Abstract
Salmonella is a major foodborne pathogen which causes widespread contamination and infection worldwide. Salmonella Typhimurium is one of the leading serovars responsible for human and animal salmonellosis, globally. The increasing rate of antibiotic resistance in Salmonella Typhimurium poses a significant global concern, and an improved understanding of the distribution of antibiotic resistance patterns in Salmonella Typhimurium is essential for choosing the suitable antibiotic for the treatment of infections. To evaluate the roles of animal and human in antibiotic resistance dissemination, this study aims to categorize 11,447 S. Typhimurium strains obtained across the food-chain, including food animals, retail meats and humans for 21 years in the United States by analyzing minimum inhibitory concentrations (MICs) values for 27 antibiotics. Random Forest Algorithm and Hierarchical Clustering statistics were used to group the strains according to their minimum inhibitory concentration values. Classification and Regression Tree analysis was used to identify the best classifier for human- and animal-populations’ isolates. We found the persistent population or multi-drug resistant strains of S. Typhimurium across the four time periods (1996∼2000, 2001∼2005, 2006∼2010, 2011∼2016). Importantly, we also detected that there was more diversity in the MIC patterns among S. Typhimurium strains isolated between 2011 and 2016, which suggests significant emergence of diversified multi-drug resistant strains. The most frequently observed (43%) antibiotic resistance patterns found in S. Typhimurium were tetra-resistant pattern ASSuT (ampicillin, streptomycin, sulfonamides, and tetracycline) and the penta-resistant pattern ACSSuT (ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline). Animals (mainly swine and bovine) are the major source for these two frequently found antibiotic resistance patterns. The occurrence of antibiotic resistant strains from humans and chicken is alarming. Strains were mostly susceptible to fluoroquinolones. Together, this study helped in understanding the expansion of dynamics of antibiotic resistance of S. Typhimurium and recommended fluoroquinolones as a possible treatment options against S. Typhimurium infection.
Keywords: Salmonella Typhimurium, foodborne pathogen, antibiotic resistance, minimum inhibitory concentration, population diversity, fluoroquinolones
Introduction
The burden of foodborne illnesses is tremendous, affecting 10% of global population with 33 million deaths annually (Havelaar et al., 2015). Numerous factors contribute to diarrheal diseases, and Salmonella enterica causes foodborne illnesses with significant public health impact. Salmonella enterica has a group of ∼2600 closely related bacteria defined by their mosaic combination of surface O and H antigens, so-called serovar. Based on the different pathogenic behaviors there are two groups of Salmonella, these are, typhoid Salmonella and non-typhoidal Salmonella. While typhoidal Salmonella can result in systemic infections with high fatal capabilities, non-typhoidal Salmonella infections are generally self-limiting (Su and Chiu, 2007; Gal-Mor et al., 2014). The emergence of pathogenic Salmonella enterica serovar Typhimurium (S. Typhimurium), armed with multiple antibiotic resistance (AR) in particular, presents a considerable threat to public health and food safety.
Salmonella Typhimurium, with its broader range of host tropism, is one of the top two serovars responsible for causing infections in human and animal worldwide (Hohmann, 2001; Herikstad et al., 2002; Foley and Lynne, 2008; Lan et al., 2009; Majowicz et al., 2010; Hendriksen et al., 2011). Throughout the last few decades, S. Typhimurium DT104 (DT104) evolved and disseminated rapidly across the globe (Helms et al., 2005; Lan et al., 2009). Genomic island (SGI-1) was suggested to be an important factor for the multi-drug resistance (MDR) phenotype in DT104 (Paul et al., 2016) and the acquisition of different resistance genes changed the bacterial fitness and virulence, which lead to emergence of highly virulent ST34 or ST313 clones (Okoro et al., 2012; Mather et al., 2018). The bacterial strains resistant to more than two antimicrobial drug classes are defined as multi-drug resistant. Importantly, DT104 isolates of animal were suggested to be different from the isolates of human population by comparative population genomic analysis. Additionally, the antibiotic resistance pattern between human and food–animal isolates population has some overlaps.
Salmonella Typhimurium is usually isolated from human, retail meats, and animal origins (Zhang et al., 2003; Kingsley et al., 2009; Izumiya et al., 2011). S. Typhimurium is an excellent model to address antibiotic resistant bacterial persistence and transmittance through the food-chain. The National Antimicrobial Resistance Monitoring System (NARMS) in the United States tracks the information about AR in Salmonella including other foodborne pathogens by comparing different bacterial isolates obtained from human infections, animals, and their retail meats. Such information is essential for understanding the effectiveness of antibiotics for both humans and animals (McDermott et al., 2016).
Severe Salmonella infections require treatment, and AR is the most serious challenge to treat these bacterial infections (CDC, 2013a, b). The misuse of antibiotics is one of the important factors responsible for high resistance to antibiotics in various pathogenic bacteria, including S. Typhimurium (CDC, 2013b). AR in Salmonella is linked with horizontal gene transfer and these genes are found on mobile genetic elements. The expansion of antibiotic resistant Salmonella serovars are efficient in worldwide dissemination (Butaye et al., 2006; Michael et al., 2006; Alcaine et al., 2007; Mather et al., 2013). In vitro antibiotic susceptibility testing is an important basis for monitoring antibiotic susceptibility and resistance trends and for guiding effective anti-infective therapy.
Due to the lack of studies on how the antibiotic resistant S. Typhimurium isolates persist and distribute across the food-chain, the objective of this study was to analyze the minimum inhibitory concentrations (MIC) data of different clinically relevant classes of antibiotics of 11,447 S. Typhimurium strains from humans, food animals and their products in the United States collected between 1996 and 2016, to investigate the AR pattern through time, food-chain, host origins and also to recommend effective therapeutic options against S. Typhimurium infections.
Materials and Methods
Salmonella Typhimurium Strains
We analyzed available data of S. Typhimurium strains obtained from the US NARMS (CDC and NARMS, 2019) database for Enteric Bacteria obtained from 1996 through 2016. Total 11,447 S. Typhimurium strains used to analyze in this study were from humans (n = 6381), animals (n = 2940), and retail meats (n = 2126). The S. Typhimurium strains from animals were from chickens (n = 1396), bovines (n = 810), porcine (n = 609) and turkeys (n = 125). No samplings from ready-to-eat foods and non-animal origin were reported in the current dataset. All the bacterial isolate has clear information of year of isolation, while only around 11% of isolate has clear location information, such as US state of isolation. The aim of this study is to understand the dynamic (time-scale) feature of S. Typhimurium across food-chain (animal, retail meat, and human).
The “food-chain” or “food-chain transmission” defined in this study is all about three steps from an infectious disease point of view. The recognized chain for transmission is from animal to animal product, and then to humans, this is how we design the study. We did not study the spread of foodborne pathogens at different stages of industrial food production and processing.
MIC Determination of Antimicrobial Agents
The minimum inhibitory concentration of the antimicrobial agents tested was recorded for each isolate and compared to breakpoints that were defined by the CLSI when available; otherwise, breakpoint interpretations from the National Antimicrobial Resistance Monitoring System were used as described in the NARMS 2012–2013 annual integrated report (FDA, 2015; Clinical and Laboratory Standards Institute [CLSI], 2017).
The MIC values interpreted according to the CLSI guidelines (Clinical Laboratory Standards Institute [CLSI], 2017) for 27 antibiotics were evaluated and analyzed in this study. These 27 antibiotics were Amikacin (AMI), Apramycin (APR), Gentamicin (GEN), Kanamycin (KAN), Streptomycin (STR), Amoxicillin-clavulanic acid (AMC), Piperacillin-tazobactam (PTZ), Cephalothin (CEP), Cefoxitin (FOX), Ceftriaxone (AXO), Ceftiofur (TIO), Ceftazidime (CAZ), Cefotaxime (CTX), Cefotaxime/clavulanic acid (CTC), Cefquinome (CEQ), Cefepime (FEP), Sulfamethoxazole (SMX), Sulfisoxazole (FIS), Sulfamethoxazole-trimethoprim (COT), Azithromycin (AZM), Aztreonam (ATM), Imipenem (IMI), Ampicillin (AMP), Chloramphenicol (CHL), Ciprofloxacin (CIP), Nalidixic acid (NAL), Tetracycline (TET).
