Abstract
Development is often coordinated by biologically active mobile compounds that move between cells or organs. Arabidopsis mutants with defects in the BYPASS1 (BPS1) gene overproduce an active mobile compound that moves from the root to the shoot and inhibits growth. Here, we describe two related Arabidopsis genes, BPS2 and BPS3. Analyses of single, double and triple mutants revealed that all three genes regulate production of the same mobile compound, the bps signal, with BPS1 having the largest role. The triple mutant had a severe embryo defect, including the failure to properly establish provascular tissue, the shoot meristem and the root meristem. Aberrant expression of PINFORMED1, DR5, PLETHORA1, PLETHORA2 and WUSCHEL-LIKE HOMEOBOX5 were found in heart-stage bps triple-mutant embryos. However, auxin-induced gene expression, and localization of the PIN1 auxin efflux transporter, were intact in bps1 mutants, suggesting that the primary target of the bps signal is independent of auxin response. Thus, the bps signal identifies a novel signaling pathway that regulates patterning and growth in parallel with auxin signaling, in multiple tissues and at multiple developmental stages.
Keywords: Long-distance signaling, Embryogenesis, Vascular meristem
INTRODUCTION
Coordination of development among distinct cell types is an underlying requirement for multicellular organisms. Numerous hormone and peptide signals are used in plants both at long-range and more locally to regulate patterning and growth of different organs. Identification of additional mobile signaling molecules is expected to elucidate further the fundamental aspects of plant development (Van Norman et al., 2011a). A putative long-distance signal was uncovered by the analysis of the Arabidopsis bypass1 (bps1) mutant. This mutant shows severe shoot growth arrest and abnormal root development, phenotypes that arise due to overproduction of a mobile compound (Van Norman et al., 2004). In bps1 mutants, this compound is synthesized in the root, and it moves to the shoot, where it is sufficient to arrest development.
The developmental defects in bps1 roots appear to be caused by the same mobile compound as the shoot defects. This connection was established from studies of bps1 mutants grown on media containing inhibitors of carotenoid biosynthesis, fluridone and 2-(4-chlorophenylthio)-triethylamine hydrochloride (CPTA). Treatment with these compounds results in albino phenotypes, which strongly reduce growth in normal plants. However, the same treatment partially rescues both the root and the shoot defects of bps1 mutants (Van Norman et al., 2004). Although carotenoids are required for synthesis of the mobile compound, genetic analysis revealed that the mobile compound was distinct from the known carotenoid-derived signaling molecules, abscisic acid and strigolactones (Van Norman and Sieburth, 2007). Here, we refer to this unidentified mobile compound as the bypass (bps) signal.
Arguably the best-known plant mobile signaling molecule is auxin. Developmental roles of auxin include establishment of the embryonic apical-basal axis, root establishment, vascular patterning, gravitropic responses and organogenesis (Jenik et al., 2005; Benjamins and Scheres, 2008; Nawy et al., 2008). Normal embryogenesis requires auxin production, as revealed by mutant combinations deficient in members of the YUCCA auxin biosynthetic enzymes, which show embryonic defects in root and cotyledon formation, and in the patterning of internal tissues (Cheng et al., 2007). Transport of auxin is also essential, as embryos with chemically induced or genetic defects in auxin transport have defects in apical/basal polarity, and root and cotyledon formation (Hadfi et al., 1998; Friml et al., 2003; Weijers et al., 2005; Mravec et al., 2008). Cellular responses to auxin include changes in transcription, and mutants with defects in auxin-responsive transcription factors display a similar range of embryo development defects (Hardtke and Berleth, 1998; Long et al., 2006; Weijers et al., 2006). Auxin transport is, in turn, under regulation by Class III homeodomain-leucine zipper (HD-ZIPIII) and KANADI (KAN) activities (Ilegems et al., 2010).
