Abstract
Understanding the causal relationship between neural activity and behavior requires the ability to perform rapid and targeted interventions in ongoing activity. Here we describe a closed-loop all-optical strategy for dynamically controlling neuronal activity patterns in awake mice. We rapidly tailor and deliver two-photon optogenetic stimulation based on online readout of activity using simultaneous two-photon imaging, thus enabling the manipulation of neural circuit activity ‘on the fly’ during behavior.
Understanding how the spatiotemporal patterns of activity in neural circuits drive behavior represents a fundamental problem in neuroscience. To define causal relationships between activity patterns and behavior we must not only read out activity patterns in identified neurons, but also make precisely targeted interventions. The ability to simultaneously read out and manipulate activity in neural circuits using an “all-optical” combination1 of imaging and photostimulation now allows optogenetic interventions to be targeted to individual neurons in the mammalian brain in vivo2–7 based on their functional signature2, 4. However, this targeting has hitherto been guided by offline analysis, and typically by averaging activity across multiple trials. Since the activity of individual neurons can be highly variable, and their contribution to population activity can vary from moment to moment8–11, it is essential to be able to target photostimulation guided by online measurements of activity. If the readout and targeting is sufficiently rapid this offers the prospect of online closed-loop control12 in which photostimulation can be targeted to the appropriate cells and titrated to enhance and transform evolving patterns of neural activity.
Here we implement an online feedback strategy to close the loop between optical readout (using a genetically encoded calcium indicator, GCaMP6) and optical stimulation (using an optogenetic actuator, C1V1). Our closed-loop module accesses the raw pixel data streaming in real time from the microscope, enabling rapid online analysis of population activity acquired through two-photon calcium imaging. This online analysis is used to reconfigure photostimulation patterns and is integrated with the hardware to deliver them (Fig. 1a and Supplementary Fig. 1). To demonstrate the range of new experiments enabled by our flexible closed-loop approach, we use it to perform different classes of dynamic, activity-guided circuit manipulations at cellular resolution in vivo.
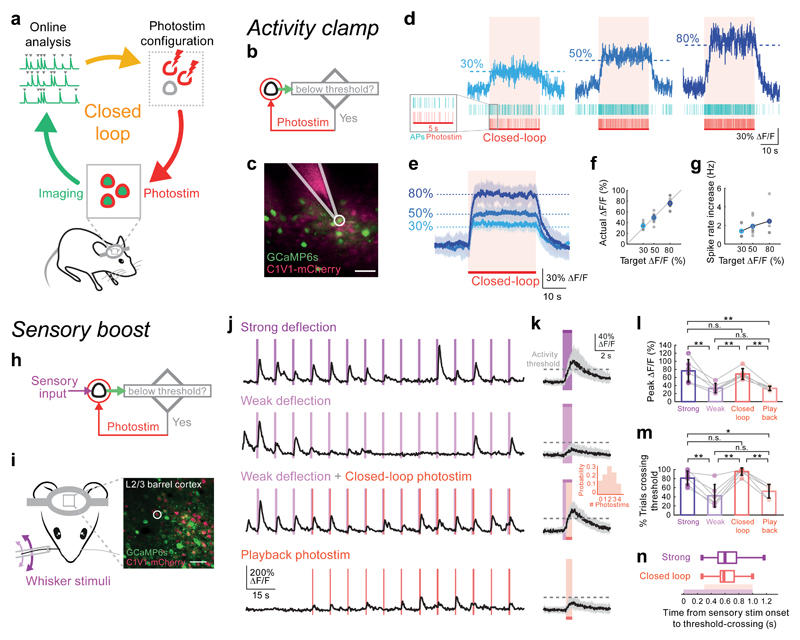
Figure 1. Targeted closed-loop all-optical readout and control in vivo.
(a) Schematic illustration of the closed-loop all-optical control system in which two-photon imaging is combined with simultaneous two-photon photostimulation. Readout and control are coupled by tailoring the photostimulation according to measurement of ongoing activity. Activity traces are extracted online from two-photon calcium imaging movies and real-time analysis detects events used to tailor the photostimulation rate and/or targets.
(b) Experimental strategy for the ‘activity clamp’ paradigm. A single neuron is photostimulated (spiral scan; 20 μm in diameter; 10-20 ms duration; 30 mW on sample, Fidelity laser or 10 mW on sample, Satsuma laser) whenever its calcium signal recorded online falls below a defined threshold.
(c) Simultaneous cell-attached recording from a neuron (white circle) that was under ‘activity clamp’ in layer 2/3 of mouse somatosensory cortex expressing GCaMP6s (green) and C1V1 (pink, scale bar 50 μm, 50 mW on sample, Chameleon laser). Alexa 594 was put in the pipette for visualization.
(d) Top: calcium transient from a single neuron clamped at 3 different activity levels (30, 50, 80% ΔF/F; 30 sec clamping period). Middle: raster plot of electrophysiologically recorded action potentials (APs) from the neuron in (c) during single trials. Bottom: photostimulation times.
(e) Average activity clamp (30 second clamp period) of 18 cells in somatosensory and visual cortex of awake animals. Shaded area is SD. Same photostimulation parameters as in (d). n = 18 cells in 6 animals.
(f) Calcium signal levels achieved during activity clamp in (e) versus the target calcium signal levels. Gray points are individual cells, colored points are average.
(g) Spike rate increase during activity clamp at three different target calcium signal levels, measured by simultaneous electrophysiological recording (n = 7 cells in 5 mice under anesthesia).
(h) Experimental strategy for closed-loop boosting of sensory responses: a sensory-responsive neuron was photostimulated if the sensory-evoked activity failed to reach a threshold.
