Abstract
Inhibitors of the bromodomain and extra-terminal domain family (BETi) offer a new approach to treating hematological malignancies, with leukemias containing MLL rearrangements being especially sensitive due to a reliance on the regulation of transcription elongation. We explored the mechanism of action of BETi in cells expressing the t(8;21), and show that these compounds reduced the size of AML cells, triggered a rapid but reversible G0/G1 arrest, and with time, cause cell death. Meta-analysis of PRO-seq data identified ribosomal genes, which are regulated by MYC, were down-regulated within 3 hr of addition of the BET inhibitor. This reduction of MYC regulated metabolic genes coincided with the loss of mitochondrial respiration and large reductions in the glycolytic rate. In addition, gene expression analysis showed that transcription of BCL2 was rapidly affected by BETi, but this did not cause dramatic increases in cell death. Cell cycle arrest, lowered metabolic activity, and reduced BCL2 levels suggested that a second compound was needed to push these cells over the apoptotic threshold. Indeed, low doses of the BCL2 inhibitor, venetoclax, in combination with the BETi was a potent combination in t(8;21) containing cells. Thus, BET inhibitors that affect MYC and BCL2 expression should be considered for combination therapy with venetoclax.
Introduction
The bromodomain and extra-terminal domain (BET) proteins consist of 4 family members including BRD2, BRD3, BRD4, and BRDT1. These BET proteins bind to acetylated lysine in the tails of histones as well as other non-histone nuclear proteins through two conserved N-terminal bromodomains1–4. BET proteins are typically associated with enhancers and associate with P-TEFb to regulate the transition from paused to elongating polymerase5–8. In fact, the binding of BRD4 appears to release P-TEFb from the inhibitory HEXIM1–7SK complex6,9,10. Small molecule inhibitors of BET proteins, such as JQ1, I-BET, and MS417 mimic the acetylated lysine moiety and competitively bind to the two bromodomains (BD1 and BD2) to displace BET proteins from chromatin11–13. JQ1 was originally identified as a tool compound to target midline carcinoma expressing a fusion protein caused by a chromosomal translocation between BRD4 and nuclear protein in testis (NUT)11. However, BET inhibitors (BETi) also show efficacy in preclinical models of acute myeloid leukemia (AML), multiple myeloma, and certain types of lymphoma as well as other cancer types8,11,14–19. RNAi screening linked the inhibitory effect of these molecules to suppressing BRD4 activity in AML15.
Gene expression studies showed that BETi induced down-regulation of mRNAs including key oncogenes important for cell cycle progression, such as MYC and E2F1, genes that control cell death such as BCL2, as well as lineage-specific oncogenes such as BCL68,11,14–16. Genomic binding studies revealed that these genes are associated with BRD4-enriched enhancers that are essential for the efficient transcription of these genes. The so-called “super-enhancers”, defined as clusters of sub-enhancers, are particularly sensitive to BETi, which rapidly displace BRD4 from chromatin causing the selective transcriptional repression of those super-enhancer-driven genes. However, as a global chromatin reader, BRD4 is also found at active promoters and typical enhancers. Moreover, inducible degradation of BRD4 indicated that it was a general factor that is required for the elongation of most expressed genes20.
AML containing chromosomal translocations involving MLL appear to be especially sensitive to BETi15, perhaps due to the translocations that fuse the N-terminal domain of MLL with components of the super elongation complex to stimulate the expression of key regulators of hematopoietic stem cell self-renewal such as Hox family members21. Intriguingly, in these early studies the t(8;21) cell line Kasumi-1 showed the most pronounced sensitivity15. The t(8,21) is one of the most common chromosomal translocations in AML and fuses the N-terminal DNA binding domain of RUNX1 to almost the entire ETO gene. While RUNX1 associates with multiple DNA binding proteins and/or histone modifying complexes to activate or repress transcription, the presence of the ETO moiety skews the activity of the fusion protein towards repression of RUNX1-regulated genes22–25. That is, ETO can associate with histone acetyltransferases26, but recruits class 1 histone deacetylases27, and global studies of t(8;21) cells suggest that this latter effect is the predominant mechanism of action28. However, ETO family members also associate with the “E proteins” HEB and Lyl1 in a complex containing LDB1, LMO2 and CDK929,30, leaving open the possibility that ETO family members regulate transcriptional elongation, which might explain the sensitivity of Kasumi-1 cells to BETi. By using precision nuclear run-on sequencing (PRO-seq), we previously identified many of the earliest targets of BET inhibitor action and demonstrated that these compounds cause transcriptional pausing of drivers of the cell cycle and metabolic activity, and affect eRNA synthesis in the MYC super-enhancer31. Here, we extended our study by showing that t(8;21) AML are not only highly sensitive to BETi, but treatment with JQ1 or MS417 dramatically reduced cell size and induced cell cycle arrest rather than cell death. Cell cycle analyses and assessment of mitochondrial function and glycolytic activity indicated that the BETi-induced cell cycle arrest was reversible. However, the metabolic stress and impaired transcription of BCL2 after JQ1 treatment provides further molecular rationale for combination therapy using BETi and venetoclax.
Materials and Methods
Cell proliferation analysis.
Cells were seeded at 2×105 cells/ml on the day of the experiment, and treated with increasing doses of JQ1, MS417 or venetoclax for 3 consecutive days. Cell proliferation rate was measured on each day using alamarBlue assay (Life Technologies, Inc.) according to the manufacturer’s instructions. Briefly, 100 μl of cell suspension was transferred to a 96-well plate before 10 μl of alamarBlue was added. Plates were incubated at 37 °C for 4 hours, and alamarBlue fluorescence was measured at 590 nm. Viable cells were also quantified by Trypan Blue exclusion. Human primary AML cells were seeded at 5×105 cells/ml in a 96-well plate and treated with 250 nM JQ1 for 3 days. On day 3, alamarBlue was added and cells were incubated for 8 hours before reading.
Assessment for cell size, cell cycle progression, and apoptosis.
The distribution of cell size was analyzed by flow cytometry and presented by forward-scatter histogram plots. For cell cycle analyses, cells were treated with JQ1 or MS417 for 24, 48, and 72 hours, fixed with 70% ethanol in the cold room for overnight, and stained with propidium iodide at room temperature for 30 minutes before flow cytometry. For BrdU analysis, 10 million cells were pulsed with 20 μM BrdU for 1 hour and fixed with 70% ethanol in the cold room for overnight. Cells were stained with FITC-conjugated BrdU antibody (Cat #556028, BD Pharmingen) and counterstained with propidium iodide before analysis by flow cytometry. Apoptosis was analyzed using a FITC-AnnexinV/PI Apoptosis Detection kit (cat # 556547, BD Pharmingen). All flow cytometry figures were generated using Flowjo software.
RNA-seq.
PolyA+ RNA was enriched for library preparation and submitted to the Vanderbilt DNA sequencing facility (VANTAGE) for library preparation and RNA sequencing. Pre-processed reads were aligned to the human transcriptome hg19 (downloaded from UCSC) using Bowtie2 (version 2.2.4)32 and TopHat (version v2.0.10)33. Data will be deposited in the NCBI Sequence Read Archive (SRA) upon acceptance.
Oxygen consumption rate and extracellular acidification rate measurements.