Statistical Analysis
In this study, we used Random Forest Algorithm and Hierarchical Clustering analysis to classify Salmonella Typhimurium population based on their MIC patterns. Recent work demonstrated the usefulness of random forest method because of its unique advantages (Pan et al., 2018). Random Forest package 4.6 was used in this study for the classification. The data set for random forest analysis was classified based on three source (animal, meat, and humans) and five hosts (chicken, bovine, swine, turkey, and human). The input variable was the data set of MIC distribution. The test feature was the MIC values of each individual strains. The strains were categorized into four groups according to the time of their isolation as period 1 (Year 1996–2000), period 2 (Year 2001–2005), period 3 (Year 2006–2010) and period 4 (Year 2011–2016). Based on these four time periods, the diversity of S. Typhimurium population was segregated using Random Forest. The MIC values of 27 different antibiotics were used to group different population. Single dot depicts individual strains. The rule for each randomly created decision trees was based on the MIC cut-off values and the vote count was denoted by present (MIC higher than cut off) or absent (MIC lower than cut off). Proximity computation was done on three scales. To identify the best classifier, we have used Classification and Regression Tree (CART) method. Both Random Forest and CART were used to identify the most crucial variables. Using multinomial logistic regression (Afema et al., 2015), here we analyzed antibiotic-susceptibility and antibiotic resistant profiles of S. Typhimurium population.
Results
Isolates From Different Hosts Behave Differently
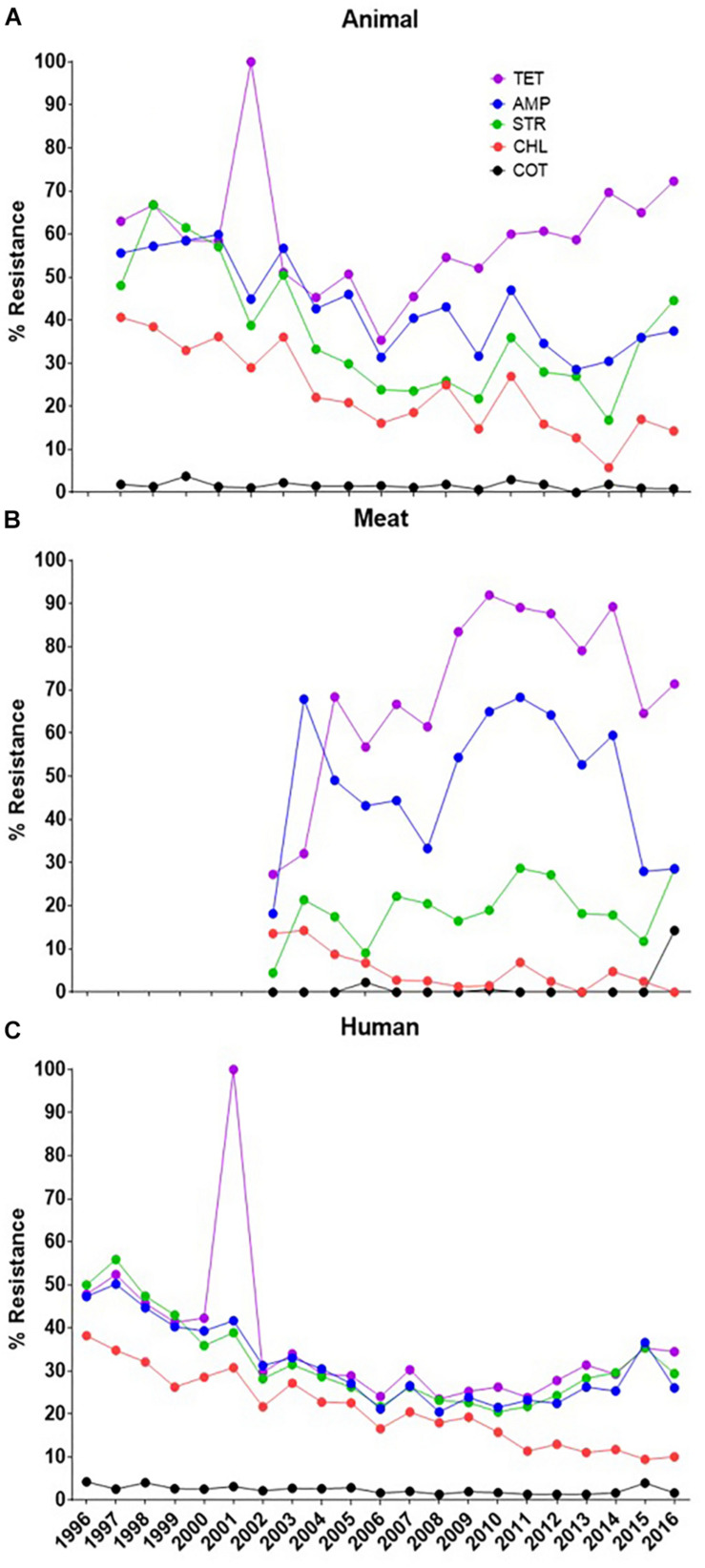
A general trend of the AR of the S. Typhimurium isolates from animals, meat and humans to some commonly used antimicrobial agents is presented in Figure 1. In the early 2000s, a surge in resistance to tetracycline by isolates from humans coincided with a similar surge in the animals’ isolates. These results apparently convince that food animals may serve as a pool of antibiotic resistant organisms to humans. But in absence of the records of the simultaneous meat samples a direct link between the animals and humans is missing. For the other years (after 2002) when data is available in all three hosts, the resistance pattern has remained in a generally steady state with a higher rate in animals/meat and a lower rate in humans. In this context, our major focus in this analysis was to show that the isolates from different hosts behave differently.
FIGURE 1.
General trend analysis of antibiotic resistance for some certain commonly used molecules (denoted by the color of the lines as given in legend). (A) The resistance pattern in animal isolates, (B) the resistance pattern in meat isolates and (C) the resistance pattern for the human isolates. The XX’ presents the time of data collection while YY’ gives the resistance percent. [Tetracycline (TET), Ampicillin (AMP), Streptomycin (STR), Chloramphenicol (CHL), Sulfamethoxazole-trimethoprim (COT)].