(i) Left: configuration of whisker stimulation and two-photon imaging. Right: a field of view (scale bar, 50 μm, 30 mW on sample, Chameleon laser) showing neurons co-expressing GCaMP6s and C1V1-2A-mCherry in the C2 barrel of mouse somatosensory cortex. A neuron that responded to single whisker deflections was selected as the target (white circle).
(j) Calcium transients from the target neuron in (i) recorded while the mouse was presented with a train of single whisker deflections (20 Hz, 1 sec sinusoidal deflection once every 10 sec). Top row: the neuron responds strongly to strong whisker deflections. Second row: the neuron responds weakly and unreliably to weak whisker stimuli. Third row: neural responses to weak whisker deflection were boosted by photostimulation (red lines; stimulation duration 50 ms, 9 mW on sample, Satsuma laser) if the activity recorded online is below a pre-defined threshold after 300 ms from the onset of the whisker deflection until the end of the deflection. Bottom row: playback photostimuli in the closed-loop trials.
(k) Calcium transients during individual sensory stimulation trials (black traces are the median value of all trials; gray shaded areas show the inter-quartile ranges (25% to 75% quantiles); n = 6 stimulus responsive neurons in 4 animals). First panel: large increase in calcium signal during a strong sensory stimulus. Second panel: weak responses to weak sensory stimuli. Third panel: photostimulation boosted the calcium signal to pass the user-defined activity threshold (30% ΔF/F) during closed-loop intervention (stimulation duration 50 - 80 ms; 6 - 12 mW on sample, Satsuma laser; inset shows that on average 2 photostimuli were delivered per trial). Fourth panel: photostimulation alone was not able to drive the calcium signal above threshold. Purple: 1 sec whisker stimulation; red: closed-loop intervention.
(l) The peak neural response to weak sensory stimuli with closed-loop intervention (Average ΔF/F: 68.6% ± 13.5%) was similar to the response to strong sensory stimuli (Average ΔF/F: 76.5% ± 27.9%); P = 0.00014, one-way ANOVA, ** P = 0.0013, strong versus weak (Average ΔF/F: 33.5% ± 11.5%); ** P = 0.0011, weak versus closed-loop; ** P = 0.007 closed-loop versus playback; n.s. P = 0.84, strong versus closed-loop; n.s. (not significant) P = 0.99, weak versus playback; ** P = 0.0011, strong versus playback; n = 6 cells in 4 animals. Measure of center is mean; error bars correspond to standard deviation).
(m) Percentage of trials that showed strong neural activity (ΔF/F response to stimulation > 30%) after the sensory stimulus. The ΔF/F response to sensory stimulation passed the activity threshold in 42.2 ± 24.7% of weak sensory stimulation trials, 81.1 ± 15.4% of strong sensory stimulation trials and 95.6 ± 8.1% of weak sensory stimulation trials with closed-loop photostimulation. When closed-loop intervention was enabled, the neural response to weak sensory stimuli was as reliable as the response to strong sensory stimuli (P = 0.00006, one-way ANOVA, ** P = 0.0037, strong versus weak; ** P = 0.0001, weak versus closed-loop; ** P = 0.0013 closed-loop versus playback; n.s. P = 0.47, strong versus closed-loop; n.s. P = 0.74, weak versus playback; * P = 0.036, strong versus playback; n = 6 cells in 4 animals. Measure of center is mean; error bars correspond to standard deviation).
(n) The time taken for sensory-evoked responses to cross the 30% ΔF/F threshold for strong sensory-evoked responses (top values) and weak sensory-evoked responses boosted by photostimulation (bottom values). Purple bar: whisker stimulation window; red: photostimulation window. Data are presented as box-and-whisker plots displaying median, interquartile and 90% ranges; the interquartile range was 500 to 767 ms after the onset of strong sensory stimulation, compared with 533 to 733 ms after the onset of weak sensory stimulation with closed-loop photostimulation (P = 0.33, Wilcoxon signed-rank test, n = 90 trials, 6 cells in 4 animals).
First, since many neural circuits are thought to use the average rate of spiking as a neural code8, 10, 11, 13, we sought to implement an all-optical ‘activity clamp’ in which a target activity level is maintained in individual neurons. We achieve this by tailoring photostimulation to the online readout of somatic calcium signals, which are correlated with spike rate14. The GCaMP6s fluorescence of a neuron also expressing C1V1 in layer 2/3 of mouse neocortex was ‘clamped’ at various user-defined levels through continuous on/off feedback control (Fig. 1b-d). During the closed-loop clamping period (30 s), two-photon optogenetic stimulation was delivered to the cell whenever its calcium signal fell below the target threshold (30%, 50% or 80% ΔF/F). The achieved activity levels were near the target values (absolute error = 5.2% ± 4.7% ΔF/F, Fig. 1d-f). Simultaneous cell-attached patch-clamp recordings confirmed that the clamp of the calcium signal reflected clamping of the average spike rate (after a transient instantaneous peak to reach the target; Fig. 1g and Supplementary Fig. 2). These experiments demonstrate that online feedback control can be used to tailor optogenetic stimulation to produce a specific pre-set level of neuronal activation. This strategy can also be applied to ensembles of neurons, efficiently mitigating the cell-to-cell and trial-to-trial variability in optogenetic responses across a neuronal population (Supplementary Fig. 3).