Cells were seeded on Cell-Tak adhesive coated Seahorse XF96 Cell Culture Microplates to prepare adherent monolayer cultures. Seahorse XFe96 Extracellular flux analyzer (Agilent, Inc.) was utilized to determine the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) by measuring the concentration of oxygen and free protons in the medium surrounding the monolayer of cells in real-time. The XF Cell Mito Stress Test Kit (Agilent, Inc.) was used to access the mitochondrial function after serially injecting oligomycin, FCCP and a mix of rotenone and antimycin A to measure ATP production, maximal respiration, and non-mitochondrial respiration, respectively. The XF Glycolysis Stress Test Kit (Agilent, Inc.) was used to access the glycolysis capacity after three sequential injections of glucose, oligomycin and 2-deoxy-glucose (2-DG).
Optical Metabolic Imaging.
Multi-photon fluorescence lifetime imaging was performed on a Nikon Ti:E inverted microscope equipped with time-correlated single photon counting electronics. NAD(P)H and FAD fluorescence were excited with a titanium:sapphire laser (Chameleon Ultra II, Coherent, Inc.) tuned to 750 nm and 890 nm, respectively. A 400–480 nm bandpass filter was used to isolate NAD(P)H fluorescence emission, and a 500 nm high pass dichroic mirror and 500–600 nm bandpass filter were used to isolate FAD fluorescence emission. Fluorescence signal was collected through a 40× oil-immersion objective across a 170 × 170 μm field of view, capturing approximately 40–60 cells. Four fields of view per sample were acquired, yielding 125–250 cells for each of three biological replicates. Images were processed with custom software previously described34. Briefly, the NAD(P)H and FAD intensities were calculated by integrating the fluorescence lifetime decay within each pixel. Then, the ratio of NAD(P)H intensity divided by FAD intensity (the optical redox ratio) was calculated at each pixel. Pixels within the cell cytoplasm were averaged and statistical differences were calculated on a per-cell level.
Statistical analyses.
For cell proliferation, apoptosis, and quantitative RT-PCR analyses, results were presented as mean ± SEM for data following normal distribution. Unpaired two-sided Student’s t-test was used to compare the means between two independent treatments. For data that did not follow a normal distribution such as read density and pausing index analyses, Wilcoxon signed-rank test was used for paired comparisons between two treatments. P values < 0.05 were considered as statistically significant. Optical redox ratio was reported as mean ± SEM for each condition. Mann-Whitney tests were used to compare optical redox ratio differences between control and treated samples.
Results
BET inhibitors affect Kasumi-1 cell proliferation without inducing apoptosis
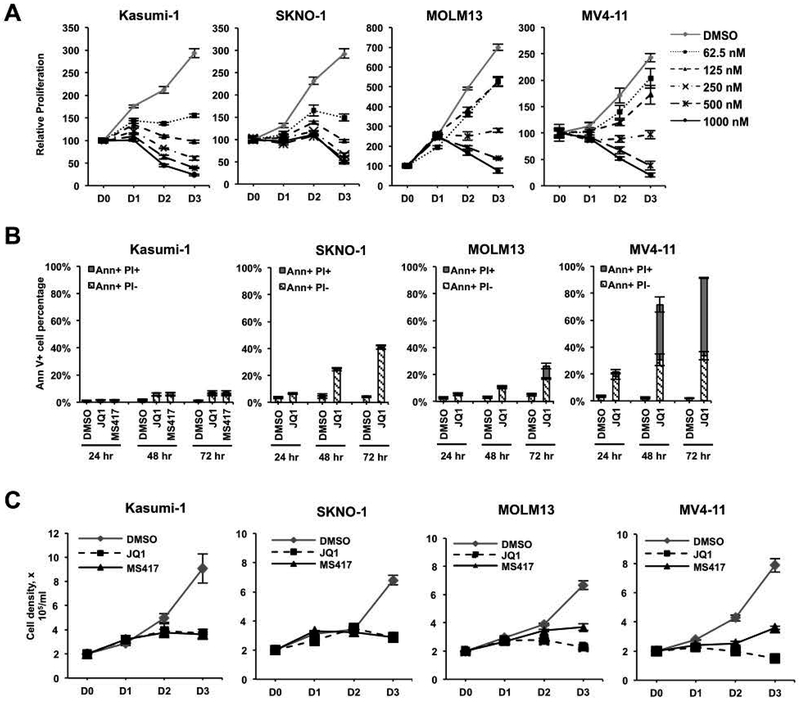
High throughput screens of cancer cell lines identified the t(8;21) cell line Kasumi-1 as the most sensitive cell type to the BET inhibitor (BETi) JQ1 using alamarBlue assays as a surrogate for cell proliferation15. Consistent with these previous studies, we found that Kasumi-1 cells, as well as the t(8;21)-containing SKNO-1 cell line that requires GM-CSF for growth35, were more sensitive than MOLM13, and MV4–11 that contain MLL disruptions (Figure 1A) when using alamarBlue assays. However, alamarBlue measures metabolic output, rather than cell death. When we tested for cell death using JQ1 and another potent BET inhibitor, MS41736, which shares molecular and transcription targets with JQ131, both JQ1 and MS417 only had minor effects on triggering apoptosis in Kasumi-1 cells, whereas SKNO-1 showed more cell death at 48 hr (Figure 1B). In addition, while MOLM13 and MV4–11 cells required higher concentrations of BETi to show an effect in alamarBlue assays, MOLM13 cells showed minimal apoptosis, while BETi more robustly induced apoptosis in MV4–11 (Figure 1B). Consistent with these observations and in contrast to the alamarBlue data, manual cell viability counts using Trypan Blue dye exclusion showed JQ1 and MS417 had a cytostatic rather than cytotoxic effect on cell growth in Kasumi-1 cells, but reduced the number of viable cells in cultures of MV4–11 cells (Figure 1C).
Figure 1. BETi inhibit proliferation, but induce variable levels of cell death in AML cell lines.
(A) AlamarBlue assays show a dose-dependent loss of cellular metabolism after JQ1 treatment of Kasumi-1, SKNO-1, MOLM13, and MV4–11 cells for 3 days (D0-D3). Data are mean ± SEM (n=3). (B) BETi induce variable levels of apoptosis. Kasumi-1 cells were treated with 250 nM JQ1 or 125 nM MS417. SKNO-1 were treated with 250 nM JQ1. MOLM13 and MV4–11 were treated with 500 nM JQ1. The levels of dying cells were quantified using Annexin V positivity (Ann+) and uptake of propidium iodide (PI). Data are mean ± SEM (n=4). (C) SKNO and Kasumi-1 cells were treated with 250 nM JQ1 or 125 nM MS417, whereas MOLM13 and MV4–11 were treated with twice these levels. Cell counts were determined by Trypan Blue dye exclusion. Data are mean ± SEM (n=4).
BETi disrupt the communication between enhancers and RNA polymerases that are paused just after transcriptional initiation. Inhibitors of pTEFb (that is, CDK9 inhibitors), such as flavopiridol, also block RNA polymerase elongation and cause DNA damage. However, we did not detect any phosphorylated H2AX in the nuclei of JQ1 or MS417 treated cells (Supplemental Figure 1). This indicated that BETi inhibited cell proliferation without DNA double strand breaks and without causing apoptosis in Kasumi-1 cells. Moreover, these compounds did not trigger a DNA damage-dependent cell cycle arrest or apoptosis of Kasumi-1 cells. These results raised the possibility that BETi directly or indirectly affected the metabolic rate as measured by alamarBlue.
