Abstract
Terpene synthases typically form complex molecular scaffolds by concerted activation and cyclization of linear starting materials in a single enzyme active site. Here we show that iridoid synthase, an atypical reductive terpene synthase, catalyzes the activation of its substrate 8-oxogeranial into a reactive enol intermediate, but does not catalyze the subsequent cyclization into nepetalactol. This discovery led us to identify a class of nepetalactol-related short-chain dehydrogenase enzymes (NEPS) from catmint (Nepeta mussinii) that capture this reactive intermediate and catalyze the stereoselective cyclisation into distinct nepetalactol stereoisomers. Subsequent oxidation of nepetalactols by NEPS1 provides nepetalactones, metabolites that are well known for both insect-repellent activity and euphoric effect in cats. Structural characterization of the NEPS3 cyclase reveals that it binds to NAD+ yet does not utilize it chemically for a non-oxidoreductive formal [4 + 2] cyclization. These discoveries will complement metabolic reconstructions of iridoid and monoterpene indole alkaloid biosynthesis.
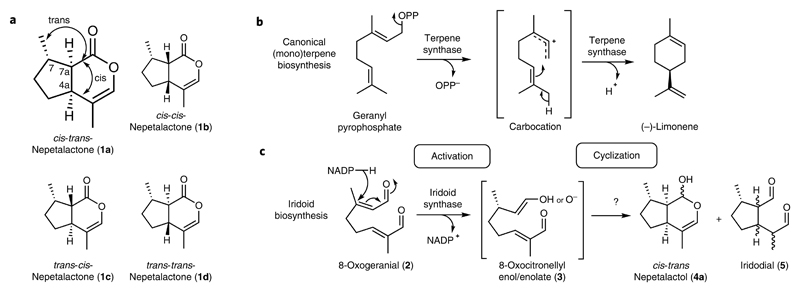
Nepetalactones (1a, 1b, 1c and 1d) are volatile terpenoids produced by plants of the genus Nepeta, notably catmint (N. mussinii syn racemosa) and catnip (Nepeta cataria) (Fig. 1a)1,2. These compounds are responsible for the stimulatory effects these plants have on cats3–5. Moreover, certain insects use nepetalactones as sex pheromones, so production of these compounds by the plant also impacts interactions with insects6. Notably, the bridgehead stereocenters (carbons 4a and 7a) vary between2 and within2,4,7 Nepeta species. N. mussinii individuals, for example, produce different ratios of cis-trans nepetalactone (1a), cis-cis nepetalactone (1b) and trans-cis-nepetalactone (1c)7. Variation in the stereoisomer ratio may influence the repellence of insect herbivores8,9. Though the ratio of stereoisomers may be responsible for important biological effects, the mechanism of stereocontrol in nepetalactone biosynthesis is not known. Nepetalactones are iridoids, compounds that are biosynthesized via a noncanonical terpene pathway. The alternative nepetalactone stereoisomers found in Nepeta provide an exemplary system to explore how the inherent chemical reactivity of a biosynthetic intermediate can be harnessed to generate stereochemical diversity.
Fig. 1. Nepetalactones and terpenoid biosynthesis.
a, Nepetalactone 1 stereoisomers observed in Nepeta species. b, Representative canonical terpene biosynthesis mechanism (limonene synthase). Typical terpene synthases activate linear precursors by removal of diphosphate or protonation. The activated carbocation intermediates undergo selective cyclization inside the same terpene synthase active site. c, Iridoid biosynthesis mechanism. Iridoid synthase activates its linear precursor (8-oxogeranial (2)) by reduction to form the 8-oxocitronellyl enol/enolate intermediate 3, which cyclizes to form a mixture of cis-trans-nepetalactol (4a) and iridodials (5).
Canonical cyclic terpenoids (for example, (–)-limonene) are bio-synthesized from linear precursors by terpene synthases (Fig. 1b)10. These enzymes activate linear precursors by either loss of pyrophosphate or protonation10–12. The resulting carbocations generated cyclize rapidly to form an array of cyclic products13. Therefore, in canonical terpenoid biosynthesis, activation and cyclization of precursors are coupled and occur in the same enzyme active site.
In plant iridoid biosynthesis, geranyl pyrophosphate is hydrolyzed and oxidized into 8-oxogeranial (2; ref. 14). This precursor then undergoes a two-step activation–cyclization process analogous to canonical terpene synthesis (Fig. 1c)15. Unlike canonical terpene synthesis, however, activation is achieved by reduction, and the intermediate is not a carbocation, but the enol or enolate species 3. Cyclization of this intermediate yields cis-trans-nepetalactol (4a) along with iridodial side products (5a, 5b, 5c and 5d; Fig. 1c).
The conversion of 2 to 4a and 5 is catalyzed by iridoid synthase (ISY)15. ISY was first discovered in Catharanthus roseus (CrISY), where it forms part of the biosynthetic route to the anticancer monoterpene indole alkaloids vincristine and vinblastine15,16. Subsequent studies revealed ISYs from other species including Olive (Olea europaea, OeISY)17 and Snapdragon (Antirrhinum majus, AmISY)18. Recently, we identified ISYs from Nepeta19.
The enzymatic control of the initial reductive activation step has been structurally characterized in CrISY; crystal structures with cofactor and inhibitor or substrate show binding modes conducive to reduction and formation of an enolate intermediate20–22. Furthermore, this reduction is stereoselective, as exemplified by the comparison of CrISY, which produces 7S-4a, with AmISY, which produces the enantiomer18.
In contrast, it is unknown how the cyclization that determines the stereochemistry of the bridgehead 4a-7a-carbons is controlled. Structures of CrISY did not provide evidence for enzyme-catalyzed cyclization; though the protein exhibited evidence of conformational flexibility near the active site, the substrates bound occupied a linear conformation not conducive to cyclization20–22. Furthermore, active site mutations failed to impact the product profile, raising the possibility that the cyclization step is not enzyme catalyzed18,20.
We have now determined the biosynthetic route to two nepetalactone stereoisomers in N. mussinii, 1a and 1b. The discovery of these genes reveals that the reduction and cyclization steps of iridoid biosynthesis in Nepeta are uncoupled and catalyzed by distinct enzymes. This process involves the diffusion of the activated intermediate 8-oxocitronellyl enol 3 between enzyme active sites, in contrast to canonical terpene biosynthesis, wherein generation of the activated intermediate and cyclization occur in the same enzyme active site. We have discovered and characterized three cyclases (NEPS1, NEPS2 and NEPS3) from N. mussinii that are responsible for the stereoselective cyclization and subsequent oxidation of activated intermediate 3 into distinct nepetalactone diastereomers. We have also determined the crystal structure of NEPS3, providing insight into its mechanism and evolution from a reductase into a redox-neutral cyclase.
Results
The mechanism of iridoid synthase (ISY)
Recently, we hypothesized that synthesis of the different nepetalactone stereoisomers in Nepeta was controlled by species-specific ISYs catalyzing both the reduction of 2 and the subsequent stereodivergent cyclizations. However, this was not the case; Nepeta ISYs produced the same stereoisomeric product profile as CrISY19. Therefore, an alternative mechanism for the control of iridoid stereochemistry was developed.
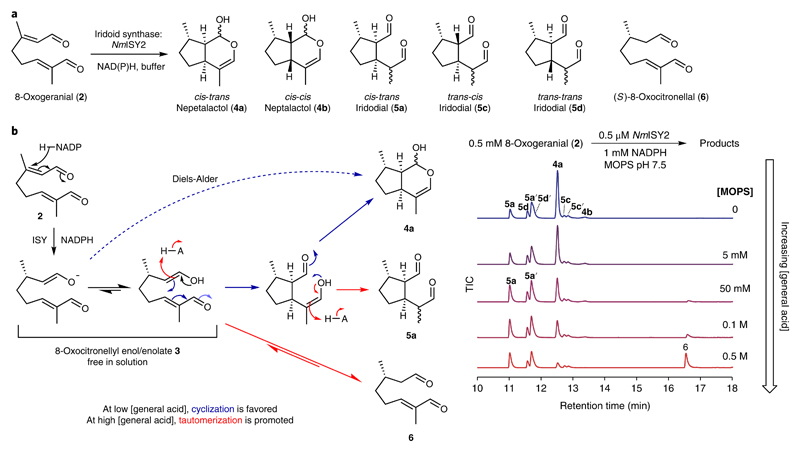
The first step toward understanding the origin of the divergent stereochemistry of nepetalactones (1) was to further explore the ISY mechanism. As observed in previous studies15,18,19, the ISY-catalyzed reduction of 2 generates a mixture of products (Fig. 2a; Supplementary Figs. 1 and 2). The ratios of different structural isomers and diastereomers formed in the reaction are largely independent of the ISY employed, despite the enzymes tested having modest sequence identities (48–65%) and different enantiose-lectivities at the reduction step (Supplementary Figs. 1 and 2). The product profile was, however, strongly influenced by the buffer conditions, both by its concentration (Fig. 2b) and its pH (Supplementary Fig. 3). This sensitivity was observed across all ISYs and buffers tested (Supplementary Figs. 3 and 4) and was not due to the decomposition of 4a in high buffer concentrations (Supplementary Fig. 5a)23,24.