Other plant hormones and signaling molecules can modulate auxin-signaling processes (Wolters and Jürgens, 2009). Interaction between auxin and the plant hormone cytokinin is crucial for establishment of the root stem cell niche in the Arabidopsis embryo (Müller and Sheen, 2008), and cytokinins can also affect PIN auxin transporter abundance (Marhavy et al., 2011; Zhang et al., 2011). Strigolactone also influences auxin transport, thereby modulating lateral organ outgrowth (Crawford et al., 2010; Domagalska and Leyser, 2011). The bps signal might be an additional molecule that affects auxin response and/or transport, as shoots of bps1 mutants, treated with a synthetic auxin, 2,4-D, are unable to induce expression of the auxin-responsive reporter DR5::GUS (Van Norman et al., 2004). Although this result implicated auxin signaling as a potential target of the bps signaling pathway, direct evidence linking the bps signal to auxin pathways is lacking.
The bps1 mutant phenotype is evident upon germination, particularly in the emerging root, and experiments involving genetic manipulation of root development revealed that the signaling pathway is also effective in older seedlings (Van Norman et al., 2011b). However, its possible function at other developmental stages has not been addressed. The BPS1 gene is conserved in plant genomes, but no function can be deduced by its amino acid sequence. The Arabidopsis genome encodes two genes highly similar to BPS1, which we call BPS2 and BPS3. Here, functional analysis of the BPS gene family uncovered a role for the bps signal in embryo patterning. Use of molecular markers during embryogenesis, and auxin-focused experiments with the bps1 single mutant suggest that auxin signaling is intact in bps mutants, and that the bps signal interferes with a developmental process common to embryogenesis, seedlings and late vegetative development.
MATERIALS AND METHODS
Plant materials
The bps1-1 and bps1-2 alleles have been described previously (Van Norman et al., 2004). Details on the bps2 and bps3 alleles and genotyping are in supplementary material Tables S1 and S2 (McCallum et al., 2000a; McCallum et al., 2000b; Till et al., 2003; Henikoff et al., 2004). Double and triple bps mutants used the bps1-2, bps2-2 and bps3-2 alleles (all Col background). Repeat analyses with independently generated double and triple mutants, and different seed batches, gave identical results. Auxin analyses used axr1-3 (Leyser et al., 1993) and rty-1 (Boerjan et al., 1995). Molecular markers included pPIN1::PIN1-GFP (Heisler et al., 2005), DR5(rev)::GFP (Friml et al., 2003), pWOX5::NLS-vYFP3 and WOX8gΔ::NLS-vYFP3 (Breuninger et al., 2008), pPLT1::CFP and pPLT2::CFP (Galinha et al., 2007). Analyses were carried out on the self-pollinated progeny of BPS1+/bps1– bps2–/– bps3–/– [Marker+/+].
Growth conditions
Plants were grown as previously described (Van Norman et al., 2004), and seedling phenotypes analyzed at either 9 days post-imbibition (dpi) (22°C) or 11 dpi (16°C). Growth media were supplemented with the auxin NAA (1 or 100 μM) or NPA (5 μM) at 22°C.
BYPASS gene family
An unrooted phylogenetic tree was based on predicted amino acid sequence identified using BLAST on the TAIR web site and constructed using phylogeny.fr, using the ‘A la Carte’ mode (Dereeper et al., 2008).
Expression analysis
Q-RT PCR used three biological and two technical replicates, and ACT2 as an internal reference (Zhang et al., 2010). Auxin-induced gene expression experiments used 4-day-old seedlings treated with 10 μM 2,4-D for 6 hours. Primer sequences are provided in supplementary material Table S2. Whole-mount in situ hybridization of BPS1 was performed as described (Hejátko et al., 2006) and used a BPS1-specific probe corresponding to the 3′ region of the gene. Figures show representative data more from than 10 embryos for each stage, and negative controls with a BPS1 sense probe.
Microscopy
Developing seeds were mounted in 70% chloral hydrate solution, 4% glycerol and examined with differential interference contrast optics on Olympus BX50 microscope. Normal sibling embryos served as an internal standard to stage embryos. Fluorescence (GFP, YFP and CFP) was visualized using a Zeiss 510 Meta laser-scanning confocal microscope and an Olympus BX50 microscope. Analysis of PIN1-GFP trafficking was conducted on 3 dpi seedlings following methods previously described (Geldner et al., 2001; Sieburth et al., 2006).