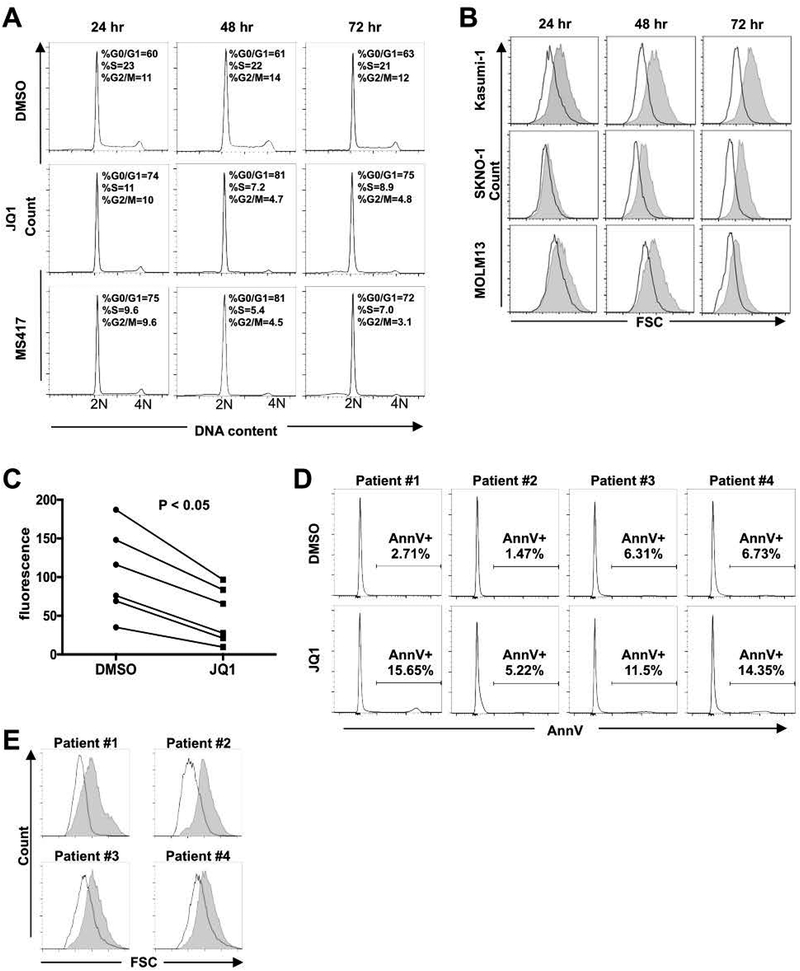
BET inhibitors reduce cell size
We extended this analysis by using propidium iodide (PI) staining of DNA, which showed that Kasumi-1 cells treated with JQ1 and MS417 rapidly accumulated in the G0/G1 phase of the cell cycle (Figure 2A). Interestingly, we noted that treatment with BETi led to dramatic reductions in cell size in Kasumi-1 and SKNO-1cells within 24–48 hr of treatment with JQ1, as assessed by diminished forward scatter in flow cytometry analysis (Figure 2B). JQ1 also reduced cell size in MOLM13 cells but the response was delayed (Figure 2B) relative to Kasumi-1 cells. While there were no apparent morphological changes toward myeloid differentiation, Wright-Giemsa staining confirmed the smaller cell size and condensed nuclei of Kasumi-1 cells (Supplemental Figure 2). Given the apparent sensitivity of t(8;21) cell lines, we extended this analysis to primary AML samples containing this translocation. Again, JQ1 inhibited cell growth as measured by alamarBlue staining without large increases in Annexin V staining. These primary AML cells also showed a reduction in cell size (Figure 2C–E).
Figure 2. BETi induce cell cycle arrest and reduce cell size in t(8;21) AML cells.
(A) Cell cycle analyses of Kasumi-1 cells treated with BETi show cell cycle arrest with modest cell death 24–72 hr after treatment. Representative graphs of DNA content (2N to 4N) are shown (n=4). (B) Flow cytometry analyses showing forward scatter plots indicate that t(8;21) cells are distinctly smaller after treatment with 250 nM JQ1. Representative graphs are shown (n=4). Shaded area represents DMSO and white plots represent JQ1. (C and D) High blast count t(8;21) AML patient samples (n=6) were treated with 250 nM JQ1 for 3 days. AlamarBlue assays show the inhibition of cell growth p < 0.05 by two-sided Wilcoxon signed-rank test (C). (D) Shows the lack of Annexin V positive cells for four of the samples in C. (E) Forward scatter plots of flow cytometry analyses show that t(8;21) AML patient cells are distinctly smaller after JQ1 treatment for 3 days. Representative flow cytometry plots are shown. Shaded area represents DMSO and empty area represents JQ1.
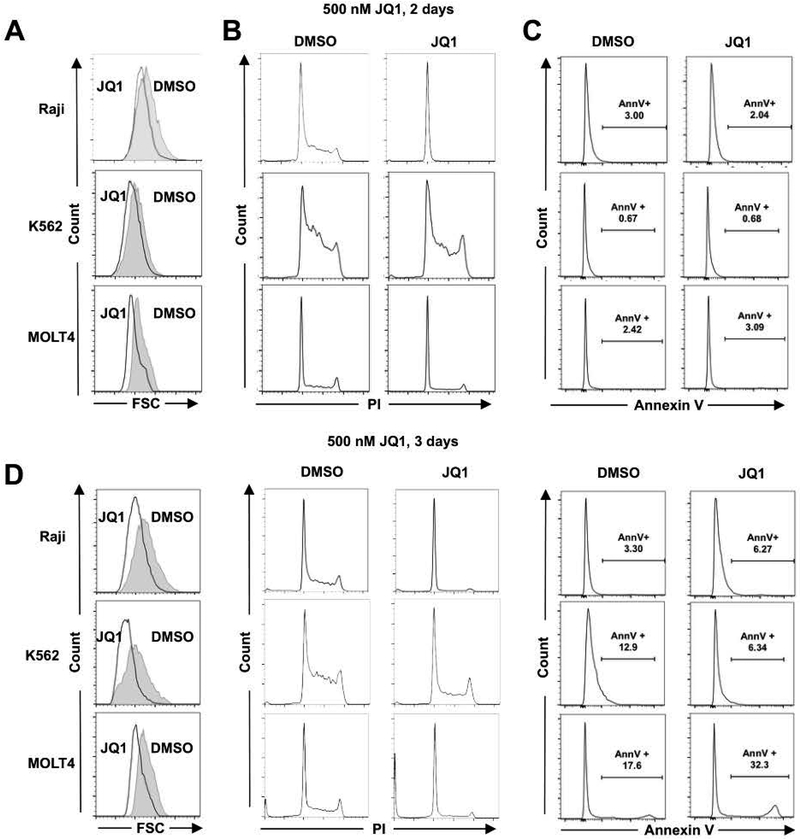
We further extended these observations to other leukemia/lymphoma cell lines, including Raji, K562, and MOLT4. JQ1 treatment reduced cell size in all cell lines tested (Figure 3A and D). This reduction in cell size was associated with robust cell cycle arrest in all cell lines while the annexin V positive population was not greatly increased (Figure 3B–C) in the first 48 hr. Taken together, our data suggested that BET proteins are required for maintaining cell metabolism and cell size if the cells do not undergo rapid cell death.
Figure 3. BETi reduces cell size in leukemia and lymphoma cell lines.