Fig. 2. Iridoid synthase (ISY) reaction mechanism.
a, Product mixture observed in the NmISY2-catalyzed reductive cyclization of 2. b, ISY-catalyzed reduction of 2 at different buffer concentrations depicted as GC–MS total ion chromatograms (TICs) and its possible mechanism. At low buffer concentrations, the main product is 4a. As buffer concentrations increase, higher quantities of cis-trans-iridodial (5a) and then 8-oxocitronellal (6) are observed. We propose that ISY reduces 2 and then releases the activated 3 into the solvent, where cyclization occurs. Buffer appears to act as a general acid catalyst, promoting tautomerization in place of cyclization. See Supplementary Figs. 1–5 for further exploration of the solvent conditions. Experiments were repeated seven time, but employing slightly different conditions by varying ISY (three different ISYs, Supplementary Fig. 3) and varying the buffer (four different buffers, Supplementary Fig. 4). Similar results were obtained for all seven experiments exploring buffer concentration effects.
The sensitivity of the product distribution to solvent conditions led us to hypothesize that ISY reduces 2 to form the activated intermediate 3, which then leaves the enzyme active site and diffuses into the solvent. In the solvent, 3 can be quenched through either cyclization or tautomerization to form a mixture of products. We propose that the buffer can act as a general acid catalyst, promoting tautomerization. In MOPS buffer, this approximates to three regimes (Fig. 2b): at low buffer concentrations (< 50 mM) two cyclizations, with possible contributions from a Diels-Alder reaction, generate the bicyclic 4a as the dominant product. In moderately buffered concentrations (50–500 mM) one cyclization followed by keto-enol tautomerization to form monocyclic 5a becomes favored. At high buffer concentrations (≥500 mM) direct keto-enol tautomerization of 3 into 6 becomes the dominant route. The promotion of tautomerization by buffer molecules mirrors simulations that have highlighted the bimolecular nature of tautomerization mechanisms25,26.
To validate that the ISY product profile was a result of the non-enzymatic cyclization of 3, we aimed to form 3 in the absence of enzyme. This was achieved by incubation of (S)-8-oxocitronellal (6) in unbuffered water at acidic (< 2) or alkaline (> 10) pHs, or in buffered water at pH 7.5 (Supplementary Figs. 6 and 7). Buffer or extreme pH promotes keto-enol tautomerization of 6 into 3, which could then undergo cyclization/tautomerization in an manner analogous to ISY reactions. In fact, formation of 4a from 6 is an established synthetic route27. In high concentrations of buffer (500 mM MOPS), the product profiles of the ISY-catalyzed reduction of 2 and the nonenzyme catalyzed cyclization of 6 are remarkably similar, supporting the hypothesis that iridoid cyclization is not enzymatically catalyzed (Supplementary Figs. 6e and 7).
Therefore, unlike canonical terpene synthases, which catalyze concerted activation and cyclization (Fig. 1b), ISY catalyzes the activation of its linear substrate (2 to 3), but does not appear to catalyze the subsequent cyclization. Instead, we hypothesize that 3 diffuses out of the ISY active site into the solvent, where it cyclizes. This mechanism is supported by the ISY crystal structure, which did not show substrate binding modes conducive to cyclization20. The notion of free 3 raised the possibility that the iridoid stereochemistry may be defined by a partner cyclase enzyme, capable of accepting 3 as a substrate and catalyzing diastereoselective cyclization.
Nepetalactol-related short-chain reductases (NEPS)
Identifying a cyclase that works in partnership with ISY presents a challenge: because this proposed reaction is unprecedented, it is difficult to predict what type of enzyme family would catalyze such a cyclization. However, nepetalactone biosynthesis in Nepeta is localized to a specific plant organ, glandular trichomes7. Therefore, we could compare the proteome of trichomes to trichome-depleted leaves to identify genes that are selectively expressed at the site of nepetalactone biosynthesis, thereby considerably narrowing the pool of potential gene candidates.
We obtained proteomes for N. mussinii trichomes, leaves and trichome-depleted leaves (Supplementary Fig. 8a, Supplementary Dataset 1). Comparison of these proteomes enabled identification of trichome-enriched proteins. This approach was validated by the identification of trichome-enriched enzymes from upstream isoprenoid biosynthesis (the 2-C-methylerythritol 4-phosphate (MEP) pathway) and iridoid biosynthesis (Supplementary Fig. 8b,c).
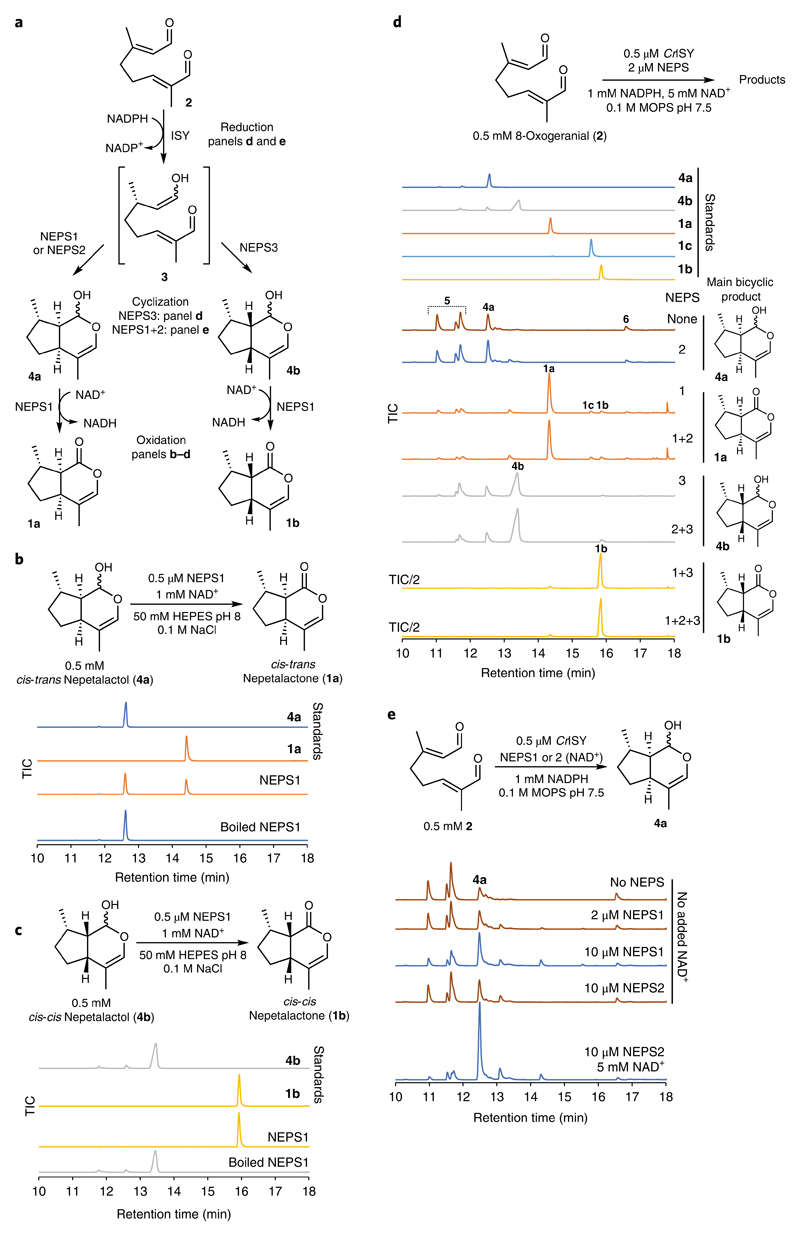
As a starting point, we initially used these proteomes to identify the enzyme that converts nepetalactol 4 to nepetalactone 1, which is an NAD+-dependent enzyme. An enzyme with such activity had previously been isolated from the trichomes of N. mussinii, but its sequence was not identified28. Six trichome-enriched dehydrogenase genes were cloned and recombinantly expressed in E. coli (Supplementary Fig. 8d; Supplementary Table 1). Of these, one demonstrated 4a dehydrogenase activity (Supplementary Fig. 8e).
The active enzyme is a short-chain dehydrogenase/reductase (SDR), part of the SDR110C family, a large and diverse family of NAD-dependent dehydrogenases often associated with plant secondary metabolism29. Consequently, it was named nepetalactol-related SDR 1 (NEPS1). NEPS1 could catalyze the NAD+-dependent dehydrogenation of either 4a or 4b to 1a and 1b, respectively (Fig. 3a–c; Supplementary Fig. 9a; Supplementary Table 2). The observed activities were in accordance with characterization of the native enzyme28.