Next, we applied our closed-loop feedback control to manipulate sensory responses in the awake mouse (Fig. 1h). In layer 2/3 of rodent somatosensory cortex, the reliability and amplitude of sensory-evoked responses to passive whisker deflections depend on deflection amplitude15. We deflected a single or multiple whiskers under two conditions: one evoked small amplitude (weak), and the other large amplitude (strong) neural responses (Fig. 1i-j). We used closed-loop optogenetic control to selectively boost the responses of an identified neuron to the weak sensory stimulus only when the whisker stimulus failed to produce a large response (evoked activity < 30% ΔF/F). This closed-loop manipulation resulted in neuronal responses to the weak sensory stimuli being boosted to a similar level as those evoked by strong sensory stimuli (Fig. 1k-l) as well as restoring the reliability of trial-to-trial responses (Fig. 1m). The boosted transients in the closed-loop trials are significantly larger than the linear sum of the weak transients and the replayed photostimuli (Supplementary Fig. 4). The latencies of the boosted responses were comparable to those of strong sensory stimulation (Fig. 1n). These experiments demonstrate that we can rapidly enhance ongoing patterns of sensory-evoked activity in neural circuits in the awake mouse.
Since the spatial and temporal patterns of activity in neural circuits are crucial elements of neural codes9, 13, 16, we next developed a strategy for flexibly manipulating neurons in a circuit based on online readout of patterns of activity in the circuit. We selectively activated groups of ‘target’ neurons according to readout of ongoing activity in a ‘trigger’ neuron in layer 2/3 of mouse neocortex (Fig. 2a-b). When the online activity trace of the ‘trigger’ neuron exceeded a dynamic threshold (see Methods; Supplementary Fig. 5), we rapidly (Supplementary Fig. 6) activated the ‘target’ neuron group by simultaneous two-photon optogenetic stimulation(Fig. 2c-d). During this closed-loop protocol, the activity of the ‘trigger’ neuron and the associated ‘target’ neurons became highly correlated (Supplementary Fig. 7a). Next, we multiplexed the approach using multiple feedback loops running concurrently, enabling multi-cell readout combined with multi-cell control. We selected multiple ‘trigger’ neurons in a single field of view each with a different corresponding set of ‘targets’ (Fig. 2e-f). As before, the ‘target’ neuron groups were selectively activated when the activity of the appropriate ‘trigger’ neuron exceeded its threshold (Fig. 2g). Activity across the ‘trigger’ and relevant ‘target’ neurons again became highly correlated during this closed-loop protocol (Fig. 2h and Supplementary Fig. 7b). These experiments demonstrate that we can use our closed-loop strategy to flexibly manipulate the temporal and spatial pattern of neural circuit activity.
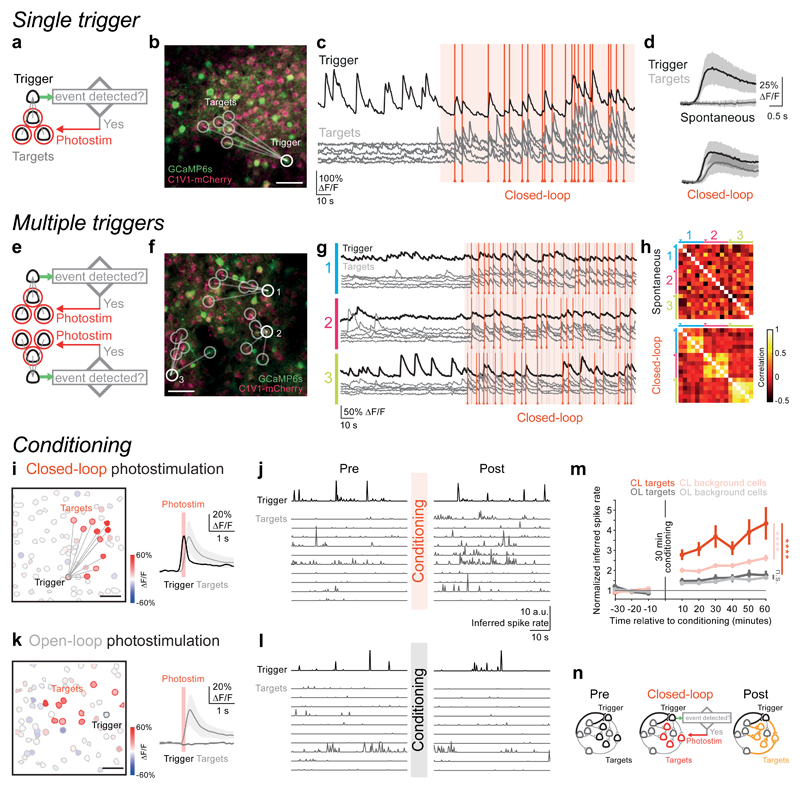
Figure 2. Online activity-guided photostimulation of neuronal ensembles can induce long-term plasticity of network activity.
(a) Experimental strategy: dynamic interrogation of ensembles with a single trigger neuron and multiple targets. A group of ‘target neurons’ was photostimulated (5 × 10 ms spiral stimulation of target neurons, 30 mW per cell, Fidelity laser) when an event was detected in the pre-defined ‘trigger neuron’.
(b) A field of view in layer 2/3 of mouse somatosensory cortex co-expressing GCaMP6s (green) and C1V1-2A-mCherry (pink; scale bar, 50 μm, 50 mW on sample, Chameleon laser) in an awake mouse. A trigger neuron (thick white circle) was selected to drive photostimulation of 5 pre-defined target neurons (thin white circles; representative example of 6 independent experiments with similar results).
(c) Top: calcium signal recorded from the trigger neuron. Bottom: calcium signal traces recorded simultaneously from the target neurons. We activated target neurons by photostimulation (red vertical lines) upon detection of an event in the trigger neuron during the closed-loop control period (140 s, shaded area).
(d) Calcium transients (mean ± SD) averaged across detected events during the spontaneous activity period (top) and closed-loop control period (bottom) (n = 6 trigger neurons, black trace; each trigger neuron was associated with a different group of 5 target neurons; grey trace).