Recent Diversification of Antibiotic Resistant Strains
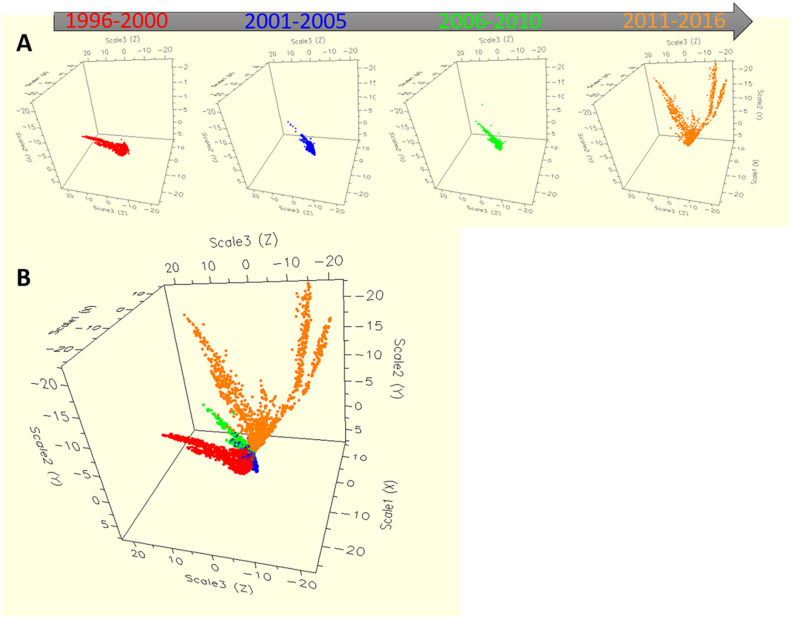
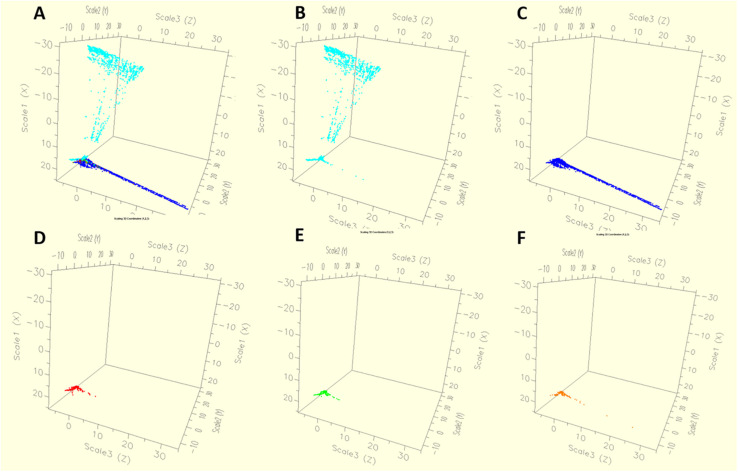
Based on the four time periods (1996–2000, 2001–2005, 2006–2010, 2011–2016), the diversity of S. Typhimurium population was segregated using RandomForest (Figure 2). The MIC values of 27 different antibiotics were used to group different population. Single dot depicts individual strains, with four different colors pointing isolates from four different periods of time. The Figure 2A showed the temporal distribution of the S. Typhimurium strains for four time periods from 1996 till 2016. While the distribution based on the MIC of individual strains for until the year 2010 was clustered, the divergence was seen after 2011. For the period 2011–2016, two distinct clusters of the strains are typically visible (Figures 2A,B). It shows that there could have been a shift in the MIC patterns of the isolates after 2011 so they started being divergent.
FIGURE 2.
The dynamics of antibiotic resistance features of Salmonella Typhimurium strains across 21 years in the United States. The multidimensional scaling plot of 11,447 Salmonella Typhimurium strains with MIC values for each bacterial isolate were used for producing the proximity matrices, where x-, y- and z- axes are the multidimensional scaling coordinates. Bacterial isolates with similar MIC values are represented by points close one to the other, whereas isolates with dissimilar antibiotic resistant MIC values are represented by separated points. Each bacterium was indicated as individual dot, where colors represented four different periods. (A) The dynamics of antibiotic resistance for four separate time periods. Time period 1 from the year 1996 to 2000, time period 2 from the year 2001 to 2005, time period 3 from the year 2006 to 2010 and time period 4 from the year 2011 to 2016. (B) The dynamics of antibiotic resistance for all four time periods describing together.
Antibiotic Susceptibility and Resistance Analysis
Antibiotic resistance analysis with high discriminatory capability allowed differentiation of the 11,447 strains of Salmonella Typhimurium. Using hierarchical clustering, population diversity of S. Typhimurium was grouped in this study (Figure 3). The log values of the MIC of different antibiotics were used to group Typhimurium population (Figure 3A) which showed the diversification within S. Typhimurium strains. The blue to yellow color of the heatmap describing the MICs of individual S. Typhimurium strain for every antibiotic. The yellow color pointing the resistance, and blue color pointing the susceptibility. Gray color pointing the strains lacking MIC value (Figure 3B). We found that the resistance was very low against the fluoroquinolones and the resistance was high against ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline, amoxicillin-clavulanic acid, ceftriaxone, ceftiofur antibiotics (Figure 3B).
FIGURE 3.
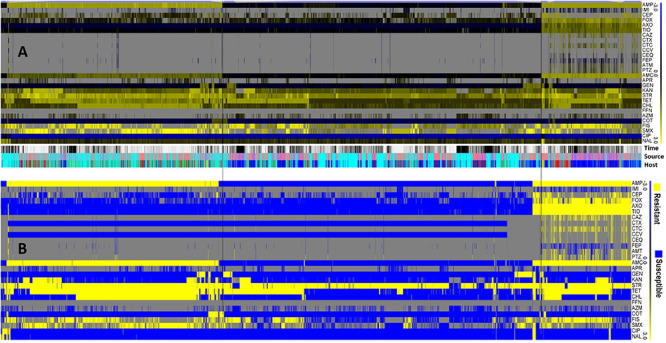
The population diversity of 11,447 S. Typhimurium strains from humans, animals, and retail meats. (A) Population diversity of S. Typhimurium strains grouped by hierarchical clustering. A hierarchical tree with 200 bootstrapping, by using the MIC value of 27 antibiotics, was used to group different population. (B) The antibiogram for individual S. Typhimurium strains were shown, with blue indicating the susceptibility, and yellow indicating the resistance, based on the MICs interpreted by the CLSI-2017 standards. Gray color indicates strains without MIC value.
ASSuT and ACSSuT Are the Most Frequently Found Antibiotic Resistance Patterns
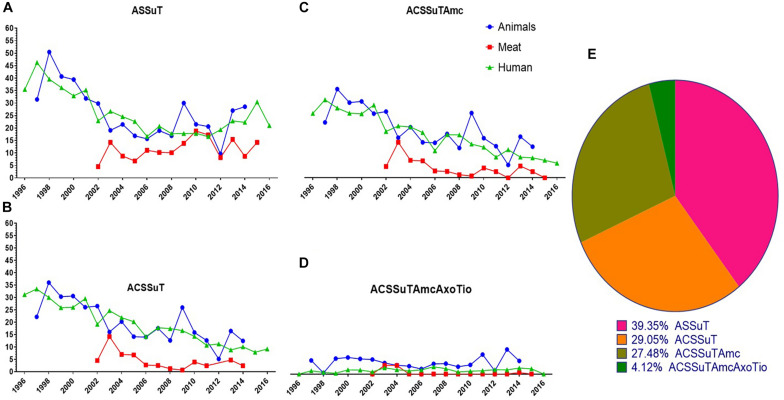
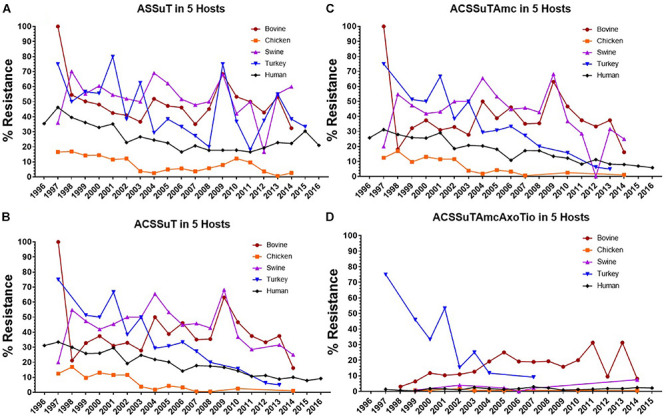
The AR patterns found in human isolates were compared with the AR patterns of isolates from animals and retail meats. Interestingly, the most frequently observed resistance patterns were tetra-resistant pattern ASSuT (Ampicillin, Streptomycin, Sulfonamides, and Tetracycline) and penta-resistant pattern ACSSuT (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, and Tetracycline). Other AR patterns such as ACSSuTAmc (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, Tetracycline, and Amoxicillin-clavulanic acid) and ACSSuTAmcAxoTio (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, Tetracycline, Amoxicillin-clavulanic acid, Ceftriaxone, and Ceftiofur) were also analyzed in this study. These four patterns comprised of 63% of the total 11,447 isolates. Among these, 39% of isolates showed ASSuT resistance pattern, 29% of isolates showed ACSSuT resistance pattern, 28% of isolates showed ACSSuTAmc and 4% of isolates showed ACSSuTAmcAxoTio resistance pattern. Four AR patterns found in S. Typhimurium isolates in this study are shown in Figure 4. Figure 5 showed the graphical representations of antibiotic resistance patterns found in S. Typhimurium isolates obtained from five different hosts (chicken, bovine, swine, turkey, and human).