Fig. 3. Formation of nepetalactones by NEPS enzymes.
a, Summary of NEPS enzyme activities described in this figure. b, cis-trans-Nepetalactol dehydrogenase activity of NEPS1. NEPS1 catalyzes the NAD+-dependent dehydrogenation of 4a to 1a. c, cis-cis-Nepetalactol dehydrogenase activity of NEPS1. NEPS1 catalyzes the NAD+-dependent dehydrogenation of 4b to 1b. NEPS1 activities on 4a and 4b are consistent with cascade activities (d), kinetics (Supplementary Table 2) and activities with 6 (Fig. 4). d, Combined activities of ISY and NEPS enzymes. Incubation of 2, CrISY, NEPS and cofactors enables the production of 4a (no NEPS or NEPS2), 1a (NEPS1), 4b (NEPS3) or 1b (NEPS1 and NEPS3). Experiments performed under nearly identical conditions also provided similar results (Supplementary Fig. 12). Observed enzyme activities are consistent with activities observed in Fig. 4b. e, NEPS-catalyzed formation of 4a. Adjusting the NAD+ and/or NEPS concentrations reveals that NEPS1 and NEPS2 can promote the formation of 4a. See Supplementary Fig. 9 for EI spectra, Supplementary Fig. 12 for NEPS cascades with NmISY2 and CrISY, and Supplementary Fig. 13 for further analysis of the NEPS3 cyclization reaction. All experiments were performed independently twice with similar results.
Sequence analysis of NEPS1 and the remaining dehydrogenase candidates revealed two additional trichome-enriched paralogs of NEPS1, NEPS2 and NEPS3 (Supplementary Fig. 10a,b). Phylogenetic analysis revealed that these three proteins have a close evolutionary relationship and are found uniquely within the Nepeta lineage (Supplementary Fig. 10c). Therefore, we hypothesized that NEPS2 and NEPS3 also play a role in nepetalactone biosynthesis. Consequently, NEPS1– NEPS3 enzymes were assayed with a variety of nepetalactone-related compounds and precursors such as nepetalactol 4, iridodials 5, and 2 (Supplementary Fig. 11). However, besides the NEPS1 activities described above, no other notable activities were observed for NEPS1–NEPS3.
NEPS activities in conjunction with ISY
As described above, mechanistic investigations of ISY led us to hypothesize that a separate cyclase enzyme may act on the activated intermediate 3 generated by ISY. To test whether NEPS acts as such cyclases, we performed one-pot cascade reactions combining NEPS enzymes with the ISY-catalyzed reduction of 2 (Fig. 3a,d,e). As anticipated, addition of NEPS1 and excess NAD+ led to the formation of 1a (Fig. 3d and Supplementary Fig. 9b), although ISY iridodial side product 5 was unexpectedly diminished. Remarkably, addition of NEPS3 to ISY and 2 led to the formation of 4b. A combination of NEPS1 and NEPS3 led to the production of 1b. Adjusting enzyme and cofactor concentrations revealed that NEPS1 and NEPS2 promoted the formation of 4a at the expense of 5 (Fig. 3e).
The products observed indicated that the NEPS enzymes are cyclases (Fig. 3a), capable of accepting 3, the product of ISY, and cyclizing it to 4a (NEPS1 and NEPS2) or 4b (NEPS3). NEPS1 can then oxidize 4a and 4b into 1a and 1b, respectively. The activities of NEPS enzymes did not appear to differ when tested with CrISY or NmISY2, suggesting that protein–protein interactions between ISYs and NEPS do not play a role in the system (Supplementary Fig. 12a). The cascade can operate at a range of total enzyme concentrations (Supplementary Fig. 12b). NEPS2 and NEPS3 show no notable oxidation activity with NAD+ or NADP+ or at different pHs (Supplementary Fig. 12c,d).
The cyclization of 3 into 4 is a non-oxidoreductive net [4 + 2] cycloaddition. The cyclase activities of NEPS also do not appear to be oxidoreductive. Investigation into the cofactor dependence of the ISY-NEPS3 reactions demonstrated that NAD+ concentrations were not limiting to overall reaction conversions, suggesting that NAD+ was not consumed (Supplementary Fig. 13). Furthermore, NEPS3 was active in the absence of supplemented NAD+, though addition did improve activity. It appeared that although NAD+ was not turned over by NEPS3 during cyclization, it may promote the enzyme’s catalytic ability, perhaps through stabilization of the protein structure.
NEPS activities with (S)-8-oxocitronellal
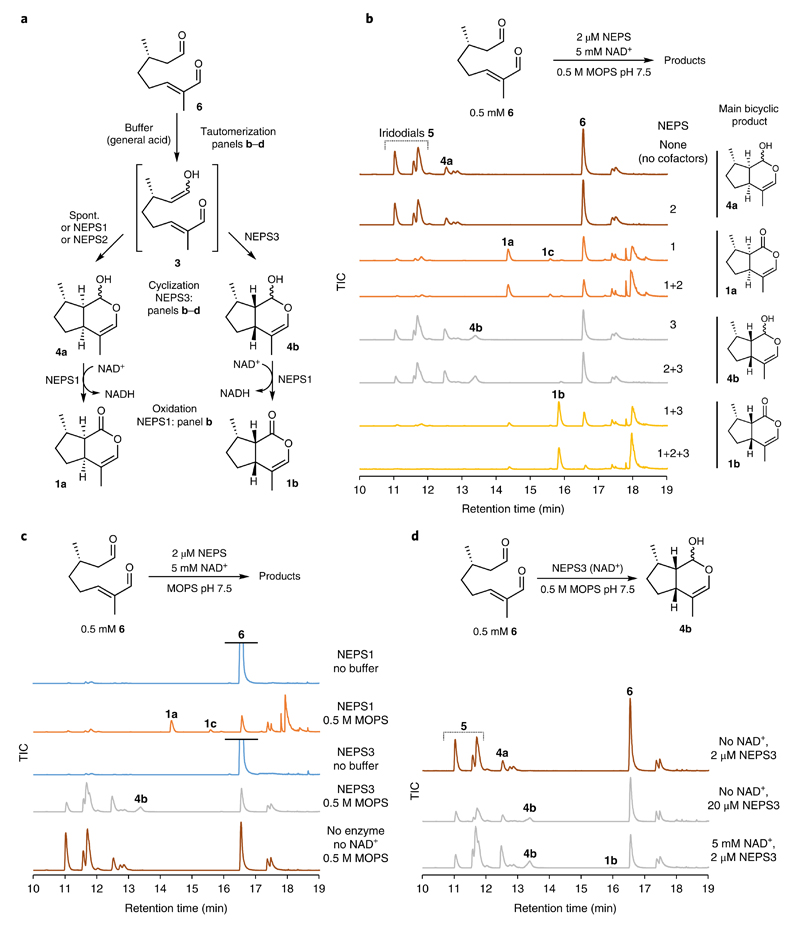
To verify the NEPS activities, reactions were conducted with 6 and without ISY (Fig. 4). High concentrations of buffer were employed to promote the formation of 3, the proposed NEPS substrate, from 6. In these conditions, the previously observed activities of NEPS were recapitulated (compare Fig. 4b to Fig. 3d).
Fig. 4. NEPS activities explored with (S)-8-oxocitronellal (6).
a, Summary of NEPS enzyme activities described in this figure. b, NEPS activities with 6, buffer and NAD+, presented as GC–MS TICs. The panel largely recapitulates observations of Fig. 3b, but in the absence of ISY. Unknown side products are formed by NEPS1. c, Buffer dependence of NEPS activity with 6. In the absence of buffer, NEPS1 and NEPS3 have no detectable activity; addition of buffer reveals enzyme activities. Buffer-catalyzed tautomerization of 6 appears to be necessary for enzyme activity, supporting the hypothesis that the activated 3, and not 6, is the key NEPS substrate. d, NEPS3-catalyzed cyclization. The addition of NAD+ is not required for NEPS3 cyclization activity, though addition does promote the reaction. We hypothesize that the cyclization is not oxidoreductive, but NAD+ acts in a nonchemical manner (that is, protein stabilization). All reactions were incubated for 16 h (in contrast to 3h for Fig. 3) and are presented as GC–MS TICs. All experiments were performed independently twice with similar results.
Neither NEPS1 nor NEPS3 were active when incubated with 6 in the absence of buffer (Fig. 4c). Buffer was necessary for activity, supporting the notion that that 6 is not the key substrate, but the tautomer 3 is. Further evidence for this was obtained by adding CrISY to reactions containing NEPS and 6; the pattern of products observed implied that NEPS were binding to 6 without turning it over (Supplementary Fig. 14).
Support for the non-oxidoreductive nature of the NEPS3 cyclization came from incubating NEPS3 with 6 and buffer in the absence of supplemented NAD+ or NADPH; formation of 4b was still observed (Fig. 4d). As noted above, addition of NAD+ does promote the reaction, though it is not necessary for activity. Interestingly, at high NAD+ concentrations, trace quantities of 1b were observed (Figs. 3d and 4d). Overall, NEPS reactions with 6 support the notion of 3 as the substrate and add further evidence the for a non-oxidoreductive cyclization (Fig. 4a).