(e) Experimental strategy: dynamic interrogation of ensembles with multiple trigger neuron and multiple targets. Multiple neurons in the field of view were selected as triggers and associated with a unique group of target neurons. The associated group of target neurons were photostimulated (same photostimulation settings as in a) when an event was detected in the corresponding trigger neuron.
(f) A field of view in layer 2/3 of mouse somatosensory cortex (scale bar, 50 μm) in an awake mouse. Three trigger neurons (numbered) were selected to drive photostimulation of three different groups of 5 pre-defined target neurons (indicated by lines from triggers to targets; representative example of 3 independent experiments with similar results).
(g) Calcium transients in which target neurons (gray traces) were photostimulated (red vertical lines show when the target group was stimulated; grey vertical lines show when the other target group(s) was stimulated) when their corresponding trigger neurons crossed an activity threshold (black traces).
(h) Pearson’s correlation matrices of the calcium transients recorded from the trigger neurons and target neurons as plotted in (g), during the spontaneous period (top) and closed-loop period (bottom). Each color bar covers the rows (or columns) that corresponds to neurons in the same trigger-targets group, with arrows pointing at the trigger neurons (n = 3 trigger neurons and 15 target neurons in this example).
(i) Closed-loop conditioning: a group of 10 target cells (red contours) were activated when an event was detected in a trigger cell (black contour) in layer 2/3 of somatosensory cortex in an awake mouse (scale bar, 50 μm, 30 mW on sample). Left: spatial map of all detected neurons in the field of view. Color shows the amplitude of each neuron’s calcium transients averaged in a 500 ms window after the onset of photostimulation (stimulation duration 12 ~ 25 × 10 ms spirals at 100 Hz; 30 mW per cell, Fidelity laser or 4 mW per cell, Satsuma laser). Right: average calcium transient of the trigger cell (black) and the target cells (grey) after photostimulation (red vertical line. Center value is mean; shaded area shows standard deviation).
(j) Example inferred spike traces from calcium imaging of the trigger cell (top row) and the target cells (bottom rows) before and after closed-loop photostimulation (30 minutes conditioning in each experiment).
(k) Open-loop conditioning: a group of 10 target-cells (red contours) were randomly activated during periods when no events were detected in a trigger cell (black contour) in layer 2/3 of somatosensory cortex in an awake mouse (scale bar, 50 μm, 30 mW on sample). Left: spatial map of all detected neurons in the field of view (colored in the same fashion as (i)). Right: average calcium transient of the trigger cell (black) and the target cells (grey) after photostimulation (red vertical line. Center value is mean; shaded area shows standard deviation).
(l) Same as (j) for open-loop photostimulation.
(m) The inferred spike rate of the target cells increased significantly after closed-loop photostimulation (red, **** P = 1.3 × 10-10, Wilcoxon signed-rank test (two-sided), n = 57 cells in 6 animals); no significant change in inferred spike rate is observed in the open-loop photostimulation targets (grey, n.s. (not significant) P = 0.11, Wilcoxon signed-rank test, n = 56 cells in 6 animals). The change in inferred spike rate after closed-loop photostimulation is significantly higher than the change after open-loop photostimulation (**** P = 3.5 × 10-8, Wilcoxon rank sum test (two-sided); center values are mean; error bars show standard error; all statistical tests in the panel were performed on the average inferred spike rates of the whole pre- or post-conditioning recording).
(n) Schematic showing potential loci of plasticity: closed-loop photostimulation may have induced changes in the connections from the upstream neurons driving the trigger and target neurons to the target neurons, the direct and indirect connections from trigger to target neurons, and between the targets themselves, in addition to possible changes in intrinsic excitability (yellow).
Finally, the ability to activate ensembles of neurons based on rapid detection of activity patterns in defined trigger neurons allows us to ‘yoke together’ the activity of user-defined neuronal ensembles. Such correlated activity is the mechanistic hallmark of many theories of synaptic plasticity, which are challenging to test directly in the intact brain17–19.To drive plasticity mechanisms in a targeted manner, we used our closed-loop strategy to repeatedly drive activity in an ensemble of target neurons based on activity in a trigger neuron and then monitored the resulting changes in activity in the local network after this conditioning period (Fig. 2i-j). This was compared with changes resulting from activation of a similar number of target neurons using an ‘open-loop’ protocol, which avoids yoking together the activity of an ensemble with that of a trigger cell (Fig. 2k-l). Following the conditioning period, there was a persistent and significant increase in the spontaneous activity of the target neurons (Fig. 2m and Supplementary Fig. 8-9) in the closed-loop protocol, which continued to increase post-conditioning. The changes were far more pronounced than in the open-loop protocol, despite the photostimulation-triggered activity in the target cells and number of stimuli being similar (Supplementary Fig. 10a-c). The spontaneous activity of background cells was also enhanced, though to a much lesser extent than in the target cells (Supplementary Fig. 11), and only after the closed-loop conditioning protocol (Fig. 2m). The inferred spike rate of the target cells specifically in episodes following detected events in the trigger cell became significantly larger post conditioning (Supplementary Fig. 12a), indicating long-lasting changes to the temporal synchrony of our artificial ensemble. These findings suggest that optically yoking together the activity of a trigger neuron with a neighboring ensemble can lead to specific and durable reconfiguration of the functional connectivity patterns in the network. This is likely to involve changes at multiple loci in the network, involving trigger, target and non-target cells (Fig. 2n), including the shared inputs of the trigger and target neurons, the direct and indirect connections from trigger to target neurons, and between the targets themselves, in addition to possible changes in intrinsic excitability20. The involvement of upstream and downstream connections may result in the global increase in activity level after conditioning. Further work will be required to examine the nature and relative importance of these changes in the network. These results demonstrate that our all-optical closed-loop strategy is a powerful approach for probing the mechanisms and function of plasticity induced by manipulating correlated activity with cellular resolution in neural circuits of awake animals.