FIGURE 4.
Graphical representations of four antibiotic resistance patterns (ASSuT, ACSSuT, ACSSuTAmc and ACSSuTAmcAxoTio) found in S. Typhimurium strains through 1996–2016 in United States. (A) ASSuT (Ampicillin, Streptomycin, Sulfonamides, and Tetracycline) resistance patterns found in S. Typhimurium isolates from animals, meat and humans. Though the ASSuT resistance in animal and human isolates of Typhimurium declined sharply during 2002–2008, the resistance is on rise in the recent years. The ASSuT resistance in meat isolates showed increasing pattern with time. (B) ACSSuT (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, and Tetracycline) resistance patterns found in S. Typhimurium isolates from animals, meat and humans. The ACSSuT resistance in human and meat isolates of Typhimurium showing decreasing pattern but the same resistance in animal isolates showing increasing trend with the time. (C) ACSSuTAmc (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, Tetracycline, and Amoxicillin-clavulanic acid) resistance patterns found in S. Typhimurium isolates from animals, meat and humans. The ACSSuTAmc resistance in human and meat isolates of Typhimurium showing decreasing pattern but in animal isolates the resistance remained high with time. (D) ACSSuTAmcAxoTio (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, Tetracycline, Amoxicillin-clavulanic acid, Ceftriaxone, and Ceftiofur) resistance patterns found in S. Typhimurium isolates from animals, meat and humans. ACSSuTAmcAxoTio resistance in animal isolates showed increasing tendency in the recent years. The XX’ presents the time of data collection while YY’ gives the percent of resistance. (E) Pie chart showing the percentage of distribution of four different resistance patterns among 7237 isolates of S. Typhimurium from animals, meat and humans.
FIGURE 5.
Graphical representations of four antibiotic resistance patterns (ASSuT, ACSSuT, ACSSuTAmc and ACSSuTAmcAxoTio) found in S. Typhimurium isolates through 1996–2016 in United States obtained from five different hosts (bovine, chicken, swine, turkey, and human). (A) ASSuT (Ampicillin, Streptomycin, Sulfonamides, and Tetracycline) resistance patterns found in S. Typhimurium isolates from bovine, chicken, swine, turkey and human. S. Typhimurium isolates from bovine, swine and turkey showed higher percentage of this resistance pattern than the isolates from chicken and human. (B) ACSSuT (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, and Tetracycline) resistance patterns found in S. Typhimurium isolates from bovine, chicken, swine, turkey and human. Swine and bovine isolates showed more ACSSuT resistance pattern than other host isolates. (C) ACSSuTAmc (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, Tetracycline, and Amoxicillin-clavulanic acid) resistance patterns found in S. Typhimurium isolates from bovine, chicken, swine, turkey and human. Swine and bovine isolates showed more percentage of resistance pattern among all host isolates. (D) ACSSuTAmcAxoTio (Ampicillin, Chloramphenicol, Streptomycin, Sulfonamides, Tetracycline, Amoxicillin-clavulanic acid, Ceftriaxone, and Ceftiofur) resistance patterns found in S. Typhimurium isolates from bovine, chicken, swine, turkey and human. Percentage of this resistance pattern found high in bovine Typhimurium isolates. The XX’ presents the time of data collection while YY’ gives the percent of resistance.
We found that, the ASSuT resistance in animal and human isolates of Typhimurium declined sharply during 2002–2008, but the resistance is on rise in the recent years. The ASSuT resistance in meat isolates showed increasing pattern with time (Figure 4A). S. Typhimurium isolates from bovine, swine, and turkey showed higher percentage of this resistance pattern than the isolates from chicken and human (Figure 5A). In this study, high resistance was recorded in S. Typhimurium strains to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (ACSSuT). The ACSSuT resistance in human and meat isolates of Typhimurium showed a decreasing but the same resistance in animal isolates showed an increasing trend with time (Figure 4B). Swine and bovine isolates showed more ACSSuT resistance pattern than other host isolates (Figure 5B). The ACSSuT pattern also found in different combinations with AMC, AXO, TIO antibiotics as additional resistances in our study. For ACSSuTAmc resistance, the isolates from human and meat showed decreasing pattern, but in animal isolates the resistance remained high with time (Figure 4C). The frequency of ACSSuTAmcAxoTio pattern remained constant over the years, though the resistance in isolates from animal showed a slight increase in the recent years (Figure 4D). ACSSuTAmc pattern was more prevalent in swine and bovine isolates and ACSSuTAmcAxoTio pattern in the bovine isolates (Figures 5C,D). Figure 4E showed the percentage of distribution of four different resistance patterns among 7237 strains of S. Typhimurium obtained from animals, meat and humans.
Analysis of the Best Classifier for Antibiotic Resistance Divergence
Classification and regression tree analysis of individual strains based on the MIC for multiple antimicrobials reveal that the MIC value toward COT (Trimethoprim-sulfamethoxazole) could be the best classifier to identify the origin of the isolates. The isolates originating in the animals and meat had a COT MIC of >0.12250 whereas those originating in the humans were having COT ≤ 0.12250. This implies that in spite of being susceptible to COT, the effective MIC is higher for the animal or meat isolate as compared to the human isolate. Similarly, if the isolates are to be classified based on their host (as humans, chickens, bovine, porcine, and turkey), the chicken isolates could be best identified as those isolates with MIC for COT > 0.12250 (biological cut off is 2), GEN ≤ 6 (biological cut off is 4), and STR ≤ 48 (biological cut off is 16). That means strains susceptible to COT but resistant to gentamicin and streptomycin are likely to have originated from chickens.
Sources for Human Infections and Dissemination of Resistance Genes
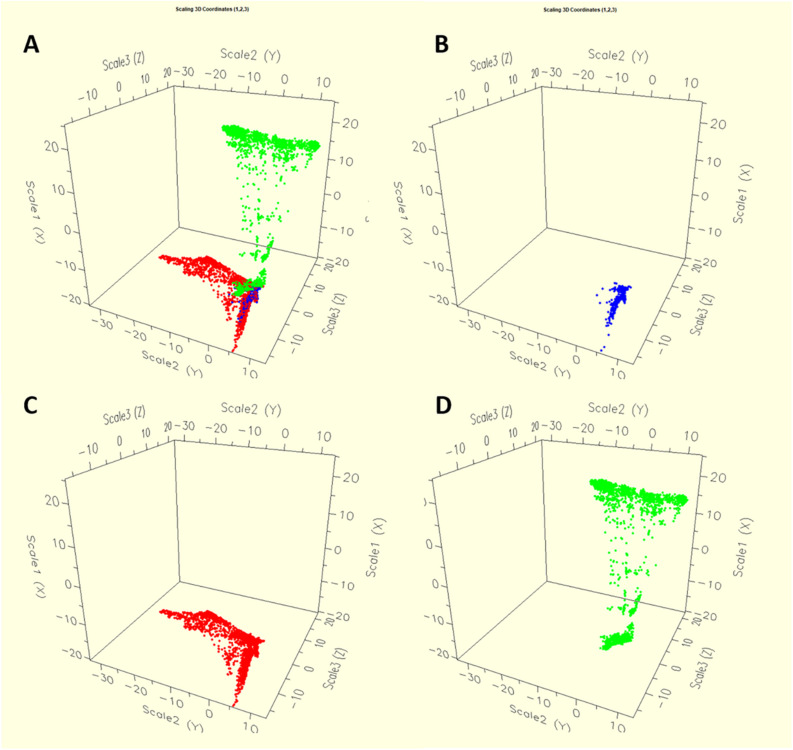
We found considerable variation in the resistance profile of Salmonella Typhimurium obtained from different sources. The AR profile abundance was higher in S. Typhimurium isolates obtained from humans. We found that S. Typhimurium strains from human and animal were emerged in easily distinguishable populations. Antibiogram of the strains derived from animals showed less diverse than the strains derived from humans. This significantly suggests that multiple sources could be involved in Salmonella infections in human. Our result also described that, epidemics of the human and animal are not similar and the diversity of AR is greater in the human isolates than the diversity of AR in isolates derived from animals and animal meat (Figure 6). We also found that S. Typhimurium strains from chicken showed higher AR diversity than the bovine, porcine or turkey strains (Figure 7).
