell function, showed what might be interpreted as beneficial changes in peripheral blood methylation which were most evident in the highest risk patients with recurrent cancers. We also observed significantly lower levels of methylation of this gene in tumor tissue compared to whole blood, similar to our findings for LINE-1. Methylation of CD1A could help regulate immunosuppressive M1 macrophage and Langerhans cell functions in tissue. Because our analysis included small subsets for several tested genes however, the trends noted for individual genes would be expected to be less significant if multiple statistical testing adjustments were made. Since low methylation of CD1A was previously shown to be associated with decreased survival (1), these findings even in a small number of patients are nonetheless intriguing for further exploration of soy isoflavone supplementation as an immunomodulatory and chemopreventive agent in HNSCC.
Conclusions
This study has several unique implications. First, our results indicate that meaningful epigenetic changes can be achieved after a short dietary intervention. Changes in the tumor tissue itself may be an early indicator of beneficial effects of a nutritional treatment intervention. However, our results should be interpreted with caution since this was a relatively small, albeit carefully performed, prospective clinical trial. Second, this is the first report of specific alterations in LINE-1 hypomethylation in response to soy isoflavone administration in HNSCC patients in vivo. The fact that a moderate dose soy intervention beneficially altered an important marker of global hypomethylation in a short time period suggests that sustained delivery could have greater clinical effects. This is particularly important for agents that may be prescribed for long term primary or secondary cancer prevention. These preliminary data suggest the need for a larger study that will carefully evaluate soy as a chemopreventive agent in HNSCC as well as consideration of other natural compounds for the epigenetic treatment of HNSCC.
Acknowledgements
This work was supported by the University of Michigan Head and Neck Specialized Program of Research Excellence (SPORE) through funding from the National Cancer Institute at the National Institutes of Health (grant number NIH/NCI P50CA097248). We also acknowledge the SPORE patients and families.
References:
- 1.Virani S, Bellile E, Bradford CR, Carey TE, Chepeha DB, et al. : NDN and CD1A are novel prognostic methylation markers in patients with head and neck squamous carcinomas. BMC Cancer 15, 825, 2015. doi: 10.1186/s12885-015-1806-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colacino JA, Dolinoy DC, Duffy SA, Sartor MA, Chepeha DB, et al. : Comprehensive analysis of DNA methylation in head and neck squamous cell carcinoma indicates differences by survival and clinicopathologic characteristics. PLoS One 8, e54742, 2013. doi: 10.1371/journal.pone.0054742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostareli E, Hielscher T, Zucknick M, Baboci L, Wichmann G, et al. : Gene promoter methylation signature predicts survival of head and neck squamous cell carcinoma patients. Epigenetics 11, 61–73, 2016. doi: 10.1080/15592294.2015.1137414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw RJ, Liloglou T, Rogers SN, Brown JS, Vaughan ED, et al. : Promoter methylation of P16, RARbeta, E-cadherin, cyclin A1 and cytoglobin in oral cancer: quantitative evaluation using pyrosequencing. Br J Cancer 94, 561–8, 2006. doi: 10.1038/sj.bjc.6602972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith IM, Mydlarz WK, Mithani SK, Califano JA: DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer 121, 1724–8, 2007. doi: 10.1002/ijc.22889 [DOI] [PubMed] [Google Scholar]