Structure and mechanism of NEPS enzymes
To understand the mechanism of the NEPS3 cis-cis cyclization reaction, we obtained an X-ray crystal structure of NEPS3 bound to NAD+ (6F9Q, Supplementary Table 3). Similarly to structural homologs, it forms a homotetramer with 222 symmetry, with the four active centers contained entirely within individual protomers (Supplementary Fig. 15a). Efforts to generate an apo or ligand-bound structure were unsuccessful, as were efforts to crystallize NEPS1. Despite the fact that NEPS3 appears to function primarily as a non-oxidoreductive cyclase, its structure is characteristic of classical SDRs, with NAD+ bound in the typical fashion (Supplementary Fig. 15b,c). Although evidence suggests that NAD+ is not turned over by NEPS3 during cyclization, the cofactor appears to be important for the enzyme structure, as it was required for crystallization, and it is bound by multiple hydrogen bonds with the protein.
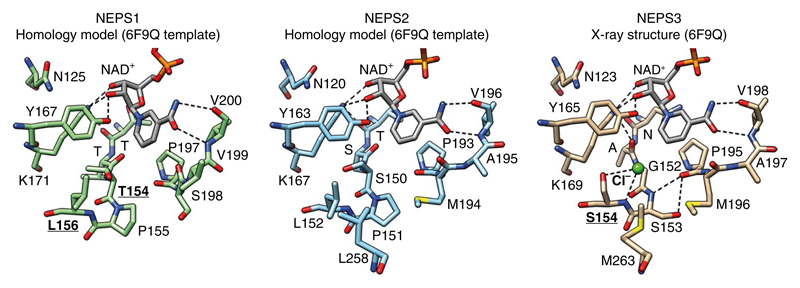
Homology models of NEPS1 and NEPS2, based on the NEPS3 structure, were generated to enable comparison of the enzyme active sites (Fig. 5). Whereas NEPS1 and NEPS2 have a typical SDR catalytic tetrad (N-Y-K-T/S), NEPS3 lacks the threonine/serine and instead has a glycine (Gly152). In addition, NEPS3 features hydrogen bonding between active site residues Ser153 and Pro190, absent from both NEPS1 and NEPS2 due to the presence of a proline in place of the serine.
Fig. 5. Structure of NEPS enzymes.
X-ray crystal structure of NEPS3 (6F9Q) and homology model structures of NEPS1 and NEPS2. Active site NAD+ and residues are depicted as sticks. Dashed lines represent proposed hydrogen bonds. The NEPS3 active site lacks the characteristic Ser/Thr of the SDR catalytic tetrad (Gly152). It also features a chloride bound to Ser154 and hydrogen bonding between Ser153 and Pro190. Ser154 is replaced by leucine in NEPS1 (Leu156) and NEPS2 (Leu152). The role of residues that are boldface and underlined have been analyzed by mutation. See Supplementary Fig. 15 for further analysis of the NEPS3 crystal structure.
Interestingly, the NEPS3 structure appeared to feature a chloride anion bound to the amide NH and side chain of Ser154. Based on the presence of the chloride, and the similarity to the substrate oxyanion binding site in CrISY, this position may be a substrate oxyanion binding site (Supplementary Fig. 15c). This site appears to be absent in NEPS1 and NEPS2 because of the steric hindrance of the leucine side chain present in the equivalent position.
Active site residue roles in NEPS1 and NEPS3 were probed via mutational screens (Supplementary Table 4). NEPS3 was especially sensitive to mutation, with several active site substitutions abolishing detectable soluble expression (A151T, G152T, S153P, Y165F). These mutations may have disrupted the hydrogen bonding network in the active site, reducing protein stability. Of the mutations yielding soluble protein, only N150T maintained native levels of activity, whereas S154L, K169M and M196S had a severe reduction or complete loss of activity (Supplementary Fig. 16a). Several NEPS1 variants had no detectable formation of 1a, signifying that these may be functionally important residues (T152N, L156S, Y167F, K171M, S198M, T202A, Supplementary Fig. 16b). Near-WT levels of 1a were identified in samples with the variants T153A and V199A, whereas trace quantities of 1a were identified with N125A and P155S (Supplementary Fig. 16c). Most interesting were variants containing the T154G substitution, which produced considerably greater quantities of 4a than WT (Supplementary Fig. 16d).
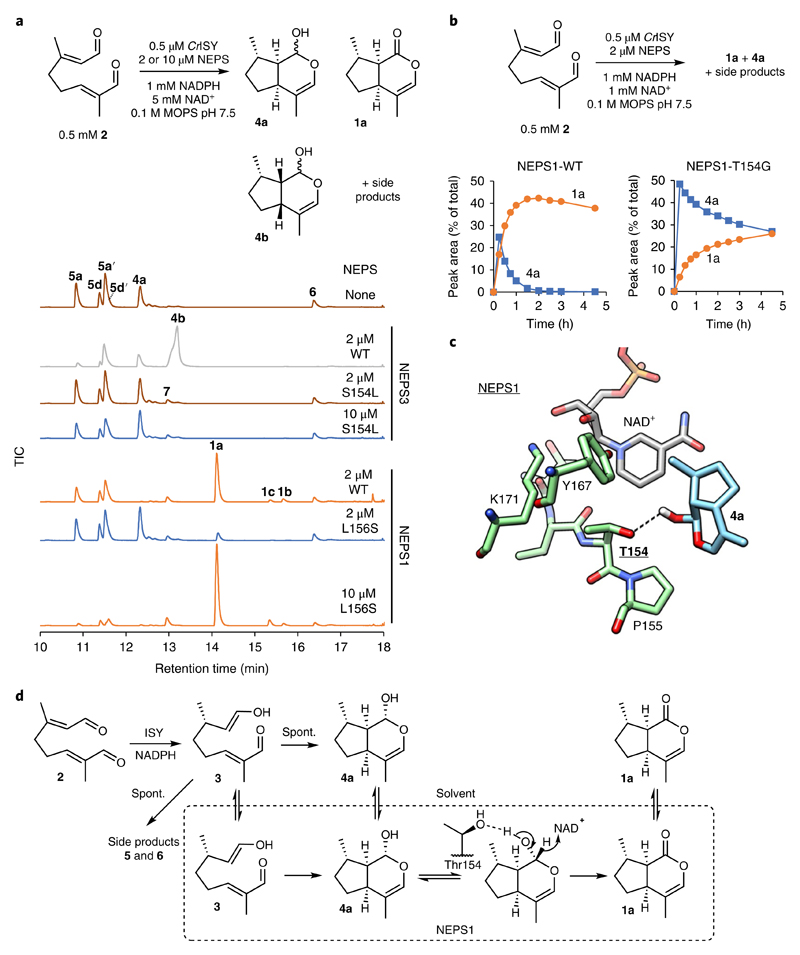
The role of the NEPS3 Ser154 putative oxyanion binding site was examined by further characterization of NEPS3-S154L and the complementary NEPS1-L156S variant. NEPS3-S154L (2 μM) demonstrated no detectable cis-cis cyclase activity, though at higher enzyme concentrations (10 μM) the variant appeared to promote the formation of 4a (Fig. 6a). The complementary substitution in NEPS1, L156S, reduced dehydrogenase activity but failed to increase the formation of 4b or 1b (Fig. 6a). The NEPS3-S154 oxyanion binding site therefore appears to be necessary for cis-cis cyclization in NEPS3, but its introduction into NEPS1 is not sufficient to establish cis-cis cyclization activity. In contrast, the removal of Ser154 from NEPS3 appears to switch the cyclization selectivity from cis-cis to cis-trans.
Fig. 6. NEPS variants.
a, Coupled assay with 8-oxogeranial (2), ISY and NEPS variants. NEPS3-S154L has no cis-cis cyclase activity (no 4b), but at 10 μM enzyme the formation of 4a is increased. NEPS1-L156S has decreased dehydrogenase activity compared NEPS1 (less 1a) but no cis-cis cyclase activity (no 4b). Experiments were performed independently twice with similar results. b, Time course with 2, ISY and NEPS1-WT or NEPS1-T154G. Quantities of 4a (blue) and 1a (orange) are reported as percent proportion of total product peak area. NEPS1-T154G has lower dehydrogenation activity compared to WT (conversion of 4a into 1a). See Supplementary Fig. 17 for all TICs of the time course and Supplementary Table 2 for kinetic analysis. Full time-course experiments were performed once, but conversions after 3 h were performed independently twice with similar results. c, Putative binding mode of 4a in the NEPS1 homology model active site. The putative hydrogen bond interaction between the lactol and Thr154 is highlighted with a dotted line. The depicted binding mode was ranked third of ten binding modes (rank 1 = -6.4 kcal.mol−1, depicted rank 3 = -6.3 kcal.mol−1). d, Scheme of NEPS1 activities and interactions. NEPS1 has distinct cyclization and dehydrogenation activities. The behavior of NEPS1-T154G suggests that Thr154 is involved in dehydrogenation. The two activities may involve different active site interactions. The slightly improved cyclase activity of T154G may be due to poor binding of 4a, which frees more enzyme for binding and cyclizing 3 into 4a.