FL and CPTA treatment
Carotenoid biosynthesis inhibitors [CPTA (2-(4-chlorophenylthio)-triethylamine hydrochloride) or Fluridone (FL) (Sigma)] were applied to siliques of soil-grown plants (bps1-2+/– bps2-2–/– bps3-2–/– and wild-type). The primary inflorescence stem was removed, and three axillary inflorescence stems of each experimental plant were marked for different treatments (100 μM FL, 5 mM CPTA or with 10% ethanol as control). Cotton-tipped applicators were used to applying the inhibitor (or control) to the outside of each silique daily. Treatments began 2 days after pollination, and continued daily until age-matched controls were starting to turn brown. Silique color was monitored to determine the effectiveness of FL and CPTA treatment; photobleaching occurred within 2 days of initiating treatment. Seeds from treated siliques were germinated on agar plates at 22°C and analyzed at 9 dpi.
RESULTS
BYPASS gene family
bypass1 (bps1) mutants show a severe growth defect that resulted from a non-cell-autonomous signal generated within the mutant root (Van Norman et al., 2004; Van Norman et al., 2011b). Although BPS1 was only required in the root, BPS1 mRNA was detected in other organs, suggesting that BPS1 might have additional functions that are masked by redundant gene activities. Indeed, the Arabidopsis genome contains two additional BYPASS (BPS) genes, BPS2 and BPS3 (Fig. 1A-B; see supplementary material Table S4). In roots, shoots and siliques, BPS1 was expressed at the highest level of these three genes, whereas BPS3 expression was almost undetectable (Fig. 1C). Expression of all three genes was highest in siliques, suggesting that BYPASS function might be particularly important during embryogenesis.
Fig. 1.
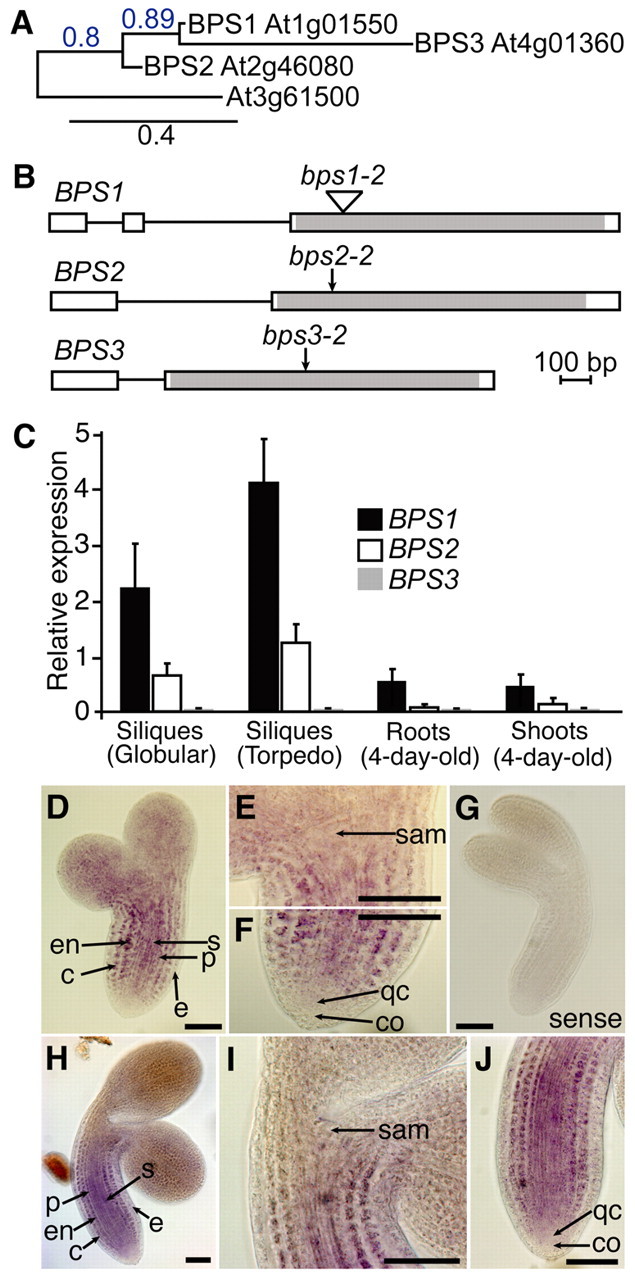
The Arabidopsis BYPASS gene family encodes conserved proteins that are predominantly expressed in siliques. (A) An unrooted phylogenetic tree based on amino acid sequences of Arabidopsis BPS gene family members reveals a close relationship between BPS1, BPS2 and BPS3. (B) The gene structure of BPS1, BPS2 and BPS3 reveals a very similar organization of exons (boxes) and introns (lines), with the coding region restricted to the 3′-most exon. Mutation sites for alleles used in this study are indicated by arrows. (C) Relative expression of BPS1, BPS2 and BPS3 in select tissues. Error bars are s.d. (D-J) In situ hybridization of BPS1 in wild-type embryos. (D-G) Torpedo-stage embryos; (D-F) hybridization with antisense probe and (G) Sense probe. The shoot apical meristem and root meristem regions are shown at higher magnification in E and F, respectively. (H-J) Bent-cotyledon stage embryos; (I) SAM and (J) RAM of embryo shown in H. Scale bars: 50 μm. c, cortex; co, columella; e, epidermis; en, endodermis; p, pericycle; qc, quiescent center; s, stele; sam, shoot apical meristem.