- 6.Park IS, Chang X, Loyo M, Wu G, Chuang A, et al. : Characterization of the methylation patterns in human papillomavirus type 16 viral DNA in head and neck cancers. Cancer Prev Res (Phila) 4, 207–17, 2011. doi: 10.1158/1940-6207.CAPR-10-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foy JP, Pickering CR, Papadimitrakopoulou VA, Jelinek J, Lin SH, et al. : New DNA methylation markers and global DNA hypomethylation are associated with oral cancer development. Cancer Prev Res (Phila) 8, 1027–35, 2015. doi: 10.1158/1940-6207.CAPR-14-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carreira PE, Richardson SR, Faulkner GJ: L1 retrotransposons, cancer stem cells and oncogenesis. FEBS J 281, 63–73, 2014. doi: 10.1111/febs.12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belancio VP, Roy-Engel AM, Deininger PL: All y’all need to know ‘bout retroelements in cancer. Semin Cancer Biol 20, 200–10, 2010. doi: 10.1016/j.semcancer.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barchitta M, Quattrocchi A, Maugeri A, Vinciguerra M, Agodi A: LINE-1 hypomethylation in blood and tissue samples as an epigenetic marker for cancer risk: a systematic review and meta-analysis. PLoS One 9, e109478, 2014. doi: 10.1371/journal.pone.0109478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodic N, Burns KH: Long interspersed element-1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet 9, e1003402, 2013. doi: 10.1371/journal.pgen.1003402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swets M, Zaalberg A, Boot A, van Wezel T, Frouws MA, et al. : Tumor LINE-1 Methylation Level in Association with Survival of Patients with Stage II Colon Cancer. Int J Mol Sci 18, 2016. doi: 10.3390/ijms18010036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Chan AT, et al. : A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 100, 1734–8, 2008. doi: 10.1093/jnci/djn359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, Dahlstrom JE, Chandra A, Board P, Rangasamy D: Prognostic value of LINE-1 retrotransposon expression and its subcellular localization in breast cancer. Breast Cancer Res Treat 136, 129–42, 2012. doi: 10.1007/s10549-012-2246-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piyathilake CJ, Badiga S, Kabagambe EK, Azuero A, Alvarez RD, et al. : A dietary pattern associated with LINE-1 methylation alters the risk of developing cervical intraepithelial neoplasia. Cancer Prev Res (Phila) 5, 385–92, 2012. doi: 10.1158/1940-6207.CAPR-11-0387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marques-Rocha JL, Milagro FI, Mansego ML, Mourao DM, Martinez JA, et al. : LINE-1 methylation is positively associated with healthier lifestyle but inversely related to body fat mass in healthy young individuals. Epigenetics 11, 49–60, 2016. doi: 10.1080/15592294.2015.1135286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueiredo JC, Grau MV, Wallace K, Levine AJ, Shen L, et al. : Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev 18, 1041–9, 2009. doi: 10.1158/1055-9965.EPI-08-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arthur AE, Peterson KE, Shen J, Djuric Z, Taylor JM, et al. : Diet and proinflammatory cytokine levels in head and neck squamous cell carcinoma. Cancer 120, 2704–12, 2014. doi: 10.1002/cncr.28778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langius JA, Zandbergen MC, Eerenstein SE, van Tulder MW, Leemans CR, et al. : Effect of nutritional interventions on nutritional status, quality of life and mortality in patients with head and neck cancer receiving (chemo)radiotherapy: a systematic review. Clin Nutr 32, 671–8, 2013. doi: 10.1016/j.clnu.2013.06.012 [DOI] [PubMed] [Google Scholar]
- 20.Arthur AE, Peterson KE, Rozek LS, Taylor JM, Light E, et al. : Pretreatment dietary patterns, weight status, and head and neck squamous cell carcinoma prognosis. Am J Clin Nutr 97, 360–8, 2013. doi: 10.3945/ajcn.112.044859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayne ST, Cartmel B, Lin H, Zheng T, Goodwin WJ Jr.: Low plasma lycopene concentration is associated with increased mortality in a cohort of patients with prior oral, pharynx or larynx cancers. J Am Coll Nutr 23, 34–42, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Morse DE, Pendrys DG, Katz RV, Holford TR, Krutchkoff DJ, et al. : Food group intake and the risk of oral epithelial dysplasia in a United States population. Cancer Causes Control 11, 713–20, 2000 [DOI] [PubMed] [Google Scholar]
- 23.De Stefani ARE, Mendilaharsu M, Deneo-Pellegrini H: Diet and risk of cancer of the upper aerodigestive tract - II. Nutrients. Oral Oncology 35, 22–26, 1999 [DOI] [PubMed] [Google Scholar]
- 24.De Stefani E D-P H, Mendilaharsu M, Ronco A: Diet and risk of cancer of the upper aerodigestive tract - I. Foods. Oral Oncology 35, 17–21, 1999 [DOI] [PubMed] [Google Scholar]
- 25.Freedman ND, Park Y, Subar AF, Hollenbeck AR, Leitzmann MF, et al. : Fruit and vegetable intake and head and neck cancer risk in a large United States prospective cohort study. Int J Cancer 122, 2330–6, 2008. doi: 10.1002/ijc.23319 [DOI] [PubMed] [Google Scholar]