(A) Cell size was assessed using forward light scatter in flow cytometry. (B) Cell cycle progression was assessed using propidium iodide staining. Representative graphs are shown. (C) Apoptosis was assessed using Annexin V (AnnV+) on viable cells. (D) Similar analyses as in (A-C), but performed at day 3 with the indicated cell lines. Statistical analysis was performed as described in Figure 2.
BET inhibitors reduce metabolic rate
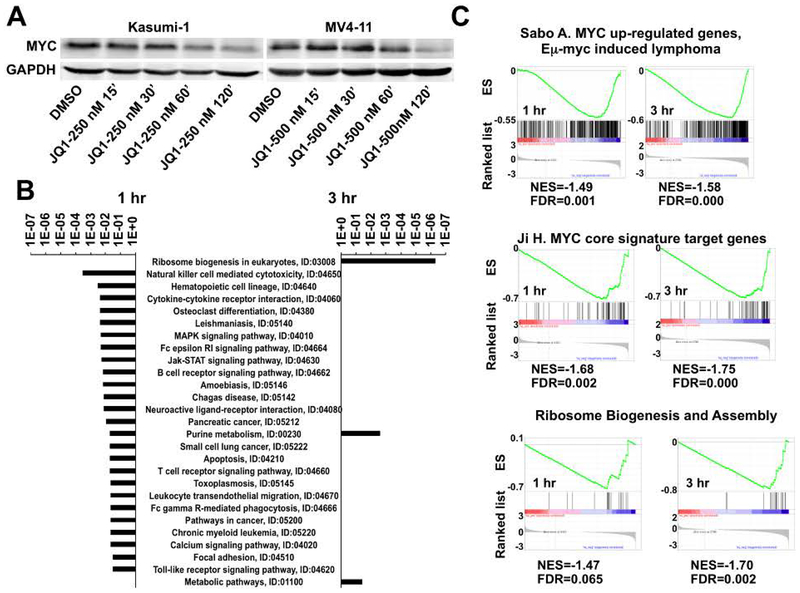
The reduction in cell size is a classical phenotype associated with the loss of MYC expression and we observed a rapid loss of MYC transcription in Kasumi-1 cells treated with JQ131. Western blot analysis confirmed that MYC levels dropped within two hr in both Kasumi-1 cells and MV4–11 cells (Figure 4A). Therefore, we used pathway analysis tools to determine whether BET inhibitors affect the expression of MYC targets and metabolic genes using our previously published precision nuclear run-on sequencing (PRO-seq) data that were generated in Kasumi-1 cells treated with JQ1 or MS417 for 1 and 3 hrs31. PRO-seq provides a high-resolution map of the actively elongating RNA polymerases, which pinpoint the genes that are directly affected by small molecule inhibitors of transcription. Genes that were inhibited by both JQ1 and MS417 (GFOLD < −0.585) at 1 hr were enriched with signaling pathways mediated by cytokines and/or growth factors regulating cell proliferation that drive the accumulation of cell mass with progression through the cell cycle (Figure 4B). Genes inhibited at 3 hr were enriched in metabolic pathways and ribosome biogenesis (Figure 4B), which is consistent with reduced metabolism and cell size (Figure 1–3). MYC controls many of these metabolism-regulating genes and ribosomal genes to control cell size37–41. Gene set enrichment analysis (GSEA) supported this idea and showed that genes inhibited by BET inhibitors at both 1 and 3 hr were enriched with previously identified MYC target genes including genes regulating ribosomal biogenesis (Figure 4C)42–44. This suggests that BET inhibitors decrease the expression of genes regulating cell metabolism and cell size by impairing the transcription of MYC.
Figure 4. Meta-analysis of PRO-seq data of Kasumi-1 cells treated with JQ1 for 1 or 3 hr.
(A) MYC levels drop rapidly upon BETi treatment. Western blot analysis of Kasumi-1 (left panel) and MV4–11 (right panel) cells treated with 250 or 500 nM JQ1 for the indicated times. (B) KEGG pathway analysis of PRO-seq data31 showing genes in which promoter-proximal pausing was increased after 1 hr (left) and 3 hr (right) treatments. (C) Gene set enrichment analysis of PRO-seq data31 showed decreased expression of MYC target genes including those genes regulating ribosomal biogenesis.
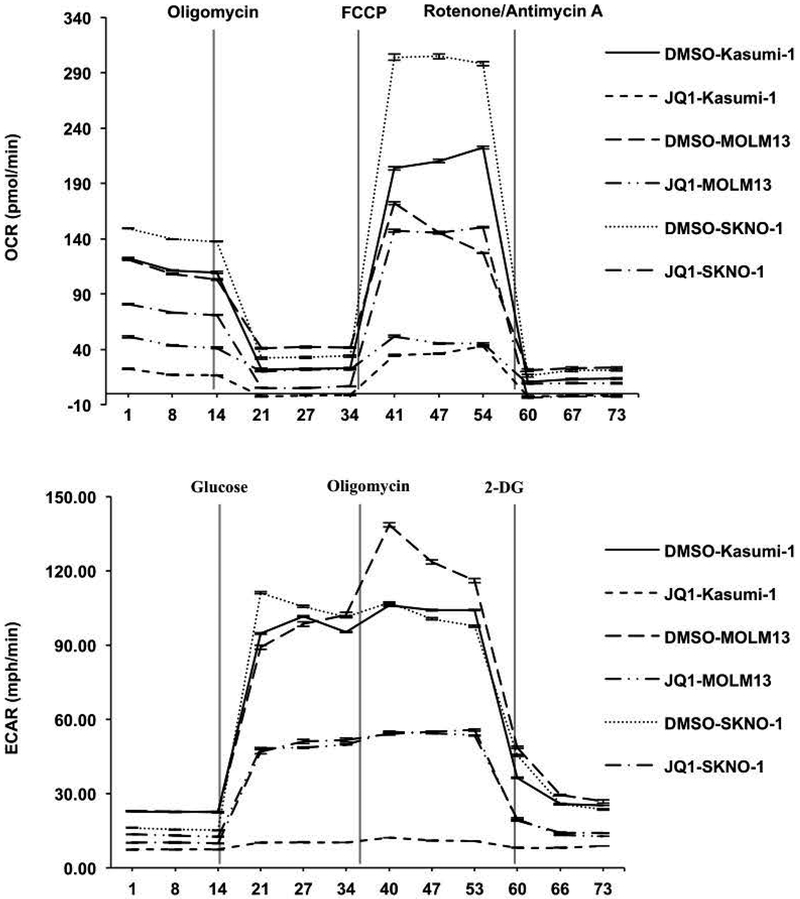
The role of BET inhibitors in reducing metabolism was further tested by directly examining bioenergetics in Kasumi-1, SKNO-1, and MOLM13 live cells by assessing the oxygen consumption rate (OCR) and extracellular acidification rates (ECAR) to track mitochondrial respiration and glycolysis using the Agilent Seahorse system (Figure 5). Cells were cultured in the absence or presence of BETi for 48 hr, and then assessed for 20 min to establish the basal respiration rates prior to injection of oligomycin to disrupt oxygen consumption and measure ATP production (Figure 5, top panel). BETi pretreatment reduced the basal respiration rate to essentially the level of control cells treated with oligomycin. Next, p-trifluoromethoxy carbonyl cyanide phenyl hydrazine (FCCP) was injected to uncouple the respiratory chain from phosphorylation and assess the maximal respiration rates. BETi treated cells showed only a marginal response compared to a robust response from the untreated control cells. Finally, Rotenone and Antimycin A were injected to block respiration (Figure 5, top panel). Overall, JQ1 dramatically impaired mitochondrial functions.