The NEPS1-T154G variant was also characterized further. Compared to wild-type NEPS1, the variant accumulated 4a and showed impaired formation of 1a (Fig. 6b and Supplementary Fig. 17a). This loss of dehydrogenase activity appears to be a result of a 150-fold increase in Km for 4a caused by the T154G substitution (Supplementary Table 2). This result indicates that Thr154 interacts with 4a during or before the dehydrogenation step; a putative binding mode is provided by docking calculations (Fig. 6c). Interestingly, the variant appears to have slightly enhanced cyclization activity: reactions with T154G show minimal formation of cis-trans-iridodial (5a) side products (Supplementary Fig. 17a), and the variant is also able to form detectable levels of 4a from 6 in buffer without supplemented cofactors (Supplementary Fig. 17b). This improved cyclization may be a consequence of poor 4a binding—if 4a is released rapidly then there is more available enzyme to bind 3 and catalyze its cyclization (Fig. 6d).
Discussion
Here we demonstrate how a mechanistic analysis of ISY led to the hypothesis that a separate cyclase is responsible for setting the stereochemistry of the iridoid framework. A comparative proteomic analysis allowed discovery of these cryptic enzymes. These NEPS enzymes demonstrate the plasticity and innovation characteristic of plant secondary metabolism enzymes30. NEPS3, for example, is an SDR by structure and sequence, yet its primary catalytic activity is non-oxidoreductive; NAD+ is utilized not as a co-substrate but as a protein structural scaffold. Interestingly, at very high NAD+ concentrations, NEPS3 can catalyze dehydrogenation with low catalytic efficiency, a phenomenon that may be a glimpse into its evolutionary past.
Due to its inherent reactivity, in situ generation of the substrate 3 was required to reveal the activity of the NEPS enzymes. Reactive non-isolable substrates appear elsewhere in plant specialized metabolism, including monoterpene indole alkaloids31 and lignan biosynthesis32,33. NEPS are reminiscent of dirigent proteins, proteins in lignan biosynthesis that control the stereoselective cyclization of a reactive intermediate that is generated by a separate enzyme32,33. There may be similar undiscovered steps in other metabolic pathways; these cannot be revealed in simple one-substrate, one-enzyme assays, but require multi-enzyme cascade strategies, which makes discovery of such enzymes challenging.
Based on the structural and mutant data, we propose that NEPS1/2 and NEPS3 catalyze cyclization in different fashions. NEPS1/2 appear to allow the enol 3 to proceed down a default ‘uncatalyzed’ path, forming the same product as that made in water (Fig. 2). Their key role in the cyclization appears to be to protect the intermediate from general acid-catalyzed tautomerization; there is no evidence to suggest that the mechanisms of NEPS1 and NEPS2 do not mirror the mechanisms operating in solution. NEPS3, on the other hand, has specific 7S-cis-cis-nepetalactol cyclase activity, binding to 3 possibly via Ser154 and exerting steric and/or electrostatic influence to enable formation of the cis-cis stereochemistry. Further analysis is required to determine whether NEPS3 catalyzes the [4 + 2] cyclization in a stepwise or concerted manner (that is, Michaelase or Diels-Alderase). The contrast between the ‘passive’ NEPS1/2 and ‘active’ NEPS3 mechanisms mirrors the dichotomy of intrinsic substrate reactivity versus enzyme influence in canonical terpene synthase mechanisms13.
Phylogenetic analysis suggests that the NEPS enzymes are unique to Nepeta. Yet ISYs from other organisms also do not catalyze iridoid cyclisation. Thus, it is likely that different cyclases, unrelated to NEPS, are operating in other iridoid-producing species. The absence of such cyclases from iridoid pathways reconstituted in microbial organisms may have negatively impacted yields34–36. The NEPS cyclases described here, especially NEPS2, a dedicated cis-trans cyclase, can be incorporated into such systems to improve yield.
This article describes how nepetalactone isomers can be formed in vitro by ISY and NEPS. The in planta roles of these enzymes in nepetalactone biosynthesis are suggested by their in vitro activities, trichome enrichment and phylogenetic distribution. However, although we have demonstrated the exquisite sensitivity of the in vitro enzyme cascades to conditions such as enzyme concentration and pH, we do not know the equivalent in planta conditions or whether other proteins are involved. Accordingly, further work is being conducted to understand the in planta biosynthesis, with a focus on how the nepetalactone stereochemistry is controlled and how the pathway has evolved.
We have revealed the enzymatic origin of cis-trans-nepetalactone and cis-cis-nepetalactone, compounds in Nepeta responsible for cat attraction and insect repellence. In doing so, we have discovered three novel enzymes: two dedicated cyclases and one multifunctional cyclase-dehydrogenase. The structure of one of these enzymes reveals it has repurposed a dehydrogenase structure for a different catalytic function. We have shown that iridoid biosynthesis involves uncoupled activation and cyclization: a reactive, non-isolable enol is formed by reduction and cyclized by separate enzymes. Such findings will contribute to synthetic biology metabolic reconstructions and inform the de novo design of (bio)synthetic pathways; they also highlight the dynamic and innovative nature of plant natural product biosynthesis.
Methods
Proteome analysis
Whole leaves from N. mussinii (1.5 g × 4, obtained from Herbal Haven, Coldhams Farm, Rickling, Saffron Walden, CB11 3YL, UK) were placed into a 50 mL centrifuge tube, and to this was added powdered dry ice (approximately 20 mL). This was vortexed (1 min, 20,000 r.p.m.), the dry material was poured through meshes (200 μm and 100 μm) and the material passing through the meshes was collected. To maximize yield, leaves and tubes were washed with cold isolation buffer (200 mM sorbitol, 25 mM HEPES, 5 mM MgCl2, 5 mM succinic acid, 1 mM EGTA, 0.5 mM NaH2PO4, 5 mM DTT, 2 mM sucrose, pH 7), and this was filtered through a mesh (100 μm) and collected. Samples were inspected by light microscopy to determine success of the extraction step. The samples were centrifuged (700 × g, 10 min), and the supernatant was removed to yield a pellet containing trichomes. Leaves and trichome-depleted leaves were frozen in liquid nitrogen and homogenized with a pestle and mortar. Trichomes were resuspended in extraction buffer (1 mL, 0.1 M Tris-HCl pH 8, 2% w/v SDS, 30% w/v sucrose, 5% v/v 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride), vortexed and homogenized by sonication (10 s ON, 10 s OFF, 20 cycles). Tris-buffered phenol (1 mL) was added to homogenized samples, then vortexed (10 min) and centrifuged (10,000 × g, 10 min). The upper layer was obtained, the extraction buffer (1 mL) was added, and the mixture was vortexed (5 min) and centrifuged (10,000 × g, 10 min). The upper layer was transferred to a new tube and to this, four volumes of methanol/0.1 M NH4Ac (1:1, −20°C) were added, and the sample was mixed and then incubated at −20°C overnight. The precipitated protein was pelleted by centrifugation, and the pellet was washed twice with methanol/0.1 M NH4Ac (1:1, −20°C) and then with acetone (−20°C). The acetone was removed, and the pellet air-dried.
Protein pellets from leaves and depleted leaves were dissolved in 5% sodium deoxycholate (SDC), 50 mM phosphate buffer pH 8, pellets from trichomes in 1% SDC, 50 mM phosphate buffer pH 8. Protein concentration was determined using the Direct Detect spectrometer (Merck Millipore, UK). The total protein amount was 400 μg for leaves, 225 μg for depleted leaves and 20 μg for trichomes. Samples were treated with DTT and iodoacetamide to reduce and alkylate cysteine residues. The total trichome sample and 50 μg of the leaf samples were digested with trypsin. SDC was removed by acid precipitation, and aliquots of approximately 1μg were used for data-dependent LC–MS/MS analysis on an Orbitrap-Fusion mass spectrometer (Thermo Fisher, Hemel Hempstead, UK) equipped with an UltiMate 3000 RSLCnano System using an Acclaim PepMap C18 column (2 μm, 75 μm × 500 mm, Thermo). The samples were loaded and trapped using a pre-column which was then switched in-line to the analytical column for separation. Peptides were eluted with a gradient of 6–38% acetonitrile in water/0.1% formic acid at a rate of 0.4% min−1. The column was connected to a 10-μm SilicaTip nanospray emitter (New Objective, Woburn, MA, USA) for infusion into the mass spectrometer. Data-dependent analysis was performed using a parallel HCD/CID fragmentation method with the following parameters: positive ion mode, orbitrap MS resolution = 60 k, mass range (quadrupole) = 300–1,500 m/z, MS2 in ion trap, threshold 2e4, isolation window 1.6 Da, charge states 2–5, inject for all available parallelizable time with 3 s cycle time, AGC target 2e3, max inject time 100 ms, dynamic exclusion 1 count within 10 s and 60 s exclusion, exclusion mass window ± 7 p.p.m. MS scans were saved in profile mode while MS/MS scans were saved in centroid mode.