- 26.Shu XO, Zheng Y, Cai H, Gu K, Chen Z, et al. : Soy food intake and breast cancer survival. JAMA 302, 2437–43, 2009. doi: 10.1001/jama.2009.1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang G, Shu XO, Li HL, Chow WH, Wen W, et al. : Prediagnosis soy food consumption and lung cancer survival in women. J Clin Oncol 31, 1548–53, 2013. doi: 10.1200/JCO.2012.43.0942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwagami S, Baba Y, Watanabe M, Shigaki H, Miyake K, et al. : LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg 257, 449–55, 2013. doi: 10.1097/SLA.0b013e31826d8602 [DOI] [PubMed] [Google Scholar]
- 29.Sasamura H, Takahashi A, Yuan J, Kitamura H, Masumori N, et al. : Antiproliferative and antiangiogenic activities of genistein in human renal cell carcinoma. Urology 64, 389–93, 2004. doi: 10.1016/j.urology.2004.03.045 [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Ahmed F, Ali S, Philip PA, Kucuk O, et al. : Inactivation of nuclear factor kappaB by soy isoflavone genistein contributes to increased apoptosis induced by chemotherapeutic agents in human cancer cells. Cancer Res 65, 6934–42, 2005. doi: 10.1158/0008-5472.CAN-04-4604 [DOI] [PubMed] [Google Scholar]
- 31.Suh KS, Koh G, Park CY, Woo JT, Kim SW, et al. : Soybean isoflavones inhibit tumor necrosis factor-alpha-induced apoptosis and the production of interleukin-6 and prostaglandin E2 in osteoblastic cells. Phytochemistry 63, 209–15, 2003 [DOI] [PubMed] [Google Scholar]
- 32.Lesinski GB, Reville PK, Mace TA, Young GS, Ahn-Jarvis J, et al. : Consumption of soy isoflavone enriched bread in men with prostate cancer is associated with reduced proinflammatory cytokines and immunosuppressive cells. Cancer Prev Res (Phila) 8, 1036–44, 2015. doi: 10.1158/1940-6207.CAPR-14-0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, et al. : Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res 11, 7033–41, 2005. doi: 10.1158/1078-0432.CCR-05-0406 [DOI] [PubMed] [Google Scholar]
- 34.Xie Q, Bai Q, Zou LY, Zhang QY, Zhou Y, et al. : Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosomes Cancer 53, 422–31, 2014. doi: 10.1002/gcc.22154 [DOI] [PubMed] [Google Scholar]
- 35.Hirata H, Hinoda Y, Shahryari V, Deng G, Tanaka Y, et al. : Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer 110, 1645–54, 2014. doi: 10.1038/bjc.2014.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colacino JA, Arthur AE, Dolinoy DC, Sartor MA, Duffy SA, et al. : Pretreatment dietary intake is associated with tumor suppressor DNA methylation in head and neck squamous cell carcinomas. Epigenetics 7, 883–91, 2012. doi: 10.4161/epi.21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett KL, Lee W, Lamarre E, Zhang X, Seth R, et al. : HPV status-independent association of alcohol and tobacco exposure or prior radiation therapy with promoter methylation of FUSSEL18, EBF3, IRX1, and SEPT9, but not SLC5A8, in head and neck squamous cell carcinomas. Genes Chromosomes Cancer 49, 319–26, 2010. doi: 10.1002/gcc.20742 [DOI] [PubMed] [Google Scholar]
- 38.Choudhury JH, Ghosh SK: Promoter Hypermethylation Profiling Identifies Subtypes of Head and Neck Cancer with Distinct Viral, Environmental, Genetic and Survival Characteristics. PLoS One 10, e0129808, 2015. doi: 10.1371/journal.pone.0129808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warta R, Herold-Mende C, Chaisaingmongkol J, Popanda O, Mock A, et al. : Reduced promoter methylation and increased expression of CSPG4 negatively influences survival of HNSCC patients. Int J Cancer 135, 2727–34, 2014. doi: 10.1002/ijc.28906 [DOI] [PubMed] [Google Scholar]
- 40.Jung AC, Job S, Ledrappier S, Macabre C, Abecassis J, et al. : A poor prognosis subtype of HNSCC is consistently observed across methylome, transcriptome, and miRNome analysis. Clin Cancer Res 19, 4174–84, 2013. doi: 10.1158/1078-0432.CCR-12-3690 [DOI] [PubMed] [Google Scholar]
- 41.Virani S, Light E, Peterson LA, Sartor MA, Taylor JM, et al. : Stability of methylation markers in head and neck squamous cell carcinoma. Head Neck 38 Suppl 1, E1325–31, 2016. doi: 10.1002/hed.24223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, et al. : A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32, e38, 2004. doi: 10.1093/nar/gnh032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, et al. : Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One 4, e4941, 2009. doi: 10.1371/journal.pone.0004941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez JG, Perez-Escuredo J, Castro-Santos P, Marcos CA, Pendas JL, et al. : Hypomethylation of LINE-1, and not centromeric SAT-alpha, is associated with centromeric instability in head and neck squamous cell carcinoma. Cell Oncol (Dordr) 35, 259–67, 2012. doi: 10.1007/s13402-012-0085-5 [DOI] [PubMed] [Google Scholar]