Figure 5. BET inhibitors reduce metabolic rate.
(A) Oxygen consumption rate (OCR) and extracellular acidification rates (ECAR) were measured after JQ1 treatment for 2 days in Kasumi-1 cells, MOLM13 cells, and SKNO-1 (lower panels) cells. Decreased OCR and ECAR indicate repression of both mitochondrial dynamics and glycolytic function. For OCR, stage I injected oligomycin (5 mM), stage II injected FCCP (1 mM), and stage III injected Rotenone/Antimycin A (0.5 mM). For ECAR, stage I injected glucose (10 mM), stage II injected oligomycin (5 mM), and stage III injected 2-Deoxyglucose (50 mM).
Cancer cells reprogram their metabolic pathways and are more dependent on glycolysis as an energy source. Therefore, we assessed glycolytic activity in the absence or presence of JQ1 (Figure 5, bottom panels). Cells were again treated with JQ1 for 48 hr prior to a 15 min assessment of the basal non-glycolytic media acidification. Glucose was then injected to assess glycolysis followed by an injection of oligomycin 20 min later to impair ATP production. Untreated control cells showed a rapid burst of glycolytic activity upon addition of glucose, and oligomycin triggered a further increase in acidification indicating that control cells had a further glycolytic reserve (Figure 5, bottom panel). In contrast, JQ1-treated AML cells showed a poor glycolytic burst with no further increase in glycolytic capacity upon oligomycin injection (Figure 5, bottom panel).
BET inhibitor-induced cell cycle arrest is reversible
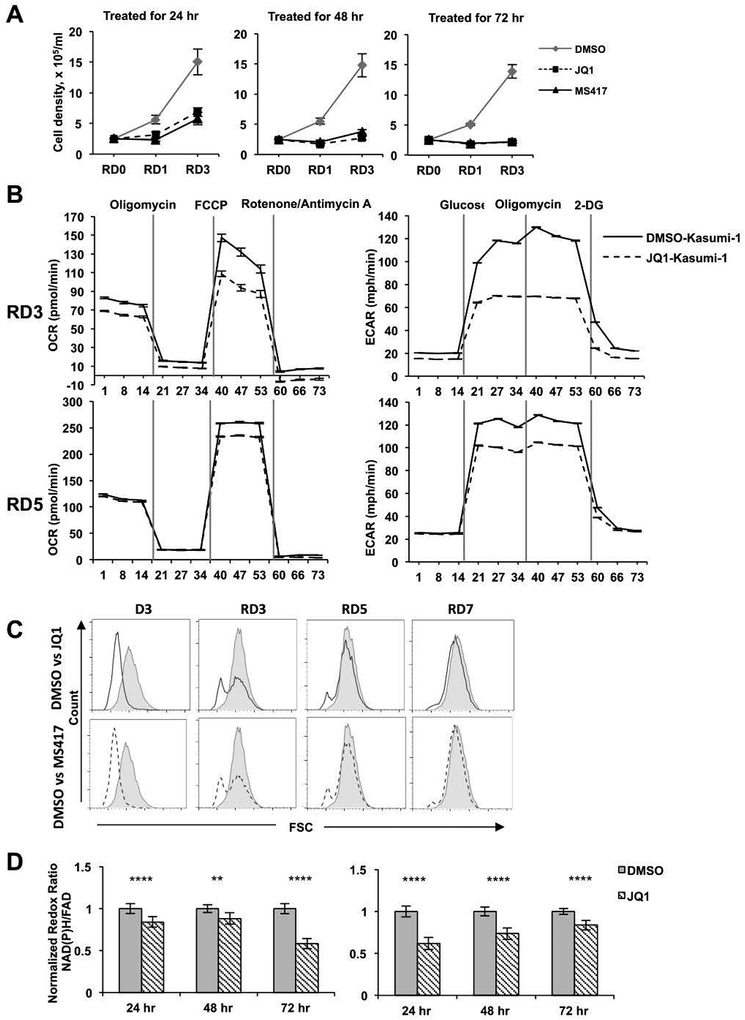
The rapid onset of cell cycle arrest without apoptosis for the first 24–48 hr after continuous drug treatment suggested that even with daily dosing in patients the trough levels might allow survival of some AML cells45. To assess whether JQ1-treated cells still possessed proliferative potential after drug removal, which could ultimately cause leukemic cell repopulation and resistance to BET inhibitor therapy, we treated Kasumi-1 cells for 24, 48, or 72 hr, washed out the drug, and then cultured them in fresh media and monitored cell growth. Consistent with these drugs inducing G0/G1 cell cycle arrest, but not senescence or cell death, Kasumi-1 cells treated for 24 or 48 hr were rescued to some degree by removing the drug (Figure 6A, left and middle panels). However, longer treatments had a stronger effect on restricting cell proliferation, as the cells treated for 3 days grew much slower during the first 3 days after drug removal (Figure 6A, right panel). The cells treated for 48 hr prior to rescue became more metabolically active three or five days after drug removal (RD3 and RD5; Figure 6B) and returned to normal size within 3 days after being transferred to fresh media (Figure 6C), and eventually began growing normally showing a rapid recovery of cell size and metabolism.
Figure 6. Metabolic effects of BETi are reversible.
(A) Kasumi-1 cells were treated with BETi for 24, 48 or 72 hr and the cells allowed to recover for 0, 1 or 3 days. Viable cell numbers were graphed over time. (B) Oxygen consumption rate (OCR) and extracellular acidification rates (ECAR) were measured after treatment of Kasumi-1 cells for 2 days and allowed to recover in the absence of JQ1 for 3 or 5 days. For OCR, stage I injected Oligomycin (5 mM), stage II injected FCCP (1 mM), and stage III injected Rotenone/Antimycin A (0.5 mM). For ECAR, stage I injected Glucose (10 mM), stage II injected Oligomycin (5 mM), and stage III injected 2-Deoxyglucose (50 mM) (C) Kasumi-1 cells treated with BETi for 3 days (D3) and then allowed to recover for 3, 5 or 7 days without BETi were tested for cell size using forward scatter in FACS. (D) Normalized optical redox ratio (NAD(P)H/FAD) was assessed using optical metabolic imaging. A minimum of 125 cells per sample in biological triplicates were measured for DMSO- and JQ1-treated Kasumi-1 cells at 24–72 hours before washout (left panel) or for the 24–72 hours after washout (recovery) of JQ1 (right panel). **, p<0.01; ****, p<0.0001; Mann-Whitney test.
Next, we performed optical metabolic imaging using two-photon fluorescence imaging to measure the optical redox ratio of Kasumi-1 cells in response to JQ1 treatment and rescue. This approach captures intrinsic fluorescence of these co-enzymes within live cells, and the optical redox ratio (NAD(P)H fluorescence intensity/FAD fluorescence intensity) informs on the relative oxidation or reduction state of the cells. This measurement distinguishes between apoptotic, proliferating, and G0/G1 cells34. JQ1 treated cells displayed a reduced redox ratio beginning at 24 hr after treatment, which was exacerbated by 72 hr (Figure 6D, left panel). Changes in redox ratio were reversed in response to recovering cells from JQ1 treatment, indicating negligible effects of apoptosis in this analysis (Figure 6D, right panel). Together, the combined effects of BET inhibitors on cell cycle arrest, cell size, and reduced metabolic rate suggested that the cells were not only being arrested in G1, but were perhaps entering a G0, quiescent-like, state.