Raw files from the Orbitrap were processed with MaxQuant (version 1.5.3.30)37 to generate recalibrated peaklist files, which were used for database searches with the Mascot search engine (version 2.4.1, Matrixscience, London). A predicted peptide library was generated from the N. mussinii transcriptome19 by translation of predicted open reading frames (minimum size 100 bp, ATG start codon, start/stop codons can be outside sequence). This predicted peptide library was annotated and used for the Mascot database search using trypsin/P with two missed cleavages, carbamidomethylation (C) as fixed and oxidation (M), acetylation (protein N terminus), and deamidation (N,Q) as variable modifications. Mass tolerances were 6 p.p.m. for precursor ions and 0.6 Da for fragment ions. Mascot search results were imported into the Scaffold software and the Scaffold quantitative value (normalized total spectra) was used for a pairwise comparison between trichome and trichome-depleted leaves samples using Fisher’s exact test with Benjamini-Hochberg multiple test correction. MEP pathway enzymes were putatively identified by functional annotation. Nepetalactol dehydrogenase candidates were selected by the following criteria: functionally uncharacterized, statistically significant trichome enrichment (P < 0.05) and enzyme class EC 1.1.1. (oxidoreductase, acting on alcohols, NAD(P)+ dependence).
Cloning
cDNA was obtained as described in Sherden et al19. Primers for dehydrogenase candidates were designed based on the transcriptome sequence, and 5′-overhangs were added for cloning into the pOPINF vector (forward: 5′-AAGTTCTGTTTCAGGGCCCG-3′, reverse: 5′-CTGGTCTAGAAAGCTTTA-3′). The primers were used to PCR amplify the genes from the cDNA. These were purified from an agarose gel and then cloned into a linearized pOPINF vector using an InFusion HD cloning kit (Clontech). Plasmid sequences were verified with Sanger sequencing. Cloned sequences NEPS2 and NEPS3 did not precisely match full-length sequences from the transcriptome but appeared to contain regions from multiple transcripts. The poor assembly of these genes is indicative of close paralogs or alternative splicing. SoluBL21 E. coli cells (Genlantis) were transformed with the plasmids for expression.
Mutations were introduced into the NEPS1 and NEPS3 genes by PCR. The mutation method is described in Kries et al20. First, two gene fragments were amplified. The first fragment was cloned using the gene-specific forward primer from the original cloning and a reverse primer ending 5′ of the mutation site (reverse mutation primer). The second fragment cloned employed a forward mutation primer encompassing the mutation site (forward mutation primer) and the reverse primer from the original cloning. The fragments were gel-purified, and then a third PCR, using the original gene-specific forward and reverse primers, was used to assemble the fragments. This gene-length amplicon was gel-purified and then cloned into the pOPINF vector using an InFusion HD cloning kit (Clontech). Plasmid sequences were verified with Sanger sequencing. SoluBL21 cells (Genlantis) were transformed with the plasmids for expression.
Enzyme expression and purification
Expression strain cells containing the plasmids of interest were grown overnight (LB media, 10 mL, 100 μg/mL carbenicillin). 2xYT media with 100 μg/mL carbenicillin was innoculated with overnight culture (5% v/v) and grown at 37 °C until OD600 = 0.5. The culture was then grown at 18 °C until OD600 = 0.6–0.8 and then protein production was induced with addition of IPTG (500 μM). The cells were incubated at 18 °C for 16 h before harvesting by centrifugation (4,000 × g, 10 min). If the pellets were not used immediately, they were washed in PBS before storage at −20°C.
For the lysate assays used for screening for dehydrogenase activity, cultures (1 mL) of each dehydrogenase candidate were grown as described above. Cell pellets were resuspended in BugBuster MasterMix (100 μL) (Merck), incubated at 4 °C for 10 min and then centrifuged (20,000 × g, 10 min). Supernatant lysate (1 μL) was added to a mixture (100 μL total) containing cis-trans-nepetalactol 4a (0.5 mM), NAD+ (1 mM) and sodium phosphate buffer (50 mM, pH 8.8). The reactions were followed for 30 min on a FLUOStar Omega multiplate reader (BMG Labtech) using absorbance at 340 nm (positioning delay 0.2 s, 22 flashes per well, 32 s cycle time, 1 s 200 r.p.m. shaking each cycle). For each candidate, two control samples lacking both NAD+ and 4a or lacking just 4a were included. Control reactions with empty vector were also measured. Averages of absorbance measurements between 0–5 min and 25–30 min were obtained and compared to identify increases in absorbance (that is, formation of NADH).
For initial screening of NEPS1 and NEPS3 variants, cultures (10 mL) of each NEPS variant were grown as described above. Cell pellets were resuspended in BugBuster MasterMix (1 mL, with cOmplete EDTA-free protease inhibitors (Roche)), incubated at 4 °C for 20 min and then centrifuged (20,000 × g, 20 min). Nickel-NTA agarose (100 μL) (Qiagen) was washed in binding buffer (50 mM Tris–HCl pH 8, 50 mM glycine, 5% v/v glycerol, 0.5 M NaCl, 20 mM imidazole, 1 mM DTT) and added to the supernatant lysate, and the mixture was incubated at 4 °C for 1 h, gently rocking. The mixture was centrifuged (1,000 × g, 1 min) and the supernatant discarded. The Ni-NTA pellet was washed twice with 1 mL binding buffer, and then elution buffer (50 mM Tris-HCl pH 8, 50 mM glycine, 5% v/v glycerol, 0.5 M NaCl, 500 mM imidazole, 1 mM DTT) was added (500 μL). The mixture was centrifuged (1,000 × g, 1 min), and the supernatant was collected and filtered (Ultrafree-MC VV Centrifugal Filter, Merck). The buffer was exchanged into sample buffer (20 mM HEPES pH 7.5, 150 mM NaCl) by four concentration-dilution steps using a centrifugal filtration (Amicon Ultra 10 kDa MWCO, (Merck)). The proteins were aliquoted, flash frozen in liquid nitrogen and stored at −80 °C. SDS–PAGE analysis and spectrophotometric analysis (absorbance at 280 nm) was used to check purity and approximate quantity of protein.
For all enzyme assays, excluding the activity screens described above, and for crystal trials, cultures (1 L) were grown as described above. Cell pellets were resuspended in lysis buffer (binding buffer plus cOmplete EDTA-free protease inhibitors (Roche) and 0.4 mg/mL lysozyme) and incubated at 4 °C for 30 min. The lysate was then centrifuged (35,000 × g, 45 min, 4 °C) and the supernatant filtered (Minisart NML Plus 0.7 μm GF (Sartorius)). The filtrate was applied to a 5 mL HisTrap HP column (GE Life Sciences) attached to a ÄKTA pure system (GE Life Sciences). The column was washed with binding buffer until no protein could be detected coming off the column (detection using absorbance at 280 nm). The protein of interest was then eluted by application of elution buffer to the column. The eluted protein was pooled, filtered (PES membrane, 0.45 μm (Starlab)) and further purified by size-exclusion chromatography on a Superdex 200 16/60 GF column (GE Life Sciences). During this process, the protein was exchanged into sample buffer. Eluant fractions were analyzed by SDS–PAGE, and those containing the protein of interest were pooled, concentrated by centrifugation (Amicon 10 kDa MWCO), flash frozen in liquid nitrogen and stored at −80 °C. Protein purity was confirmed by SDS-PAGE analysis. Protein concentration was determined by spectrophotometric analysis at 280 nm using extinction coefficients calculated by ProtParam (http://web.expasy.org/protparam/).
Enzyme assays
Kinetics measurements were conducted spectrophotometrically on a PerkinElmer Lambda 35 instrument at a wavelength of 340 nm and in cuvettes with 1 cm path length. The reactions were conducted at 25 °C and with 50 mM HEPES pH 8.0, and 100 NaCl. Reactions were initiated by addition of the enzyme, and absorbance values were recorded at a rate of 1 Hz. Enzyme concentration was varied from 0.025–0.25 μM to maintain a linear rate. The R software environment was used to fit linear initial rates over the first 20 s of the enzyme reaction. For Michaelis–Menten experiments, sufficient data points were collected to ensure fit parameters were estimated with P < 0.001 (this was n = 9–11). The Michaelis–Menten equation was fitted to the data points in R by the NLS (nonlinear least-squares) function to obtain values for the kinetic parameters. Kinetic parameters are reported as a best fit estimate ± SE, with the exact P value for each parameter. Activity measurements for cofactor and pH dependence were performed on a on a FLUOStar Omega multiplate reader (details above). Relative rates were calculated over the initial 10 min of reaction. End-point assays with purified enzymes were conducted at a total volume of 100 μL. Reactions using either 8-oxogeranial 2 or 8-oxocitronellal 6 as a substrate contained 1% v/v MeCN. In experiments examining the effect of buffer concentrations, enzymes were diluted with water. Trace residual concentrations of sample buffer in the enzyme sample were too low to effect product profiles. All reactions were conducted at 30 °C. Reactions containing 8-oxogeranial 2 as a substrate were incubated for 3 h, whereas reactions with 8-oxocitronellal 6 were incubated for 16 h, unless noted. At reaction termination, a camphor standard (10 μL, stock 1 mM in MeCN) and EtOAc (100 μL) were added. The reactions were vortexed, centrifuged (10,000 × g, 10 min) and the organic layer was used in GC–MS analysis. The concentrations of components in reactions varied extensively, and so for each reaction the figure or figure legend describes the exact conditions employed.