FIGURE 6.
The dynamics of antibiotic resistance features of Salmonella Typhimurium isolates according to the all sources (A) such as animal (C), meat (B) and human (D) used in this study. The diversity of antimicrobial resistance is greater in the isolates from the human population than in those from animals and animal meat.
FIGURE 7.
The dynamics of antibiotic resistance features of Salmonella Typhimurium isolates by host population (A) such as Human (B), Chicken (C), Bovine (D), Porcine (E), Turkey (F). S. Typhimurium strains from human and chicken showed high antibiotic resistance diversity more than bovine, porcine and turkey strains.
Discussion
Continuous monitoring of the emergence of any bacterial serotype to detect in the food-chain is very important for public health point of view globally. The Salmonella Typhimurium strains from the recent years (2011–2016) showed much AR diversification as compared to previous year ranges (Figure 2). This is interesting and it could be explained by the fact that, the use as well as misuse of antibiotics is on rise in recent years and as a result antibiotic resistant strains of S. Typhimurium increased sharply and transmission of these strains occurred in the food-chain which ultimately affects humans. The immense use of antibiotics could be the main reason behind the emergence of AR in S. Typhimurium strains.
The reduced susceptibility pattern showed by S. Typhimurium population (Figure 3) could be responsible for treatment failures in some clinical situations. Streptomycin is not regularly used for the treatment of salmonellosis; but it is commonly used as a growth promoter in animals. Due to this reason, streptomycin could serve as a marker for resistant isolates moving through the food-chain (McDermott et al., 2016). Among the multiple factors that confer the emergence of antibiotic resistant bacteria, the extensive and overuse of antibiotics in medical system and agriculture is believed as the most pivotal (NARMS, 2019). The mechanism of the AR in Salmonella at the cellular level is complex (Penesyan et al., 2015). Salmonella develops AR mechanisms by the production of enzymes that can destroy action of antibiotics, by activating efflux pumps, and by producing β-lactamase which can degrade the structure of antibiotic molecules (Foley and Lynne, 2008; Andino and Hanning, 2015). AR can also be achieved by biofilm production which can cause increased risk of food safety (Steenackers et al., 2012; Corona and Martinez, 2013).
A very important feature of DT104 is that it is bearing Salmonella genomic island 1 (SGI-1). SGI-1 contains different AR genes identified in several Salmonella enterica serovars (Mossoro-Kpinde et al., 2015). Mulvey et al. (2006) reported that, variation in SGI-1 of the DT104 strains occurs through recombination. Another study using 359 strains demonstrated frequent and recurrent loss or gain of AR genes from the entire SGI-1 to single anti-microbial resistance genes (Mather et al., 2013). One of the characteristics of DT104 is that the strain typically shows resistance to ampicillin, chloramphenicol, streptomycin, sulfonamide, and tetracycline and it has the capability to gain resistance to other clinically important antibiotics (Helms et al., 2005; Mulvey et al., 2006). During 1990s MDR DT104 rapidly emerged globally and became the most common foodborne pathogen found in humans and animals (Ridley and Threlfall, 1998; Threlfall, 2000; Helms et al., 2005). Integrons, which contain AR genes, acquisition is also a very effective mechanism for DT104 to acquire resistance to different antibiotic classes (Stokes and Hall, 1989; Collis and Hall, 1995; Levesque et al., 1995).
Our analysis reveals that resistance was very low to an important group of antibiotics, the fluoroquinolones (Figure 3B). Due to its broad-spectrum activity, ciprofloxacin, which is a fluoroquinolone compound, is used for the treatment of Salmonella infections (Fàbrega and Vila, 2013). This same antibiotic is commonly recommended in sub-Saharan Africa, and prescribed with other important fluoroquinolone compounds such as levofloxacin, ofloxacin, and norfloxacin for patients with enteric diseases (Kagambèga et al., 2018). The antibiotic treatment regimen recommended by clinicians against Salmonella infection in human includes third generation cephalosporins, quinolones and macrolides (Guerrant et al., 2001). However, as a result of the high frequency of AR of animal-origin Salmonella isolates against cephalosporins, the recommended usage of cephalosporins became a great concern for the treatment of Salmonella derived from animals which can infect humans as well as other healthy animals. Another two antibiotics such as tetracycline and streptomycin showed high level of AR in this study, suggesting that clinicians should consider avoiding these two antibiotics for clinical use as well as in animal husbandry. The AR analysis in this study suggested fluoroquinolones (i.e., Ciprofloxacin) still can be used as a therapeutic agent of choice against Salmonella Typhimurium infection.
Four important AR patterns (ASSuT, ACSSuT, ACSSuTAmc, and ACSSuTAmcAxoTio) were analyzed in S. Typhimurium strains in this study (Figures 4, 5). ACSSuT pattern generally used when bacteria become resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline. A resembling resistance pattern ASSuT is used to show resistance to four antibiotics. Other important phenotypes (ACSSuTAmc and ACSSuTAmcAxoTio) which we found to be more prevalent along with ACSSuT and ASSuT were also analyzed here. The ASSuT resistance pattern was found related with S. Typhimurium DT193 strains which caused human infection in countries such as Spain, England and Wales (Prats et al., 2000; Threlfall et al., 2000). This same AR pattern was found associated with porcine population in United Kingdom (Maguire et al., 1993; Hampton et al., 1995). The ASSuT became more usual in Italy in S. Typhimurium since 2000 (Graziani et al., 2008; Dionisi et al., 2009). Here, this ASSuT is linked with S. Typhimurium strains of animals as well as strains of human origin. Busani et al. (2004) demonstrated a lower frequency of resistance to ACSSuT pattern in S. Typhimurium isolates obtained from humans than from non-human isolates. The ACSSuT is the typical resistance pattern factors of the DT104 (Adhikari et al., 2010; Mateva et al., 2018). The MDR S. Typhimurium strains in other studies showed similar AR pattern (ACSSuT); (Ribot et al., 2002; De Vito et al., 2015; Mossoro-Kpinde et al., 2015). Some reports described that the AR type ACSSuT may have evolved in Asia around early 1980s in other pathogens and were then horizontally transferred to DT104 (Angulo et al., 2000; Ribot et al., 2002).
Salmonella Typhimurium strains obtained from animal, meat and human were found to originate in distinct clusters. In our study, trimethoprim-sulfamethoxazole was found as the best classifier for AR differentiation of different population, which is also evidenced in other serovar such as S. Newport (Pan et al., 2018).