Our all-optical closed-loop interrogation strategy allows flexible manipulation of neural circuits guided by ongoing activity. Previous approaches to closed-loop control of neural activity21–24, based on electrophysiological readout and stimulation, or a combination of electrophysiological readout and optogenetic stimulation, either lacked the ability to read out activity from genetically and anatomically targeted neurons, or the ability to target photostimulation to particular neurons. Our approach solves these problems by harnessing the spatial precision of two-photon imaging and two-photon photostimulation to enable targeting of readout and manipulation to visualized neurons with unprecedented flexibility. Our strategy can in principle be implemented using a range of genetically encoded sensors and optogenetic probes, enabling multiplexed imaging and control of different populations by selective expression of different sensors and actuators. Enhancement of the speed of the online feedback loop using future generations of genetically encoded calcium or voltage sensors and optogenetic probes with faster kinetics will further extend the reach of this approach. Closed-loop all-optical control will enable groundbreaking new experiments in which the temporal and rate codes of neural circuits can be manipulated in a targeted manner as they evolve during behavior. This will allow direct tests of models of circuit connectivity, dynamics, and plasticity by enabling on-the-fly manipulation of activity patterns. It could also be used to correct aberrant activity patterns in disease conditions such as epilepsy21, 22 and Alzheimer’s disease25, and can provide the basis for a new generation of optical brain-machine interfaces.
Online Methods
All experimental procedures were carried out under license from the UK Home Office in accordance with the UK Animals (Scientific Procedures) Act (1986).
Virus injections
Surgeries were performed as described previously4. Female adult mice (approximately 4 weeks to 8 weeks old) were anaesthetized with isoflurane (5% induction, 1.5% maintenance). A craniotomy was made above barrel cortex or visual cortex and 1 μl of a mixture of AAVdj-CaMKIIa-C1V1(E162T)-TS-P2A-mCherry-WPRE (Stanford Neuroscience Gene Vector and Virus Core, genomic titer, 7.22 × 1012) and AAV1-hSyn-GCaMP6s-WPRE-SV40 (Penn Vector Core, genomic titer, 2.04 - 3.25 × 1013) or AAV1-hSyn-GCaMP6f-WPRE-SV40 (Penn Vector Core, genomic titer, 3.45 × 1013) virus (dilution factor 10:1) was injected into layer 2/3 (~300 μm deep)4. In most experiments (Figs. 1, 2a-h), GCaMP6s was chosen for its higher reliability in reporting spikes26 (Supplementary Fig. 13a) to optimize online and offline analysis accuracy. For the plasticity experiments in Fig. 2i-n, we used the faster and lower-affinity GCaMP6f to maximize the speed of the feedback loop and minimize any disruption of calcium buffering. For chronic imaging, a metal headplate with a 5 mm circular imaging well was fixed to the skull with dental cement (Super-Bond C&B, Sun-Medical) prior to virus injection before performing a larger craniotomy and installing a glass coverslip ‘window’.
Imaging and photostimulation
Simultaneous ‘all-optical’ imaging and photostimulation2–7, 27–35 was performed using a custom in vivo dual beam path microscope (Ultima, Bruker Corporation). Two-photon imaging (512 × 512 pixel resolution per frame, 30 frames per second) of layer 2/3 mouse cortex (~100 to 300 μm deep) was performed by resonant-galvanometer raster scanning a femtosecond-pulsed laser beam (Chameleon Ultra II, Coherent). A 25x/0.95-NA objective (Leica) was used for all experiments. GCaMP6s (or GCaMP6f) was imaged with excitation wavelength of 920 nm and mCherry was imaged at 765 nm (field-of-view size, 268 × 268 μm, frame rate, 30 Hz, power on sample, 30 to 50 mW, Chameleon laser). These imaging parameters were chosen to be similar to those that have been used for imaging GCaMP6 signals in layer 2/3 in vivo with sufficient imaging quality4, 7, 26, 36, and to minimize imaging “crosstalk”, i.e. activation of C1V1 with the imaging laser2, 4, 7 (Supplementary Table). To experimentally optimize the imaging parameters, we previously performed two-photon imaging of a neuron with simultaneous electrophysiological recording26 and increased the power and/or dwell time per neuron to determine when imaging evoked spikes4 (see also refs. 5–7 and ref 37). We then chose imaging conditions that gave sufficient imaging quality while not inducing a significant change in the spike rate. Imaging at a higher power, or lower scanning speed and/or using a smaller field-of-view could lead to photoactivation4–7, 37. Therefore, imaging power needs to be carefully assessed for each experimental configuration, as the optimum parameters and thus the tradeoff between imaging quality and crosstalk are expected to depend on expression levels, kinetics and sensitivities of the opsin and the activity sensor, as well as field of view size and imaging dwell times. The excitation source for spiral-scanning photostimulation4, 7, 19, 38 was a femtosecond-pulsed laser fixed at 1070 nm (Fidelity, Coherent; average output, 2 W; pulse width, 55 fs; repetition rate, 70 ± 10 MHz; 7 experiments in Fig. 1c-g, Fig. 2a-h and 6 experiments in Fig. 2i-m) or at 1030 nm (Satsuma HP2, Amplitude Systemes; average output, 20 W; pulse width, 280 fs; repetition rate, 2 MHz; 11 experiments in Fig. 1e-g, Fig. 1h-n, Supplementary Fig. 3 and 6 experiments in Fig. 2i-m). The plasticity protocol did not lead to changes in nuclear fluorescence or the response to photostimulation across days (Supplementary Fig. 14). A reflective multilevel spatial light modulator (SLM; OverDrive Plus SLM, Meadowlark Optics/Boulder Nonlinear Systems; 7.68 × 7.68 mm active area, 512 × 512 pixels, optimized for 1064 nm) was coupled to the microscope, with its active area relayed to the back aperture of the objective as described previously4. The weighted Gerchberg-Saxton (GS) algorithm39 was used to calculate holograms to be displayed on the SLM. The weights were adjusted to compensate for the difference in diffraction efficiency between holographic spots. The maximum number of neurons that can be photostimulated simultaneously is mainly limited by the laser power. While here we demonstrated stimulation of groups of ~10 neurons, given sufficient laser power, the method can be extended to other applications where addressing more neurons (more than 100) is desired. The closed-loop approach can in principle be combined with other photostimulation strategies, such as temporal focusing2, 40–47. The spatial resolution of photostimulation can be further improved using the somatic targeting opsins that are currently under active development in the field 5, 6, 48, 49. All experiments were performed in awake mice except those with electrophysiological recordings in Fig. 1g and Supplementary Fig. 2 where the animals were anesthetized.