Gas chromatography–mass spectroscopy
Samples were injected in split mode (2μL, split ratio 5:1) at an inlet temperature of 220 °C on a Hewlett Packard 6890 GC–MS equipped with a 5973 mass selective detector (MSD) and an Agilent 7683B series injector and autosampler. Separation was performed on a Zebron ZB5-HT-INFERNO column (5% phenyl methyl siloxane; length: 35 m; diameter: 250μm) with guard column. Helium was used as mobile phase at a constant flow rate of 1.2 mL/min and average velocity 37 cm/s. After 5 min at 80 °C, the column temperature was increased to 110 °C at a rate of 2.5 K/min, then to 280 °C at 120 K/min, and kept at 280 °C for another 4 min. A solvent delay of 5 min was allowed before collecting MS spectra at a fragmentation energy of 70 eV. An internal standard of (+)-camphor was used for retention time calibration. Chemically characterized standards verified by NMR were used to identify compounds by retention time and electron impact spectra. All chromatograms presented in figures are total ion chromatograms (TICs).
Crystallization and data collection
NEPS3 was purified as described above. The protein (7 mg/mL) was thawed and incubated with C3 protease (0.36 mg/mL) for 1 h at room temperature. The protein was filtered (Ultrafree-MC VV Centrifugal Filter) and passed through Nickel-NTA agarose, removing the cleaved His-Tag and tagged C3-protease. The eluant (NEPS3 without tag) was concentrated to 7 mg/mL (Amicon 10 kDa MWCO) in sample buffer. Crystals were formed using the hanging-drop method (3 μL total, 1:2 protein:buffer, 1 mM NAD+ final concentration), using a precipitant comprised of 29% w/v PEG 4000 with 0.1 M MES pH 6.5. The crystals were cryo-protected with crystallization buffer containing 20% v/v ethylene glycol, then flash-cooled in liquid nitrogen using LithoLoops (Molecular Dimensions) and stored in Unipuck cassettes (MiTeGen) before data collection. Crystals were transferred robotically to the goniostat on beamline I03 (wavelength 0.9763 Å) at the Diamond Light Source (Oxfordshire, UK), and maintained at −173 °C with a Cryojet cryocooler (Oxford Instruments). X-ray diffraction data were recorded to 1.4 Å resolution using a Pilatus 6 M hybrid photon counting detector (Dectris), then integrated and scaled using XDS38, via the XIA2 expert system39, and then merged in a primitive monoclinic unit cell using AIMLESS;40 the resultant data collection statistics are summarized in Supplementary Table 3.
Structure solution and refinement
The majority of the downstream analysis was performed through the CCP4i2 graphical user interface (http://www.ccp4.ac.uk/)41. A molecular replacement template for the NEPS3 subunit was prepared from PDB entry 2BGK 42, with which it shares 42% sequence identity, using SCULPTOR43. The high-resolution of the X-ray data was indicative of a low solvent content, and a value of 40% was estimated for four copies of the ~28 kDa subunit in the asymmetric unit (ASU). Moreover, inspection of a self-rotation function revealed several noncrystallographic two-fold axes, in addition to the crystallographic two-fold. The functional unit of the template structure, secoisolariciresinol dehydrogenase, is a homotetramer with 222 symmetry, which prompted us to prepare an equivalent tetramer from the SCULPTOR output. The latter was used as the input for PHASER44 which produced a very convincing solution (TFZ score = 15) in space group P21. This model was rebuilt with BUCCANEER45 and then improved with iterations of manual editing in COOT46 and refinement with REFMAC5 (ref. 47). Initially, we had modeled elongated peaks of electron density, adjacent to the nicotinamide ring in each of the four independent active sites in the tetramer, as discretely disordered water molecules. However, after refinement, these were associated with significant amounts of positive difference electron density and they had refined temperature factor values significantly lower than those of the surrounding atoms, indicative of more electron dense species. Given that in each case the atom was within 3.6 Å of two backbone amides, it was re-assigned as a discretely disordered chloride ion (NaCl was present at 150 mM in the sample buffer). After refinement, the chloride ion temperature factors were comparable to, or slightly higher than, those of the surrounding atoms. The statistics of the final refined model are shown in Supplementary Table 3. Ramachandran statistics, as calculated using MolProbity48, are: 97.6/2.4/0.0 (favored/allowed/disallowed, %). This model was deposited in the Protein Data Bank with accession code 6F9Q. Figures and structural alignments were made using UCSF-Chimera (http://www.rbvi.ucsf.edu/chimera/). Homology models of NEPS1 and NEPS2 were constructed using the iTasser server49. NAD+ was added to the structures (using NEPS3 as a model) and then the complexes were energy minimized using the Yasara energy minimization server50. Docking calculations were performed with AutoDock Vina (exhaustiveness = 8)51.
Phylogenetics and sequence analysis
Protein multiple sequence alignment NEPS and MpIPDH was performed with ClustalW2 (ref. 52) and depicted with ESPript53. For phylogenetic analysis, Lamiaceae and outgroup sequences were obtained from the Mint Genome Project (http://mints.plantbiology.msu.edu/, NCBI BioProject PRJNA359989). Sequences with homology to NEPS were identified by Orthogroup analysis54. Mentha x piperita isopiperitenol dehydrogenase (MpIPDH, AY641428) was also included in the analysis55. Prior to alignment, incomplete sequences and duplicated sequences were removed. The codon alignment was performed with MUSCLE56, and then manually reviewed and curated to remove ragged ends. Phylogenetic tree inference was performed with IQ-Tree 1.5.4 (ref. 57). First, the substitution model (TIM + 1 + G4) was determined by ModelFinder58. The maximum likelihood tree was then determined heuristically over 146 iterations, and branch support values were calculated using ultrafast bootstrapping59 (1,000 replicates). The tree was visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Statistics
The statistical significance of protein abundance between proteomes was calculated using Fisher's exact test (n = 2) with Benjamini–Hochberg multiple test correction, implemented within Scaffold software. For Michaelis–Menten experiments, sufficient initial rate data points were collected to ensure fitted parameters were estimated with P < 0.001 (n = 9–11). Michaelis–Menten equation was fitted to the data points in R by the NLS (nonlinear least-squares) function to obtain values for the kinetic parameters. Kinetic parameters are reported as a best fit parameter estimate ± parameter estimate fit SE, with the exact P value for each parameter.
Supplementary Material
Supplementary information is available for this paper at https://doi.org/10.1038/s41589-018-0185-2.
Acknowledgements
We acknowledge funding from UK Biotechnological and Biological Sciences Research Council (BBSRC) and Engineering and Physical Sciences Research Council (EPSRC) joint-funded OpenPlant Synthetic Biology Research Centre (BB/L014130/1) and from the National Science Foundation Plant Genome Research Program (IOS- 1444499). For the X-ray data collection, we acknowledge Diamond Light Source for access to beamline I03 under proposal MX13467, with support from the European Community’s Seventh Framework Program (FP7/2007–2013) under grant agreement 283570 (BioStruct-X). We are grateful to: P. Brett for assistance with GC–MS analysis, M. Vigoroux for assistance with proteome annotations, and N. Sherden and H. Kries for providing chemical and genetic material. We also thank K. Houk, J. Fell, H. Kries and D. Whitaker for discussions concerning the iridoid synthase and cyclisation mechanisms.
Footnotes
Benjamin R. Lichman: 0000-0002-0033-1120
Gabriel R. Titchiner: 0000-0002-6378-9884
Clare E. M. Stevenson: 0000-0001-6695-8201
David M. Lawson: 0000-0002-7637-4303
Sarah E. O’Connor: 0000-0003-0356-6213
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The sequences of N. mussinii NEPS enzyme have been deposited in GenBank/EMBL/DDBJ with the accession codes: MG677124 (NmNEPS1), MG677125 (NmNEPS2) and MG677126 (NmNEPS3). The NAD+ bound NmNEPS3 (7S-cis-cis-nepetalactol cyclase) X-ray structure has been deposited in the PDB with the accession code 6F9Q. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD008704. Detailed experimental procedures and can be found in the supplementary information. The authors declare that all other data supporting the findings of this study are available within this article and its Supplementary Information or from the authors upon reasonable request.
Author contributions
S.E.O.’C. designed and supervised the project; B.R.L. performed molecular cloning, protein purification, enzyme assays, trichome isolation, chemical synthesis, phylogenetic analysis, homology modeling and computational docking; G.S. performed proteome analysis; M.O.K. assisted with protein purification, compound isolation and chemical synthesis; B.R.L., G.R.T. and C.E.M.S. performed crystallization trials and obtained crystals; B.R.L., G.R.T. and D.M.L. refined structures; B.R.L. and S.E.O.’C. wrote the manuscript.
Competing interests
A UK patent application has been submitted based on the work reported here (GB1808663.7).