Based on antimicrobial susceptibility testing, our result suggests that multiple sources could be involved in Salmonella infections in human (Figure 6). Certain clusters of isolates act as the bridge between these human and animal isolates. From this perspective, the isolates from meat samples are at the interface of both animal and human (Figure 6). A study by Sun et al. (2014) demonstrated that S. Typhimurium infection in humans is linked with different clades than those which are prevalent in pet animals. Contaminated meat, milk and eggs are the probable source of Salmonella infection in humans. Humans are having
different diverse source of infections as compared to the local animals and it is possible that the AR diversity is more in humans than in animals (Figure 6). In their study, Mather et al. (2012) used epidemiological as well as ecological methods to analyze phenotypic AR data in DT104. The study found greater AR diversity in isolates derived from humans as compared to isolates obtained from animals. In another study, Wiesner et al. (2009), using MLST and PFGE methods found the existence of highly distributed S. Typhimurium strains that were derived in Mexico from human and food-animals, and from diverse geographic locations during different years.
The occurrence of antibiotic resistant strains in humans and chicken (Figure 7) warns of the possible risk of consumption of contaminated food. Through contamination of food, poultry may act as a very effective vector for transmission of Salmonella to human (Bemrah et al., 2003; Vandeplas et al., 2010; Paine et al., 2014; Li et al., 2018). Periodic outbreaks of live poultry-associated salmonellosis (LPAS) pointed the need of new alternative potential strategies to control human illnesses. LPAS outbreaks in the United States have been documented since 1955 (Anderson et al., 1955) and report showed that, the number of LPAS incidents is on rise significantly in last few years. Most of the LPAS outbreaks start in February (Basler et al., 2016). The DT104 strain which was correlated with cattle gradually became prevalent in poultry, pigs, and wild animals and showed a typical penta-resistance pattern of ACSSuT (Glynn et al., 1998). Study by Zhu et al. (2017) suggested the role of broiler chicken as the reservoir for multi-drug resistant Salmonella and in the processing plants cross-contamination is frequent between chicken and the environment including the workers working in the plants. This cross-contamination is of importance and these leads to the enhance of MDR Salmonella strains in the final stages which ultimately facilitate the spread of the AR genes widely to the consumers. Bovine salmonellosis is a significant threat to beef industry (Wray and Davies, 2000). Pigs are also responsible for the transmission of Salmonella to humans (Barilli et al., 2018). As the S. Typhimurium sometimes asymptomatic in pigs, most of the time the pathogen does not cause illness in pigs, but pigs could be an important source of contamination of the dead animals in the slaughter houses (Davies and Wray, 1997). This bacterial contamination could not be detected in the pigs in the farm, but this could finally cause the transmission to human (Kich et al., 2011; Almeida et al., 2016). Very few reports detected the antibiotic resistant Salmonella in turkey meat (Fakhr et al., 2006; Khaitsa et al., 2007) which is supporting our findings.
Conclusion
Our results revealed some significant observations for Salmonella Typhimurium epidemiology describing AR pattern dynamics across the food-chain. The recent diversification of AR found among the S. Typhimurium isolates is of great importance and significantly draw attention about the misuse of the antibiotics in the recent years. Major resistance patterns found were tetra-resistance and penta-resistance. The population diversity analysis calculated based on the MIC value revealed typical patterns of host segregation suggesting the distinction between the isolates from different host and sources but with some cross-overs and common shared population. This study helped in understanding the expansion of dynamics of antibiotic resistance of S. Typhimurium and recommended fluoroquinolone antibiotics as a possible potential therapeutic option against S. Typhimurium infection. The antimicrobial susceptibility, resistance profiles and different AR patterns in Salmonella strains including other potential pathogenic and virulence factors is expanding globally and this phenomenon must be monitored cautiously and continuously for the betterment of the treatment of salmonellosis. Nevertheless, this study has some limitations. The sampling bias is the one, with over sampling in human and less in animals, including disequilibrium in samples of animal origins. Most of the data presenting in this study is based on the quantitative MICs data. Future study based on genetic information, particularly whole genome sequence, can accelerate our understanding of S. Typhimurium population diversity and transmission along the food-chain.
Author Contributions
XW and SB contributed equally to this work, conducted the data analysis, and wrote the manuscript. HP and NP made significant contribution to refine and reorganized the data used in the manuscript and its presentation. XL and WF provided critical comments and aided with the edition of the manuscript. MY conceived the idea, collected the data, and suggested the statistical interpretation of the data analyzed and aided with the writing. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
All the individuals involved in the United States NARMS are acknowledged for their contribution in data collection and deposition of those, on the World Wide Web.
Footnotes
Funding. This study was supported by grants from National Program on Key Research Project (2017YFC1600103, 2018YFD0500501, and 2018YFD0701001), Fundamental Research Funds for the Central Universities (2-2050205-18-237), Zhejiang Provincial Natural Science Foundation of China (LR19C180001), Zhejiang University “Hundred Talent Program” (2016111), and the Recruitment Program of Global Youth Experts (13-313).
References
- Adhikari B., Besser T. E., Gay J. M., Fox L. K., Hancock D. D., Davis M. A. (2010). Multilocus variable-number tandem-repeat analysis and plasmid profiling to study the occurrence of blaCMY-2 within a pulsed-field gel electrophoresis-defined clade of Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 76 69–74. 10.1128/AEM.00210-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afema J. A., Mather A. E., Sischo W. M. (2015). Antimicrobial resistance profiles and diversity in Salmonella from humans and cattle, 2004-2011. Zoonoses Public Health 62 506–517. 10.1111/zph.12172 [DOI] [PubMed] [Google Scholar]
- Alcaine S. D., Warnick L. D., Wiedmann M. (2007). Antimicrobial resistance in nontyphoidal Salmonella. J. Food Prot. 70 780–790. 10.4315/0362-028x-70.3.780 [DOI] [PubMed] [Google Scholar]
- Almeida F., Medeiros M. I., Kich J. D., Falcão J. P. (2016). Virulence-associated genes, antimicrobial resistance and molecular typing of Salmonella typhimurium strains isolated from swine from 2000 to 2012 in Brazil. J. Appl. Microbiol. 120 1677–1690. 10.1111/jam.13110 [DOI] [PubMed] [Google Scholar]
- Anderson A. S., Bauer H., Nelson C. B. (1955). Salmonellosis due to Salmonella typhimurium with Easter chicks as likely source. J. Am. Med. Assoc. 158 1153–1155. [DOI] [PubMed] [Google Scholar]
- Andino A., Hanning I. (2015). Salmonella enterica: survival, colonization, and virulence differences among serovars. ScientificWorldJournal 2015:520179. 10.1155/2015/520179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angulo F. J., Johnson K. R., Tauxe R. V., Cohen M. L. (2000). Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb. Drug Resist. 6 77–83. 10.1089/mdr.2000.6.77 [DOI] [PubMed] [Google Scholar]