Online analysis and closed-loop stimulation control
Raw data from the imaging acquisition card was made available to our custom closed-loop interface (Supplementary Fig. 1a-b) written in .NET Framework (Microsoft Visual Basic) using the PrairieLink API which allows communication between external, custom written software and the standard microscope control software (Prairie View, Bruker Corporation). The raw data consisted of a continuous stream of intensity samples which required interpretation using knowledge of the following acquisition parameters: samples per pixel, pixels per line and lines per frame, in addition to the acquisition bit-depth and bidirectional scan pattern. When online motion correction is enabled (used in Fig. 1i-n and Supplementary Fig. 3) to correct the lateral shifts in the field of view (Supplementary Fig. 15), the raw data was converted into 512 × 512 frames and then registered to a pre-loaded reference image using a discrete Fourier transform (DFT)–based algorithm50. The data conversion and motion correction process was accelerated by using a graphics processing unit (GPU; NVIDIA GeForce GTX 750 Ti). The average signal from each pre-defined region-of-interest (ROI) was extracted from the data stream, and then normalized either by the average value during spontaneous activity or the 60 frames before sensory stimulation was delivered. The frames recorded during photostimulation were not used for online decision-making and were interpolated in offline processing. The activity threshold used for event detection in the ‘trigger’ cells in Fig. 2 was continuously updated to be the mean plus 2 standard deviations and mean plus 3 standard deviations of the previous 60 frames (2 seconds), respectively. A higher threshold was used in the closed-loop plasticity protocol such that photostimulation of the target cells was paired with large burst-like events in the trigger cell (the ΔF/F threshold of photostimulation is 39% ± 4.4%, Mean ± SE). More active cells were selected to be the trigger cells in order to obtain a large number of trials during the pairing period (the baseline activity levels of the trigger cells were higher than 93.1% ± 4.0% of the other cells detected in the fields of view in the closed-loop experiments and 90.8% ± 4.0% in the open-loop experiments; P = 0.51, Wilcoxon rank sum test). In the open-loop experiment in Fig. 2, photostimulation was disabled in a 500 ms window after an event was detected in the trigger cell to avoid pairing between the trigger and target cells. Each plasticity experiment consists of 30-minute imaging of spontaneous activity followed by 30-minute photostimulation conditioning and then 60-minute recording of spontaneous activity. Custom SLM control software (Supplementary Fig. 1d) using the Blink SDK (Meadowlark Optics) written in C++ (Microsoft Visual Studio 2013) was used to load the pre-computed phase masks that could generate different combinations of the beamlets targeting the pre-defined target-neuron groups into the SLM controller’s buffer before the start of the experiment. When an online decision of the photostimulation pattern was made, the index of the required phase mask was sent from the closed-loop interface to the SLM control software via Transmission Control Protocol (TCP) communication (see Supplementary Note), and the required hologram (if different from the current one) was displayed on the SLM via the SLM driver electronics (Meadowlark Optics/Boulder Nonlinear Systems). Spiral stimulation (driven by Prairie View) was triggered by sending TTL pulses to the microscope control electronics via an analog output device (USB-6212, National Instruments). For sensory stimulation the closed-loop interface sent a command via TCP to custom software written in MATLAB (2016a, MathWorks) which then output one of multiple different waveforms via an analog-output device (USB-6343, National Instruments) to control a piezoelectric actuator for whisker stimulation.
Sensory stimulation
For single-whisker stimulation, C2 whisker was inserted into a glass capillary attached to a one-dimensional piezoelectric actuator (PL127.11, Physik Instrumente). For multi-whisker stimulation, whiskers were deflected using a star-shaped paddle attached to a two-dimensional piezoelectric actuator (NAC2710-A01, Noliac). Weak and strong whisker stimuli were delivered by driving the piezo with sinusoidal waveforms (20 Hz, 1 sec duration) of two different amplitudes (1.5 ~ 4 V for strong deflections and 0.5 ~ 2 V for weak deflections; the amplitude was adjusted based on the response of the target cell to produce significant differences in the response reliability; Fig. 1m). During whisker stimulation, animals were awake and head-fixed while sitting in a tube. Barrels were located by combining whisker stimulation with widefield fluorescence imaging using one-photon blue excitation provided by an LED (Thorlabs) together with a 5x/0.1-NA air objective (Olympus) and a CMOS camera (ORCA-Flash4.0, Hamamatsu; approximate field-of-view 1.5 mm × 1.5 mm; 512 × 512 pixel resolution, 10 frames per second).