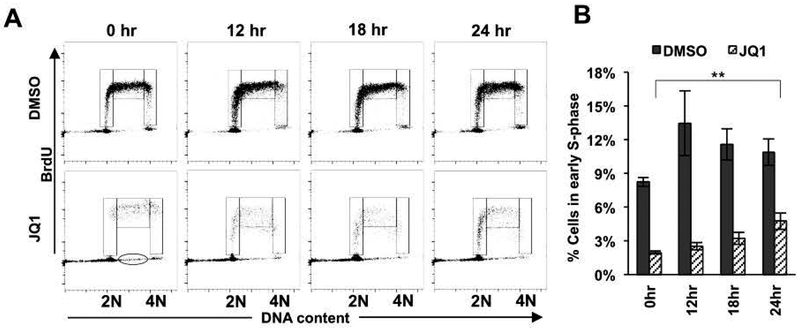
Finally, we arrested cells with JQ1 for 72 hr (a time where the cells were beginning to die, Figure 1 and 2), washed the cells to remove the compound, and cultured them in fresh media lacking JQ1 for 12–24 hr before quantifying the number of cells entering S-phase using BrdU incorporation. By 12 hr after drug removal, cells were beginning to re-enter early S phase and this trend continued through 24 hr (Figure 7A, JQ1 panels; and 7B, hashed bars). Intriguingly, it appears that some of the S phase-arrested cells began incorporating BrdU. Notably, cells already synthesizing DNA do not incorporate as much BrdU leading to a broadening of the band of cells in the 12, 18 and 24 hr samples (Figure 7A, JQ1 panels). Conversely, some cells with between 2N and 4N DNA content did not incorporate BrdU (Figure 7A, oval in the 0 hr panel), suggesting that these cells were actually arrested in the S phase or were dying. Nevertheless, even after a 72 hr treatment with JQ1, there were cells that appeared to be quiescent, but still capable of re-entering the cell cycle after drug removal, indicating a potential mechanism of resistance to BETi.
Figure 7. BETi induce reversible cell cycle arrest in Kasumi-1 cells.
(A) Flow cytometry plots of incorporated BrdU versus propidium iodide show that Kasumi-1 cells treated with 250 nM JQ1 for 72 hr can recover and re-enter the cell cycle after drug removal for the indicated times. Cells were gated as early, middle, and late S phases from left to right. Oval indicates BrdU− cells in the S phase. (B) Bar graph displays the percentage of cells in the early S phase. Data are presented as mean ± SEM (n=4). ** P < 0.001 by two-sided Student’s T-test when 0 hr and 24 hr levels were compared.
BET inhibitors sensitize cells to BCL2 inhibitors
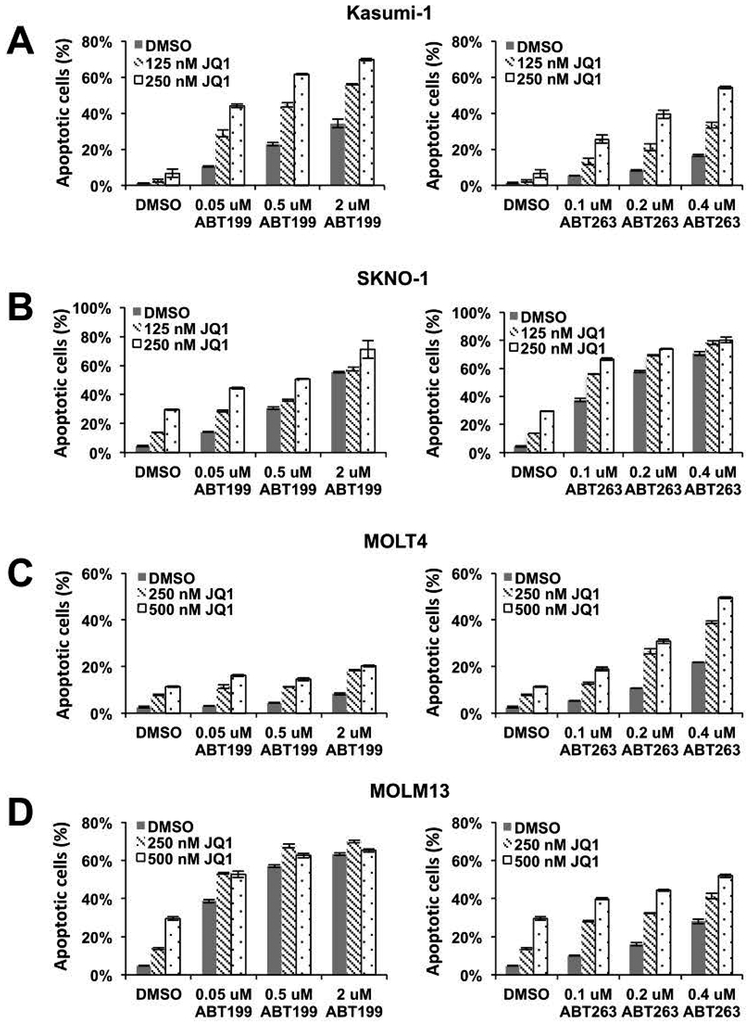
The finding that BET inhibitors can arrest the cell cycle without dramatic AML cell killing suggested the need for combination therapy. The anti-apoptotic protein BCL2 is often expressed in leukemia cells and our earlier study showed that BET inhibitors caused promoter-proximal pausing of RNA polymerase II and reduced the rate of transcription of BCL2 and BCL-xL in Kasumi-1 cells31. Therefore, we tested the combination of BET inhibitor and BCL2 inhibitors to target the residual BCL2. We pre-treated Kasumi-1 cells with JQ1 for 2 days to downregulate expression of BCL2 and BCL-xL and found that the addition of BCL2/BCL-xL (ABT-263) or BCL2-selective (venetoclax; ABT-199) inhibitors quickly triggered apoptosis as measured by Annexin V and PI staining (Figure 8A). We next extended this co-treatment analysis to other cell lines (SKNO-1, MOLM13, and MOLT4) that also showed cell cycle arrest with little apoptosis (Figure 1–3) and found that inhibiting BCL2 family proteins following BET inhibitor pre-treatment also induced cell death (Figure 8B–D). This suggests that entering a reversible cell cycle arrest induced by BET inhibitors may protect leukemia cells from rapid cell death and increase the chance of relapse, which necessitates a second drug (e.g. BCL2 inhibitor) to induce cell death for efficient killing of leukemia cells.
Figure 8. BET inhibitors sensitize AML cells to BCL2 inhibitor-induced cell death.
AML cell lines were treated with BCL2 selective (ABT199) or BCL2/BCLxL (ABT263) inhibitors for 6 hours after pre-treating with BETi for 2 days. Apoptotic cell population was detected by Annexin V positivity (AnnV+) and the absence of propidium iodide (PI) staining. Data are mean ± SEM (n=3). (A) Kasumi-1 cells and (B) SKNO-1 cells were incubated with 125 nM or 250 nM JQ1 before BCL2 inhibitor treatment. (C) MOLT4 cells, (D) MOLM13 cells were incubated with 250 nM or 500 nM JQ1 before BCL2 inhibitor treatment.