- Barilli E., Bacci C., StellaVilla Z., Merialdi G., D’Incau M., Brindani F., et al. (2018). Antimicrobial resistance, biofilm synthesis and virulence genes in Salmonella isolated from pigs bred on intensive farms. Ital. J. Food Saf. 7:7223. 10.4081/ijfs.2018.7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler C., Nguyen T. A., Anderson T. C., Hancock T., Behravesh C. B. (2016). Outbreaks of human Salmonella infections associated with live poultry, United States, 1990-2014. Emerg. Infect. Dis. 22 1705–1711. 10.3201/eid2210.150765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemrah N., Bergis H., Colmin C., Beaufort A., Millemann Y., Dufour B., et al. (2003). Quantitative risk assessment of human salmonellosis from the consumption of a turkey product in collective catering establishments. Int. J. Food Microbiol. 80 17–30. 10.1016/s0168-1605(02)00145-9 [DOI] [PubMed] [Google Scholar]
- Busani L., Graziani C., Battisti A., Franco A., Ricci A., Vio D., et al. (2004). Antibiotic resistance in Salmonella enterica serotypes Typhimurium, Enteritidis and Infantis from human infections, food stuffs and farm animals in Italy. Epidemiol. Infect. 132 245–251. 10.1017/s0950268803001936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye P., Michael G. B., Schwarz S., Barrett T. J., Brisabois A., White D. G. (2006). The clonal spread of multidrug-resistant nontyphi Salmonella serotypes. Microbes Infect. 8 1891–1897. 10.1016/j.micinf.2005.12.020 [DOI] [PubMed] [Google Scholar]
- CDC (2013a). National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2011. Atlanta, GA: Centers for Disease Control and Prevention. [Google Scholar]
- CDC (2013b). Office of Infectious Disease. Antibiotic resistance threats in the United States. April 2013. Available at: http://www.cdc.gov/drugresistance/threat-report-2013. (accessed January 28, 2015). [Google Scholar]
- CDC, and NARMS (2019). National Antimicrobial Resistance Monitoring System (NARMS) Now:Human Data. Atlanta, GA: Centers for Disease Control and Prevention. [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standardsfor Antimicrobial Susceptibility Testing; Supplement M100, 27th Edn Wayne,PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Collis C. M., Hall R. M. (1995). Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39 155–162. 10.1128/aac.39.1.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona F., Martinez J. L. (2013). Phenotypic resistance to antibiotics. Antibiotics 2 237–255. 10.3390/antibiotics2020237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R., Wray C. (1997). Study of multi-resistant Salmonella typhimurium infection in pig herds: preliminary findings. Pig J. 40 80–88. [Google Scholar]
- De Vito D., Monno R., Nuccio F., Legretto M., Oliva M., Coscia M. F., et al. (2015). Diffusion and persistence of multidrug resistant Salmonella typhimurium strains phage type DT120 in southern Italy. Biomed. Res. Int. 2015:265042. 10.1155/2015/265042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisi A. M., Graziani C., Lucarelli C., Filetici E., Villa L., Owczarek S., et al. (2009). Molecular characterization of multidrug-resistant strains of Salmonella enterica serotype typhimurium and monophasic variant (S. 4,[5],12:i:-) isolated from human infections in Italy. Foodborne Pathog. Dis. 6 711–717. 10.1089/fpd.2008.0240 [DOI] [PubMed] [Google Scholar]
- Fàbrega A., Vila J. (2013). Salmonella enterica serovar typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26 308–341. 10.1128/CMR.00066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhr M. K., Sherwood J. S., Thorsness J., Logue C. M. (2006). Molecular characterization and antibiotic resistance profiling of Salmonella isolated from retail Turkey meat products. Foodborne Pathog. Dis. 3 366–374. 10.1089/fpd.2006.3.366 [DOI] [PubMed] [Google Scholar]
- FDA. (2015). National Antimicrobial Resistance Monitoring System-EntericBacteria (NARMS): 2012-2013 integrated report. Rockville, MD: US Department of Health and Human Services. [Google Scholar]
- Foley S. L., Lynne A. M. (2008). Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance. J. Anim. Sci. 86 E173–E187. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O., Boyle E. C., Grassl G. A. (2014). Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 5:391. 10.3389/fmicb.2014.00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn M. K., Bopp C., Dewitt W., Dabney P., Mokhtar M., Angulo F. J. (1998). Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N. Engl. J. Med. 338 1333–1338. [DOI] [PubMed] [Google Scholar]
- Graziani C., Busani L., Dionisi A. M., Lucarelli C., Owczarek S., Ricci A., et al. (2008). Antimicrobial resistance in Salmonella enterica serovar typhimurium from human and animal sources in Italy. Vet. Microbiol. 128 414–418. [DOI] [PubMed] [Google Scholar]
- Guerrant R. L., Van Gilder T., Steiner T. S., Thielman N. M., Slutsker L., Tauxe R. V., et al. (2001). Infectious diseases society of america. Practice guidelines for the management of infectious diarrhea. Clin. Infect. Dis. 32 331–351. 10.1093/cid/civ482 [DOI] [PubMed] [Google Scholar]
- Hampton M. D., Threfall E. J., Frost J. A., Ward L. R., Rowe B. (1995). Salmonella typhimurium DT 193: differentation of an epidemic phage type by antibiogram, plasmid profile, plasmid fingerprint and salmonella plasmid virulence (spv) gene probe. J. Appl. Bacteriol. 78 402–408. [DOI] [PubMed] [Google Scholar]
- Havelaar A. H., Kirk M. D., Torgerson P. R., Gibb H. J., Hald T., Lake R. J., et al. (2015). World health organization foodborne disease burden epidemiology reference group. world health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12:e1001923. 10.1371/journal.pmed.1001923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms M., Ethelberg S., Mølbak K. (2005). International Salmonella typhimurium DT104 infections, 1992–2001. Emerg. Infect. Dis. 11 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriksen R. S., Vieira A. R., Karlsmose S., Lo Fo Wong D. M., Jensen A. B., et al. (2011). Global monitoring of Salmonella serovar distribution from the world health organization global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog. Dis. 8 887–900. 10.1089/fpd.2010.0787 [DOI] [PubMed] [Google Scholar]
- Herikstad H., Motarjemi Y., Tauxe R. V. (2002). Salmonella surveillance: a global survey of public health serotyping. Epidemiol. Infect. 129 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann E. L. (2001). Nontyphoidal salmonellosis. Clin. Infect. Dis. 32 263–269. 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- Izumiya H., Sekizuka T., Nakaya H., Taguchi M., Oguchi A., Ichikawa N., et al. (2011). Whole-genome analysis of Salmonella enterica serovar typhimurium T000240 reveals the acquisition of a genomic island involved in multidrug resistance via IS1 derivatives on the chromosome. Antimicrob. Agents Chemother. 55 623–630. 10.1128/AAC.01215-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagambèga A., Lienemann T., Frye J. G., Barro N., Haukka K. (2018). Whole genome sequencing of multidrug-resistant Salmonella enterica serovar Typhimurium isolated from humans and poultry in Burkina Faso. Trop. Med. Health 46:4. 10.1186/s41182-018-0086-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitsa M. L., Kegode R. B., Doetkott D. K. (2007). Occurrence of antimicrobial-resistant Salmonella species in raw and ready to eat Turkey meat products from retail outlets in the Midwestern United States. Foodborne Pathog. Dis. 4 517–525. 10.1089/fpd.2007.0010 [DOI] [PubMed] [Google Scholar]
- Kich J. D., Coldebella A., MoreÂs N., Nogueira M. G., Cardoso M., Fratamico P. M., et al. (2011). Prevalence, distribution, and molecular characterization of Salmonella recovered from swine finishing herds and a slaughter facility in Santa Catarina. Brazil. Int. J. Food Microbiol. 151 307–313. 10.1016/j.ijfoodmicro.2011.09.024 [DOI] [PubMed] [Google Scholar]