- 45.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, et al. : Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 16, 108–14, 2007. doi: 10.1158/1055-9965.EPI-06-0636 [DOI] [PubMed] [Google Scholar]
- 46.Roh JL, Wang XV, Manola J, Sidransky D, Forastiere AA, et al. : Clinical correlates of promoter hypermethylation of four target genes in head and neck cancer: a cooperative group correlative study. Clin Cancer Res 19, 2528–40, 2013. doi: 10.1158/1078-0432.CCR-12-3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilir B, Sharma NV, Lee J, Hammarstrom B, Svindland A, et al. : Effects of genistein supplementation on genomewide DNA methylation and gene expression in patients with localized prostate cancer. Int J Oncol 51, 223–234, 2017. doi: 10.3892/ijo.2017.4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, et al. : Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res 62, 2370–7, 2002 [PubMed] [Google Scholar]
- 49.Jones PA, Baylin SB: The fundamental role of epigenetic events in cancer. Nature Reviews Genetics 3, 415, 2002. doi: 10.1038/nrg816 [DOI] [PubMed] [Google Scholar]
- 50.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, et al. : Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene 23, 8841–6, 2004. doi: 10.1038/sj.onc.1208137 [DOI] [PubMed] [Google Scholar]
- 51.Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, et al. : Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 67, 876–80, 2007. doi: 10.1158/0008-5472.CAN-06-2995 [DOI] [PubMed] [Google Scholar]
- 52.Xing C, Wang QF, Li B, Tian H, Ni Y, et al. : Methylation and expression analysis of tumor suppressor genes p15 and p16 in benzene poisoning. Chem Biol Interact 184, 306–9, 2010. doi: 10.1016/j.cbi.2009.12.028 [DOI] [PubMed] [Google Scholar]
- 53.Kim KY, Kim DS, Lee SK, Lee IK, Kang JH, et al. : Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect 118, 370–4, 2010. doi: 10.1289/ehp.0901131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, et al. : Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect 116, 1547–52, 2008. doi: 10.1289/ehp.11338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, et al. : Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 118, 790–5, 2010. doi: 10.1289/ehp.0901429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, et al. : Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr 141, 1165–71, 2011. doi: 10.3945/jn.110.134536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hillemacher T, Frieling H, Moskau S, Muschler MA, Semmler A, et al. : Global DNA methylation is influenced by smoking behaviour. Eur Neuropsychopharmacol 18, 295–8, 2008. doi: 10.1016/j.euroneuro.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 58.Pramio DT, Pennacchi PC, Maria-Engler SS, Campos AH, Duprat JP, et al. : LINE-1 hypomethylation and mutational status in cutaneous melanomas. J Investig Med 64, 899–904, 2016. doi: 10.1136/jim-2016-000066 [DOI] [PubMed] [Google Scholar]
- 59.Mima K, Nowak JA, Qian ZR, Cao Y, Song M, et al. : Tumor LINE-1 methylation level and colorectal cancer location in relation to patient survival. Oncotarget 7, 55098–55109, 2016. doi: 10.18632/oncotarget.10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Block KI, Gyllenhaal C, Lowe L, Amedei A, Amin A, et al. : Designing a broad-spectrum integrative approach for cancer prevention and treatment. Semin Cancer Biol 35 Suppl, S276–S304, 2015. doi: 10.1016/j.semcancer.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delgado-Cruzata L, Zhang W, McDonald JA, Tsai WY, Valdovinos C, et al. : Dietary modifications, weight loss, and changes in metabolic markers affect global DNA methylation in Hispanic, African American, and Afro-Caribbean breast cancer survivors. J Nutr 145, 783–90, 2015. doi: 10.3945/jn.114.202853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eden A, Gaudet F, Waghmare A, Jaenisch R: Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300, 455, 2003. doi: 10.1126/science.1083557 [DOI] [PubMed] [Google Scholar]