Electrophysiological recording
For electrophysiological validation of the ‘activity clamp’ experiments, acute craniotomies were performed approximately 3 weeks post virus injection and two-photon targeted patch-clamp recordings51, 52 were made using glass pipettes pulled from borosilicate glass (~5 MΩ pipette resistance) and filled with ACSF (150 mM NaCl, 2.5 mM KCl, 10 mM HEPES, 1.5 or 2 mM CaCl2, 1 mM MgCl2). Alexa 594 (20 μM) was included in the pipette solution for visualization. Signals were recorded using a MultiClamp 700A amplifier (Molecular Devices), filtered with a low-pass Bessel filter (4 kHz) and sampled at 20 kHz using PackIO53. In target cells with extremely low firing rates the effectiveness of the activity clamp can drop (Supplementary Fig. 2k). Recordings were aborted when the baseline firing rate of the target cell went above 4 Hz during the experiment (2 cells in 2 animals). Trials in which the patched cells showed signs of a significant decline in health (i.e. the baseline spike rates and/or amplitudes decreased by more than 80%, and/or the spike widths, measured as the full width at half maximum, increased by more than 80% during the recording) were not included in the analysis.
Offline data analysis
Data were analyzed using toolboxes and custom scripts written in MATLAB and Python. PackIO53 recordings made during all the experiments enabled precise post-hoc synchronization of stimulus delivery times and individual imaging frame times. Motion correction was performed on calcium imaging movies using a discrete Fourier transform (DFT)–based algorithm50. Neuropil-subtracted fluorescence traces were extracted using the Suite2P package54. We defined GCaMP6s events based on fluorescence traces using a template matching algorithm55 (Supplementary Fig. 5). For GCaMP6f, we calculated the inferred spike rate (Fig. 2j, l-m and Supplementary Fig. 12), which is defined as the time average of the spike train (analogue) returned by Suite2P 54, and is given in arbitrary units. As an alternative measure of activity in GCaMP6f traces, we defined GCaMP6f events using an iterative routine, in which the event times were adjusted iteratively to obtain the onsets and amplitudes of bursting events (Supplementary Fig. 8). The event onset times were first initialized as the time when the inferred spike amplitude from Suite2P was greater than the mean plus one standard deviation. Then, in one iteration, the events were discarded if they occurred within a 150 ms window after the preceding event and/or if the amplitude (90% reference level) of the fluorescence signal (measured in a 2 sec window around the detected event time, normalized to the average value in 20 frames before the event onset frame) was below zero. The calculated onset times (10% rise time) were adjusted to be the new event onset times, and the iterative procedure was repeated three times (Supplementary Fig. 8b). Event rates are given in Hz and event amplitudes in ΔF/F (Supplementary Fig. 9). Neuropil-subtracted calcium traces were smoothed by a low-pass filter (1.5 Hz) and then normalized to the average of the 3% of data points with the lowest calcium fluorescence values before taking the Pearson’s correlation coefficients. For plasticity experiments, the normalized calcium transients were high-pass filtered above 0.02 Hz to correct for slow drifts in baseline in the longer recordings before calculating correlation and the population coupling factors56. Fig. 2i-m only includes experiments in which greater than 100 trigger-targets pairings were performed.
Data presentation and statistical analyses
All values are given as mean ± SD, unless otherwise noted. Statistical analyses were performed using different tests as appropriate, as stated in the figure legends.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article
Supplementary Material
Acknowledgments
We thank I. Bianco, H.W. Dalgleish, M. Fisek, B. Judkewitz, P. Latham, D. Selviah and Arnd Roth for helpful discussions and comments on the manuscript; K. Deisseroth (Stanford University) for access to AAVdj virus; and Bruker Corporation for software support. We are particularly grateful to D. Selviah and the Faculty of Engineering at UCL for support and training of Z.Z. This work was supported by grants from the Wellcome Trust, MRC, the Gatsby Charitable Foundation, the European Commission (Marie Curie International Incoming Fellowship grant no. 328048), EMBO, ERC, and the BBSRC.
Footnotes
Data and code availability
The original code, together with detailed instructions, is available at the following Github link: https://github.com/alloptical/ClosedLoop. The authors will keep the repository operating and freely accessible. The datasets generated and/or analyzed in this study are available from the corresponding author upon reasonable request. Further information on experimental design is available in the Nature Research Reporting Summary linked to this paper.