- Kingsley R. A., Msefula C. L., Thomson N. R., Kariuki S., Holt K. E., Gordon M. A., et al. (2009). Epidemic multiple drug-resistant Salmonella typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19 2279–2287. 10.1101/gr.091017.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan R., Reeves P. R., Octavia S. (2009). Population structure, origins and evolution of major Salmonella enterica clones. Infect. Genet Evol. 9 996–1005. 10.1016/j.meegid.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Levesque C., Piche L., Larose C., Roy P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39 185–191. 10.1128/aac.39.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Yang X., Tan H., Ke B., He D., Wang H., et al. (2018). Whole genome sequencing analysis of Salmonella enterica serovar Weltevreden isolated from human stool and contaminated food samples collected from the Southern coastal area of China. Int. J. Food Microbiol. 266 317–323. 10.1016/j.ijfoodmicro.2017.10.032 [DOI] [PubMed] [Google Scholar]
- Maguire H. C. F., Codd A. A., MacKay V. E., Rowe B., Mitchell E. (1993). A large outbreak of human salmonellosis traced to a local pig farm. Epidemiol. Infect. 110 239–246. 10.1017/s0950268800068151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz S. E., Musto J., Scallan E., Angulo F. J., Kirk M., O’Brien S. J., et al. (2010). The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 50 882–889. 10.1086/650733 [DOI] [PubMed] [Google Scholar]
- Mateva G., Pedersen K., Sørensen G., Asseva G., Daskalov H., Petrov P., et al. (2018). Use of multiple-locus variable- number of tandem repeats analysis (MLVA)to investigate geneticdiversity of Salmonella enterica subsp. enterica serovar Typhimurium isolates from human, food, and veterinary sources. Microbiologyopen 7:e00528. 10.1002/mbo3.528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather A. E., Matthews L., Mellor D. J., Reeve R., Denwood M. J., Boerlin P., et al. (2012). An ecological approach to assessing the epidemiology of antimicrobial resistance in animal and human populations. Proc. Biol. Sci. 279 1630–1639. 10.1098/rspb.2011.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather A. E., Phuong T. L. T., Gao Y., Clare S., Mukhopadhyay S., Goulding D. A., et al. (2018). New variant of multidrug-resistant Salmonella enterica serovar Typhimurium associated with invasive disease in immunocompromised patients in Vietnam. MBio. 9:e1056-18. 10.1128/mBio.01056-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather A. E., Reid S. W., Maskell D. J., Parkhill J., Fookes M. C., Harris S. R., et al. (2013). Distinguishable epidemics of multidrug-resistant Salmonella typhimurium DT104 in different hosts. Science 341 1514–1517. 10.1126/science.1240578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott P. F., Tyson G. H., Kabera C., Chen Y., Li C., Folster J. P., et al. (2016). Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother. 60 5515–5520. 10.1128/AAC.01030-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael G. B., Butaye P., Cloeckaert A., Schwarz S. (2006). Genes and mutations conferring antimicrobial resistance in Salmonella: an update. Microbes. Infect. 8 1898–1914. 10.1016/j.micinf.2005.12.019 [DOI] [PubMed] [Google Scholar]
- Mossoro-Kpinde C. D., Manirakiza A., Mbecko J. R., Misatou P., Le Faou A., Frank T. (2015). Antimicrobial resistance of enteric Salmonella in Bangui, central African republic. J. Trop. Med. 2015:483974. 10.1155/2015/483974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Boyd D. A., Olson A. B., Doublet B., Cloeckaert A. (2006). The genetics of Salmonella genomic island 1. Microbes. Infect. 8 1915–1922. 10.1016/j.micinf.2005.12.028 [DOI] [PubMed] [Google Scholar]
- NARMS. (2019). NARMS Integrated Report 2014. Available at: https://www.fda.gov/downloads/AnimalVeterinary/SafetyHealth/AntimicrobialResistance/NationalAntimicrobialResistanceMonitoringSystem/UCM528861.pdf (accessed April 23, 2017). [Google Scholar]
- Okoro C. K., Kingsley R. A., Connor T. R., Harris S. R., Parry C. M., Al-Mashhadani M. N., et al. (2012). Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat. Genet. 44 1215–1221. 10.1038/ng.2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine S., Thornley C., Wilson M., Dufour M., Sexton K., Miller J., et al. (2014). An outbreak of multiple serotypes of Salmonella in New Zealand linked to consumption of contaminated tahini imported from Turkey. Foodborne Pathog. Dis. 11 887–892. 10.1089/fpd.2014.1773 [DOI] [PubMed] [Google Scholar]
- Pan H., Paudyal N., Li X., Fang W., Yue M. (2018). Multiple food-animal-borne route in transmission of antibiotic-resistant salmonella newport to humans. Front. Microbiol. 9:23. 10.3389/fmicb.2018.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Sokurenko E. V., Chattopadhyay S. (2016). Corrected genome annotations reveal gene loss and antibiotic resistance as drivers in the fitness evolution of Salmonella enterica serovar typhimurium. J. Bacteriol. 198 3152–3161. 10.1128/jb.00545-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penesyan A., Gillings M., Paulsen I. T. (2015). Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20 5286–5298. 10.3390/molecules20045286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats G., Mirelis B., Llovet T., Munoz C., Miro E., Navarro F. (2000). Antibiotic resistance trends in enteropathogenic bacteria isolated in 1985–1987 and 1995–1998 in Barcelona. Antimicrob. Agents Chemother. 44 1140–1145. 10.1128/aac.44.5.1140-1145.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot E. M., Wierzba R. K., Angulo F. J., Barrett T. J. (2002). Salmonella enterica serotype typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8 387–391. 10.3201/eid0804.010202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A., Threlfall E. J. (1998). Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb. Drug Resist. 4 113–118. 10.1089/mdr.1998.4.113 [DOI] [PubMed] [Google Scholar]
- Steenackers H., Hermans K., Vanderleyden J., De Keersmaecker S. C. J. (2012). Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 45 502–531. 10.1016/j.foodres.2011.01.038 [DOI] [Google Scholar]
- Stokes H. W., Hall R. M. (1989). A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 3 1669–1683. 10.1111/j.1365-2958.1989.tb00153.x [DOI] [PubMed] [Google Scholar]
- Su L. H., Chiu C. H. (2007). Salmonella: clinical importance and evolution of nomenclature. Chang Gung Med. J. 30 210–219. [PubMed] [Google Scholar]
- Sun J., Ke B., Huang Y., He D., Li X., Liang Z., et al. (2014). The molecular epidemiological characteristics and genetic diversity of Salmonella typhimurium in Guangdong, China, 2007-2011. PloS One. 9:e113145. 10.1371/journal.pone.0113145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall E. J. (2000). Epidemic Salmonella typhimurium DT 104-a truly international multiresistant clone. J. Antimicrob. Chemother. 46 7–10. [DOI] [PubMed] [Google Scholar]
- Threlfall E. J., Ward L. R., Skinner J. A., Graham A. (2000). Antimicrobial drug resistance in nontyphoidal Salmonellas from humans in England and Wales in 1999: decrease in multiple resistance in Salmonella enterica serotypes typhimurium, Virchow, Hadar. Microb. Drug Resist. 4 319–325. 10.1089/mdr.2000.6.319 [DOI] [PubMed] [Google Scholar]
- Vandeplas S., Dubois Dauphin R., Beckers Y., Thonart P., Théwis A. (2010). Salmonella in chicken: current and developing strategies to reduce contamination at farm level. J. Food Prot. 73 774–785. [DOI] [PubMed] [Google Scholar]
- Wiesner M., Zaidi M. B., Calva E., Fernandez-Mora M., Calva J. J., Silva C. (2009). Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar typhimurium strains. BMC microbiology 9:131. 10.1186/1471-2180-9-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray C., Davies R. H. (2000). “Salmonella infections in cattle,” in Salmonella in Domestic Animals, eds Wray C., Wray A. (Wallingford: CABI Publishing; ), 169–190. [Google Scholar]
- Zhang S., Kingsley R. A., Santos R. L., Andrews-Polymenis H., Raffatellu M., Figueiredo J., et al. (2003). Molecular pathogenesis of Salmonella enterica serotype typhimurium induced diarrhea. Infect. Immun. 71 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Lai H., Zou L., Yin S., Wang C., Han X., et al. (2017). Antimicrobial resistance and resistance genes in Salmonella strains isolated from broiler chickens along the slaughtering process in China. Int. J. Food Microbiol. 259 43–51. 10.1016/j.ijfoodmicro.2017.07.023 [DOI] [PubMed] [Google Scholar]