Discussion
While there is justifiable excitement about the therapeutic efficacy of BETi in AML, the therapeutic window appears to be due to the loss of MYC expression8,11,14–16,18. However, as observed in Kasumi-1 cells, the rapid loss of MYC was accompanied by a rapid cell cycle arrest, loss of metabolic activity, and reduced cell size, which is reminiscent of quiescence rather than cell death. In standard assays that use metabolism as a surrogate marker for cell viability, these compounds appear to work well, yet these cells recovered after BETi removal, suggesting that cell cycle arrest could be a mechanism of resistance to these compounds. The addition of a BCL2 inhibitor could provide the additional push toward apoptosis needed, and this could be an extremely safe and efficacious combination.
We identified this effect in t(8;21) containing Kasumi-1 cells, yet the effect was not limited to the t(8;21), as cells that express MLL fusion proteins were similarly slow to die in response to BETi. In addition, in a panel of diffuse large B cell lymphoma cell lines, JQ1 strongly induced G1 cell cycle arrest, but only caused minor levels of apoptosis in several of these cell types (e.g. Ly7, Toledo, and Ly19)8. Poor pharmacokinetic profile has been proposed as the cause of the greatly reduced efficacy of BETi in mouse models compared to in vitro studies with cell lines, but reversible cell cycle arrest likely contributes. This may be a limiting factor for the usefulness of these compounds as MYC activation is a later event in myeloid leukemias that are not triggered by a translocation or amplification directly affecting MYC. This is an important consideration because when additional oncogenes together with MYC drove AML development, removal of MYC expression failed to prevent tumor recurrence46,47.
Mechanistically, MYC is perhaps the best known transcriptional regulator of metabolism, cell cycle, and cell size48, as it controls the production of ribosomes that control translational outputs. Thus, it is most likely that suppression of MYC is the key event in the regulation of cell size in our studies. It is notable that our PRO-seq analysis did not identify RNA polymerase II promoter-proximal pausing associated with ribosomal genes or pathways linked to metabolism until 3 hr after JQ1 treatment (Fig. 4A), even though large numbers of genes were affected within 15–30 min after treatment, including MYC31. Thus, the loss of metabolic activity (Figures 5–6) is likely a secondary consequence, which is consistent with the down regulation of MYC in the first hour followed by the loss of MYC targets beginning at1 hr and increasing 3 hr post drug treatment (Fig. 4). However, we also noted the loss of expression of cyclin D1 (CCND1) and D2 (CCND2), CDK4, and CDK6. In addition, we detected an “E2F/retinoblastoma (RB) signature” of increased pausing ratio with E2F2 and E2F8 being directly affected within the first hour of treatment with BETi31. Conversely, we noted a rapid upregulation of CDKN1A (p21CIP), but this was not associated with DNA damage (Fig. S1). Given that RB family-dependent repression is associated with G1 arrest and quiescence and E2F family members control nucleotide metabolism, these data suggest that the effects on metabolism and cell cycle control are multi-factorial in nature.
In addition to controlling cell size, MYC is a direct regulator of genes that control glycolysis and mitochondrial biogenesis (Figures 5–649). MYC stimulates the expression of glucose transporter-1 while also stimulating the genes that directly control glycolysis50–52. Therefore, the loss of MYC expression is consistent with the loss of glycolytic burst and glycolytic capacity in AML cells treated with JQ1 (Figure 5). At the same time, JQ1 treatment induced a loss of mitochondrial functions including reductions in basal respiration, ATP-linked respiration, and maximal respiration capacity (Figure 5). Analysis of transcription at the early time points suggested only defects in growth factor signal transduction, so these effects could also be traced to reduced MYC expression by three hours post BET inhibitor treatment.
Given the loss of MYC expression and the large effects on metabolism, it is somewhat surprising that AML cells survived and could re-enter the cell cycle (Figures 6–7). An intriguing hypothesis is that the cells have entered a quiescent-like state in which metabolic needs are greatly reduced. This possibility is consistent with the gene expression profiles, the reduced cell size, and the ability of these cells to begin cycling again after removal of JQ1. Nevertheless, the increased apoptosis upon addition of venetoclax, was more than an additive effect in Kasumi-1 cells, indicating that the suppression of BCL2 levels sets the stage for the induction of apoptosis. While the effects were not as dramatic in other cell types (Figure 8), this could be due to induction of MCL1, whose levels increase after JQ1 treatment, possibly as a stress response. Given that venetoclax is beginning to gain traction as a component of the standard of care for AML53–56, these data support a rational combination approach to the utilization of BET inhibitors with venetoclax in the clinic to avoid the use of genotoxic agents such as hypomethylating agents or Cytarabine.
Supplementary Material
Acknowledgements
We thank all the members of Hiebert lab for helpful discussions, reagents and advice. We thank the Translational Pathology, the Hematological Sample Repository, Flow Cytometry, and VANTAGE Shared Resources for services and support. This work was supported by the T. J. Martell Foundation, the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, National Institutes of Health grants (RO1-CA109355, RO1-CA164605 and R01-CA64140) and core services performed through Vanderbilt Digestive Disease Research grant (NIDDK P30DK58404) and the Vanderbilt-Ingram Cancer Center support grant (NCI P30CA68485). KS was supported by 5 T32 CA009582-26 and a postdoctoral fellowship (PF-13-303-01-DMC) from the American Cancer Society. The project described was also supported by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. This work was supported by grant 1S10OD018015-01A1 for the Seahorse Extracellular Flux Analyzer and Prep Station, housed in the Vanderbilt High-Throughput Screening Facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Wu SY, Chiang CM. 2007. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J Biol Chem 282:13141–5. [DOI] [PubMed] [Google Scholar]
- 2.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100:8758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, Knapp S. 2012. The bromodomain interaction module. FEBS Lett 586:2692–704. [DOI] [PubMed] [Google Scholar]
- 4.Zhang G, Plotnikov AN, Rusinova E, Shen T, Morohashi K, Joshua J, Zeng L, Mujtaba S, Ohlmeyer M, Zhou MM. 2013. Structure-guided design of potent diazobenzene inhibitors for the BET bromodomains. J Med Chem 56:9251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–34. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19:535–45. [DOI] [PubMed] [Google Scholar]
- 7.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, Reichert E, Kung AL, Rodig SJ, Young RA, Shipp MA, Bradner JE. 2013. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 24:777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. 2012. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem 287:36609–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Xu Y, He M, Zhang M, Cui F, Lu L, Yao M, Tian W, Benda C, Zhuang Q, Huang Z, Li W, Li X, Zhao P, Fan W, Luo Z, Li Y, Wu Y, Hutchins AP, Wang D, Tse HF, Schambach A, Frampton J, Qin B, Bao X, Yao H, Zhang B, Sun H, Pei D, Wang H, Wang J, Esteban MA. 2014. Transcriptional pause release is a rate-limiting step for somatic cell reprogramming. Cell Stem Cell 15:574–88. [DOI] [PubMed] [Google Scholar]