Author Contributions
Z.Z. and L.E.R developed the custom closed-loop software. Z.Z. carried out the all-optical experiments, analyzed data and wrote the first draft of the manuscript. O.M.G., L.E.R and A.M.P. performed animal surgeries. A.M.P. performed cell-attached recordings. O.M.G. prepared the sensory stimulation setup. L.E.R., A.M.P. and M.H. conceived the project. Z.Z, L.E.R. A.M.P and O.M.G. designed detailed experimental protocols. Z.Z, L.E.R., A.M.P. and M.H. wrote the paper with input from all authors. All authors discussed the results and contributed to revision of the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Emiliani V, Cohen AE, Deisseroth K, Häusser M. J Neurosci. 2015;35:13917–13926. doi: 10.1523/JNEUROSCI.2916-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rickgauer JP, Deisseroth K, Tank DW. Nat Neurosci. 2014 doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V. Neuron. 2014;84:1157–1169. doi: 10.1016/j.neuron.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Packer AM, Russell LE, Dalgleish HW, Hausser M. Nat Methods. 2015;12:140–146. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forli A, et al. Cell Reports. 2018;22:3087–3098. doi: 10.1016/j.celrep.2018.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardinly AR, et al. Nat Neurosci. 2018 doi: 10.1038/s41593-018-0139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Carrillo-Reid L, Bando Y, Peterka DS, Yuste R. eLife. 2018;7:e32671. doi: 10.7554/eLife.32671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shadlen MN, Newsome WT. Curr Opin Neurobiol. 1994;4:569–579. doi: 10.1016/0959-4388(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 9.Rieke F. Spikes: exploring the neural code. The MIT Press; Cambridge: 1999. [Google Scholar]
- 10.Pouget A, Dayan P, Zemel R. Nat Rev Neurosci. 2000;1:125–132. doi: 10.1038/35039062. [DOI] [PubMed] [Google Scholar]
- 11.Averbeck BB, Latham PE, Pouget A. Nat Rev Neurosci. 2006;7:358–366. doi: 10.1038/nrn1888. [DOI] [PubMed] [Google Scholar]
- 12.Grosenick L, Marshel JH, Deisseroth K. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.deCharms RC, Zador A. Annu Rev Neurosci. 2000;23:613–647. doi: 10.1146/annurev.neuro.23.1.613. [DOI] [PubMed] [Google Scholar]
- 14.Helmchen F, Imoto K, Sakmann B. Biophys J. 1996;70:1069–1081. doi: 10.1016/S0006-3495(96)79653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons DJ. J Neurophysiol. 1978;41:798. doi: 10.1152/jn.1978.41.3.798. [DOI] [PubMed] [Google Scholar]
- 16.Bruno RM. Curr Opin Neurobiol. 2011;21:701–708. doi: 10.1016/j.conb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hebb DO. The Organization of Behavior. Wiley; New York: 1949. [DOI] [PubMed] [Google Scholar]
- 18.Martin SJ, Grimwood PD, Morris RG. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- 19.Carrillo-Reid L, Yang W, Bando Y, Peterka DS, Yuste R. Science. 2016;353:691–694. doi: 10.1126/science.aaf7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Linden DJ. Nat Rev Neurosci. 2003;4:885–900. doi: 10.1038/nrn1248. [DOI] [PubMed] [Google Scholar]
- 21.Paz JT, et al. Nat Neurosci. 2013;16:64–70. doi: 10.1038/nn.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krook-Magnuson E, Armstrong C, Oijala M, Soltesz I. Nat Commun. 2013;4 doi: 10.1038/ncomms2376. 1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman JP, et al. eLife. 2015;4:e07192. doi: 10.7554/eLife.07192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prsa M, Galinanes GL, Huber D. Neuron. 2017;93:929–939.e926. doi: 10.1016/j.neuron.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iaccarino HF, et al. Nature. 2016;540:230–235. doi: 10.1038/nature20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen TW, et al. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolenko V, Poskanzer KE, Yuste R. Nat Methods. 2007;4:943–950. doi: 10.1038/nmeth1105. [DOI] [PubMed] [Google Scholar]
- 28.Nikolenko V, et al. Front Neuroal Circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo ZV, Hart AC, Ramanathan S. Nat Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dal Maschio M, et al. Opt Express. 2010;18:18720–18731. doi: 10.1364/OE.18.018720. [DOI] [PubMed] [Google Scholar]
- 31.Anselmi F, Ventalon C, Begue A, Ogden D, Emiliani V. Proc Natl Acad Sci U S A. 2011;108:19504–19509. doi: 10.1073/pnas.1109111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prakash R, et al. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Packer AM, et al. Nat Methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akerboom J, et al. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochbaum DR, et al. Nat Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theis L, et al. Neuron. 2016;90:471–482. doi: 10.1016/j.neuron.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ronzitti E, et al. J Neurosci. 2017;37:10679–10689. doi: 10.1523/JNEUROSCI.1246-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rickgauer JP, Tank DW. Proc Natl Acad Sci U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerchberg RW, Saxton WO. Optik. 1972;35 [Google Scholar]
- 40.Oron D, Tal E, Silberberg Y. Opt Express. 2005;13:1468–1476. doi: 10.1364/opex.13.001468. [DOI] [PubMed] [Google Scholar]
- 41.Zhu G, van Howe J, Durst M, Zipfel W, Xu C. Opt Express. 2005;13:2153–2159. doi: 10.1364/opex.13.002153. [DOI] [PubMed] [Google Scholar]
- 42.Papagiakoumou E, de Sars V, Oron D, Emiliani V. Opt Express. 2008;16:22039–22047. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- 43.Papagiakoumou E, de Sars V, Emiliani V, Oron D. Opt Express. 2009;17:5391–5401. doi: 10.1364/oe.17.005391. [DOI] [PubMed] [Google Scholar]
- 44.Papagiakoumou E, et al. Nat Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Proc Natl Acad Sci U S A. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandez O, et al. Nat Commun. 2016;7 11928. [Google Scholar]
- 47.Pégard NC, et al. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01031-3. 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker CA, Elyada YM, Parra A, Bolton MM. Elife. 2016;5 doi: 10.7554/eLife.14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shemesh OA, et al. Nat Neurosci. 2017;20:1796. doi: 10.1038/s41593-017-0018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guizar-Sicairos M, Thurman ST, Fienup JR. Opt Letters. 2008;33:156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 51.Kitamura K, Judkewitz B, Kano M, Denk W, Hausser M. Nat Methods. 2008;5:61–67. doi: 10.1038/nmeth1150. [DOI] [PubMed] [Google Scholar]
- 52.Margrie TW, et al. Neuron. 2003;39:911–918. doi: 10.1016/j.neuron.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 53.Watson BO, Yuste R, Packer AM. bioRxiv. 2016 doi: 10.1101/054080. 054080. [DOI] [Google Scholar]
- 54.Pachitariu M, et al. bioRxiv. 2016 doi: 10.1101/061507. 061507. [DOI] [Google Scholar]
- 55.Clements JD, Bekkers JM. Biophys J. 1997;73:220–229. doi: 10.1016/S0006-3495(97)78062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Okun M, et al. Nature. 2015;521:511–515. doi: 10.1038/nature14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.