Additional information
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McElvain SM, Bright RD, Johnson PR. The constituents of the volatile oil of catnip. I. Nepetalic acid, nepetalactone and related compounds. J Am Chem Soc. 1941;63:1558–1563. [Google Scholar]
- 2.Formisano C, Rigano D, Senatore F. Chemical constituents and biological activities of Nepeta species. Chem Biodivers. 2011;8:1783–1818. doi: 10.1002/cbdv.201000191. [DOI] [PubMed] [Google Scholar]
- 3.Todd NB. Inheritance of the catnip response in domestic cats. J Hered. 1962;53:54–56. doi: 10.1093/oxfordjournals.jhered.a107121. [DOI] [PubMed] [Google Scholar]
- 4.Bates RB, Sigel CW. Terpenoids. cis-trans- and trans-cis- nepetalactones. Experientia. 1963;19:564–565. [Google Scholar]
- 5.Waller GR, Price GH, Mitchell ED. Feline attractant, cis,trans- nepetalactone: metabolism in the domestic cat. Science. 1969;164:1281–1282. doi: 10.1126/science.164.3885.1281. [DOI] [PubMed] [Google Scholar]
- 6.Dawson G, et al. Identification of an aphid sex pheromone. Nature. 1987;325:614–616. [Google Scholar]
- 7.Clark LJ, Hamilton JGC, Chapman JV, Rhodes MJC, Hallahan DL. Analysis of monoterpenoids in glandular trichomes of the catmint Nepeta racemosa. Plant J. 1997;11:1387–1393. [Google Scholar]
- 8.Eisner T. Catnip: its raison d’etre. Science. 1964;146:1318–1320. doi: 10.1126/science.146.3649.1318. [DOI] [PubMed] [Google Scholar]
- 9.Birkett MA, Hassanali A, Hoglund S, Pettersson J, Pickett JA. Repellent activity of catmint, Nepeta cataria, and iridoid nepetalactone isomers against Afro-tropical mosquitoes, ixodid ticks and red poultry mites. Phytochemistry. 2011;72:109–114. doi: 10.1016/j.phytochem.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Rajaonarivony JIM, Gershenzon J, Croteau R. Characterization and mechanism of (4 S)-limonene synthase, a monoterpene cyclase from the glandular trichomes of peppermint (Mentha x piperita) Arch Biochem Biophys. 1992;296:49–57. doi: 10.1016/0003-9861(92)90543-6. [DOI] [PubMed] [Google Scholar]
- 11.Poulter CD, Argyle JC, Mash EA. Letter: Prenyltransferase. New evidence for an ionization-condensation-elimination mechanism with 2-fluorogeranyl pyrophosphate. J Am Chem Soc. 1977;99:957–959. doi: 10.1021/ja00445a056. [DOI] [PubMed] [Google Scholar]
- 12.Baunach M, Franke J, Hertweck C. Terpenoid biosynthesis off the beaten track: unconventional cyclases and their impact on biomimetic synthesis. Angew Chem Int Ed Engl. 2015;54:2604–2626. doi: 10.1002/anie.201407883. [DOI] [PubMed] [Google Scholar]
- 13.Tantillo DJ. Importance of inherent substrate reactivity in enzyme-promoted carbocation cyclization/rearrangements. Angew Chem Int Ed Engl. 2017;56:10040–10045. doi: 10.1002/anie.201702363. [DOI] [PubMed] [Google Scholar]
- 14.Miettinen K, et al. The seco-iridoid pathway from Catharanthus roseus. Nat Commun. 2014;5 doi: 10.1038/ncomms4606. 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geu-Flores F, et al. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- 16.O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 17.Alagna F, et al. Identification and characterization of the iridoid synthase involved in oleuropein biosynthesis in olive (Olea europaea) fruits. J Biol Chem. 2016;291:5542–5554. doi: 10.1074/jbc.M115.701276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kries H, Kellner F, Kamileen MO, O’Connor SE. Inverted stereocontrol of iridoid synthase in snapdragon. J Biol Chem. 2017;292:14659–14667. doi: 10.1074/jbc.M117.800979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherden NH, et al. Identification of iridoid synthases from Nepeta species: Iridoid cyclization does not determine nepetalactone stereochemistry. Phytochemistry. 2018;145:48–56. doi: 10.1016/j.phytochem.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kries H, et al. Structural determinants of reductive terpene cyclization in iridoid biosynthesis. Nat Chem Biol. 2016;12:6–8. doi: 10.1038/nchembio.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, et al. Structures of iridoid synthase from Cantharanthus roseus with bound NAD+, NADPH, or NAD+ /10-oxogeranial: reaction mechanisms. Angew Chem Int Ed Engl. 2015;54:15478–15482. doi: 10.1002/anie.201508310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin L, et al. Structure of iridoid synthase in complex with NADP+/8-oxogeranial reveals the structural basis of its substrate specificity. J Struct Biol. 2016;194:224–230. doi: 10.1016/j.jsb.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Dawson GW, Pickett JA, Smiley DWM. The aphid sex pheromone cyclopentanoids: synthesis in the elucidation of structure and biosynthetic pathways. Bioorg Med Chem. 1996;4:351–361. doi: 10.1016/0968-0896(96)00012-0. [DOI] [PubMed] [Google Scholar]
- 24.Liblikas I, et al. Simplified isolation procedure and interconversion of the diastereomers of nepetalactone and nepetalactol. J Nat Prod. 2005;68:886–890. doi: 10.1021/np049647d. [DOI] [PubMed] [Google Scholar]
- 25.Cucinotta CS, Ruini A, Catellani A, Stirling A. Ab initio molecular dynamics study of the keto-enol tautomerism of acetone in solution. Chemphyschem. 2006;7:1229–1234. doi: 10.1002/cphc.200600007. [DOI] [PubMed] [Google Scholar]
- 26.Alagona G, Ghio C, Nagy PI. The catalytic effect of water on the keto-enol tautomerism. Pyruvate and acetylacetone: a computational challenge. Phys Chem Chem Phys. 2010;12:10173–10188. doi: 10.1039/c003999c. [DOI] [PubMed] [Google Scholar]
- 27.Schreiber SL, Meyers HV, Wiberg KB. Stereochemistry of the intramolecular enamine/enal (enone) cycloaddition reaction and subsequent Transformations. J Am Chem Soc. 1986;108:8274–8277. [Google Scholar]
- 28.Hallahan DL, West JM, Smiley DWM, Pickett JA. Nepetalactol oxidoreductase in trichomes of the catmint Nepeta racemosa. Phytochemistry. 1998;48:421–427. [Google Scholar]
- 29.Moummou H, Kallberg Y, Tonfack LB, Persson B, van der Rest B. The plant short-chain dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns. BMC Plant Biol. 2012;12:219. doi: 10.1186/1471-2229-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng JK, Noel JP. The remarkable pliability and promiscuity of specialized metabolism. Cold Spring Harb Symp Quant Biol. 2012;77:309–320. doi: 10.1101/sqb.2012.77.014787. [DOI] [PubMed] [Google Scholar]
- 31.Tatsis EC, et al. A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00154-x. 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davin LB, et al. Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science. 1997;275:362–366. doi: 10.1126/science.275.5298.362. [DOI] [PubMed] [Google Scholar]
- 33.Pickel B, et al. An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew Chem Int Ed Engl. 2010;49:202–204. doi: 10.1002/anie.200904622. [DOI] [PubMed] [Google Scholar]
- 34.Brown S, Clastre M, Courdavault V, O’Connor SE. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc Natl Acad Sci USA. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell A, et al. Engineering of a nepetalactol-producing platform strain of Saccharomyces cerevisiae for the production of plant seco-iridoids. ACS Synth Biol. 2016;5:405–414. doi: 10.1021/acssynbio.5b00289. [DOI] [PubMed] [Google Scholar]
- 36.Billingsley JM, et al. Engineering the biocatalytic selectivity of iridoid production in Saccharomyces cerevisiae. Metab Eng. 2017;44:117–125. doi: 10.1016/j.ymben.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch WXDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winter G. Xia2: An expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 40.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youn B, Moinuddin SGA, Davin LB, Lewis NG, Kang C. Crystal structures of apo-form and binary/ternary complexes of Podophyllum secoisolariciresinol dehydrogenase, an enzyme involved in formation of health-protecting and plant defense lignans. J Biol Chem. 2005;280:12917–12926. doi: 10.1074/jbc.M413266200. [DOI] [PubMed] [Google Scholar]
- 43.Bunkóczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krieger E, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins. 2009;77(Suppl. 9):114–122. doi: 10.1002/prot.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mint Evolutionary Genomics Consortium.. Electronic address: buell@msu.edu. Phylogenomic Mining of the Mints Reveals Multiple Mechanisms Contributing to the Evolution of Chemical Diversity in Lamiaceae. Mol Plant. 2018;11:1084–1096. doi: 10.1016/j.molp.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Ringer KL, Davis EM, Croteau R. Monoterpene metabolism. Cloning, expression, and characterization of (-)-isopiperitenol/(-)-carveol dehydrogenase of peppermint and spearmint. Plant Physiol. 2005;137:863–872. doi: 10.1104/pp.104.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edgar RC. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.