- 11.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A. 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468:1119–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ, Thiessen N, Pettersson S, Jones SJ, Knapp S, Yang H, Chin KC. 2012. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem 287:43137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, Chesi M, Schinzel AC, McKeown MR, Heffernan TP, Vakoc CR, Bergsagel PL, Ghobrial IM, Richardson PG, Young RA, Hahn WC, Anderson KC, Kung AL, Bradner JE, Mitsiades CS. 2011. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146:904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM. 2012. BET bromodomain inhibition targets both c-MYC and IL7R in high-risk acute lymphoblastic leukemia. Blood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJ, Kouzarides T. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478:529–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. 2012. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A 109:19408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Q, Zhang Z, Shea MJ, Creighton CJ, Coarfa C, Hilsenbeck SG, Lanz R, He B, Wang L, Fu X, Nardone A, Song Y, Bradner J, Mitsiades N, Mitsiades CS, Osborne CK, Schiff R, O’Malley BW. 2014. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res 24:809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, Roberts JM, Zeid R, Scott TG, Paulk J, Lachance K, Olson CM, Dastjerdi S, Bauer S, Lin CY, Gray NS, Kelliher MA, Churchman LS, Bradner JE. 2017. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol Cell 67:5–18 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohan M, Lin C, Guest E, Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10:721–8. [DOI] [PubMed] [Google Scholar]
- 22.Meyers S, Lenny N, Hiebert SW. 1995. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol 15:1974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutterbach B, Sun D, Schuetz J, Hiebert SW. 1998. The MYND motif is required for repression of basal transcription from the multidrug resistance-1 promoter by the t(8;21) fusion protein. Mol. Cell. Biol 18:3601–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG, Glass C, Seto E, Hiebert SW. 1998. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol 18:7176–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linggi B, Muller-Tidow C, van de Locht L, Hu M, Nip J, Serve H, Berdel WE, van der Reijden B, Quelle DE, Rowley JD, Cleveland J, Jansen JH, Pandolfi PP, Hiebert SW. 2002. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med 8:743–50. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G, Hatlen MA, Vu L, Liu F, Xu H, Asai T, Xu H, Deblasio T, Menendez S, Voza F, Jiang Y, Cole PA, Zhang J, Melnick A, Roeder RG, Nimer SD. 2011. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 333:765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol 21:6470–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ptasinska A, Assi SA, Martinez-Soria N, Imperato MR, Piper J, Cauchy P, Pickin A, James SR, Hoogenkamp M, Williamson D, Wu M, Tenen DG, Ott S, Westhead DR, Cockerill PN, Heidenreich O, Bonifer C. 2014. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep 8:1974–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. 2004. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science 305:1286–9. [DOI] [PubMed] [Google Scholar]
- 30.Meier N, Krpic S, Rodriguez P, Strouboulis J, Monti M, Krijgsveld J, Gering M, Patient R, Hostert A, Grosveld F. 2006. Novel binding partners of Ldb1 are required for haematopoietic development. Development 133:4913–23. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Liu Q, Acharya P, Stengel KR, Sheng Q, Zhou X, Kwak H, Fischer MA, Bradner JE, Strickland SA, Mohan SR, Savona MR, Venters BJ, Zhou MM, Lis JT, Hiebert SW. 2016. High-Resolution Mapping of RNA Polymerases Identifies Mechanisms of Sensitivity and Resistance to BET Inhibitors in t(8;21) AML. Cell Rep 16:2003–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heaster TM, Walsh AJ, Zhao Y, Hiebert SW, Skala MC. 2018. Autofluorescence imaging identifies tumor cell-cycle status on a single-cell level. J Biophotonics 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matozaki S, Nakagawa T, Kawaguchi R, Aozaki R, Tsutsumi M, Murayama T, Koizumi T, Nishimura R, Isobe T, Chihara K. 1995. Establishment of a myeloid leukaemic cell line (SKNO-1) from a patient with t(8;21) who acquired monosomy 17 during disease progression. British Journal of Haematology 89:805–11. [DOI] [PubMed] [Google Scholar]
- 36.Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L, Rusinova E, Gerona-Nevarro G, Moshkina N, Joshua J, Chuang PY, Ohlmeyer M, He JC, Zhou MM. 2012. Down-regulation of NF-kappaB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem 287:28840–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ 8:1039–48. [PubMed] [Google Scholar]
- 38.Mateyak MK, Obaya AJ, Sedivy JM. 1999. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol 19:4672–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graves JA, Wang Y, Sims-Lucas S, Cherok E, Rothermund K, Branca MF, Elster J, Beer-Stolz D, Van Houten B, Vockley J, Prochownik EV. 2012. Mitochondrial structure, function and dynamics are temporally controlled by c-Myc. PLoS One 7:e37699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA. 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7:295–302. [DOI] [PubMed] [Google Scholar]
- 41.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, Versteeg R. 2001. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20:1383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, Doan HM, Fan J, Cheadle C, Fallahi M, Cleveland JL, Dang CV, Zeller KI. 2011. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One 6:e26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabo A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, Tonelli C, Faga G, Bianchi V, Ronchi A, Low D, Muller H, Guccione E, Campaner S, Amati B. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, Rycak L, Dumay-Odelot H, Karim S, Bartkuhn M, Roels F, Wustefeld T, Fischer M, Teichmann M, Zender L, Wei CL, Sansom O, Wolf E, Eilers M. 2014. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511:483–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odore E, Lokiec F, Cvitkovic E, Bekradda M, Herait P, Bourdel F, Kahatt C, Raffoux E, Stathis A, Thieblemont C, Quesnel B, Cunningham D, Riveiro ME, Rezai K. 2016. Phase I Population Pharmacokinetic Assessment of the Oral Bromodomain Inhibitor OTX015 in Patients with Haematologic Malignancies. Clin Pharmacokinet 55:397–405. [DOI] [PubMed] [Google Scholar]
- 46.Rakhra K, Bachireddy P, Zabuawala T, Zeiser R, Xu L, Kopelman A, Fan AC, Yang Q, Braunstein L, Crosby E, Ryeom S, Felsher DW. 2010. CD4(+) T cells contribute to the remodeling of the microenvironment required for sustained tumor regression upon oncogene inactivation. Cancer Cell 18:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi PS, Li Y, Felsher DW. 2014. Addiction to multiple oncogenes can be exploited to prevent the emergence of therapeutic resistance. Proc Natl Acad Sci U S A 111:E3316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Riggelen J, Yetil A, Felsher DW. 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10:301–9. [DOI] [PubMed] [Google Scholar]
- 49.Dang CV. 2016. A Time for MYC: Metabolism and Therapy. Cold Spring Harb Symp Quant Biol 81:79–83. [DOI] [PubMed] [Google Scholar]
- 50.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. 2000. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem 275:21797–800. [DOI] [PubMed] [Google Scholar]
- 51.Kim JW, Zeller KI, Wang Y, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. 2004. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol Cell Biol 24:5923–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, Kurhanewicz J, Vigneron DB, Goga A. 2011. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 14:131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiNardo CD, Pratz KW, Letai A, Jonas BA, Wei AH, Thirman M, Arellano M, Frattini MG, Kantarjian H, Popovic R, Chyla B, Xu T, Dunbar M, Agarwal SK, Humerickhouse R, Mabry M, Potluri J, Konopleva M, Pollyea DA. 2018. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol 19:216–228. [DOI] [PubMed] [Google Scholar]
- 54.Konopleva M, Letai A. 2018. BCL-2 inhibition in AML - an unexpected bonus? Blood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konopleva M, Pollyea DA, Potluri J, Chyla B, Hogdal L, Busman T, McKeegan E, Salem AH, Zhu M, Ricker JL, Blum W, DiNardo CD, Kadia T, Dunbar M, Kirby R, Falotico N, Leverson J, Humerickhouse R, Mabry M, Stone R, Kantarjian H, Letai A. 2016. Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov 6:1106–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma P, Pollyea DA. 2018. Shutting Down Acute Myeloid Leukemia and Myelodysplastic Syndrome with BCL-2 Family Protein Inhibition. Curr Hematol Malig Rep 13:256–264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.