We analyzed embryonic expression of BPS1 by whole-mount in situ hybridization (Fig. 1D-J). Strong expression was found in the central hypocotyl (cortex, endodermis and vascular) tissues in both torpedo-stage (Fig. 1D-F) and mature embryos (Fig. 1H-J), but we found no expression in the epidermis. There was very low expression in the shoot apical meristem (SAM) and the root stem cell region, despite these tissues showing the most severe defects in bps1 mutants (Van Norman et al., 2004). The lack of BPS1 expression in these tissues is consistent with bps1 phenotypes arising from a non-cell-autonomous signal, and the presence of BPS1 RNA in cortex, endodermis and vascular tissues suggests that these tissues could all serve as locations for bps1 signal synthesis.
BPS1, BPS2 and BPS3 carry out partially redundant functions
To investigate biological functions of BPS2 and BPS3, we used reverse genetic resources to isolate mutant alleles. For BPS2, we isolated one T-DNA insertion allele (bps2-1), two nonsense alleles (bps2-2 and bps2-3) and 10 alleles with missense mutations (bps2-4 to bps2-13) (see supplementary material Table S1) (McCallum et al., 2000b; Alonso et al., 2003; Till et al., 2003; Li et al., 2007). The bps2-1 T-DNA integrated at W96, and the bps2-2 and bps2-3 mutations introduced premature stop codons at W63 and W96, respectively. For BPS3, we isolated one nonsense allele (bps3-2) and 14 alleles (bps3-3 to bps3-16) with missense mutations (see supplementary material Table S1). Neither bps2 nor bps3 alleles showed detectable abnormal phenotypes at any developmental stage. We selected bps2-2 and bps3-2 for generating double and triple mutants as they were predicted to be null alleles.
We generated all three double mutant combinations and the triple mutant. The bps2-2 bps3-2 double mutant showed no detectable abnormal phenotype, and bps1-2 bps3-2 double mutants were indistinguishable from bps1-2 single mutants. However, the bps1-2 bps2-2 double mutant showed additional phenotypes, and the bps1-2 bps2-2 bps3-2 triple mutant showed severe seedling phenotypes. Among the mutant combinations, one phenotypic difference was seedling size; the bps triple mutants were less than half the size of bps1 bps2 double mutants, which were similar in size to the bps1-2 single mutant (Fig. 2; see supplementary material Fig. S1). Second, the basal pole of the bps triple mutants was blunt, with no sign of a normal root, and with white hypocotyl-like tissue. An organized root was also missing from the bps1 bps2 double mutant, but the hypocotyl was longer and included both green and white tissue (Fig. 2). Third, germination rates were progressively reduced in double and triple mutants (see next section).
Fig. 2.

Seedling phenotypes of bps1, and double and triple bps mutants. Each column of this figure shows 9-day-old seedlings of the same genotype, and used the following alleles: bps1-2, bps2-2 and bps3-2.(A) Comparison of seedling sizes and formation of root and hypocotyl. (B) Leaf initiation and the SAM region. The first leaf pair of the wild type (Col-0) extended out of the field shown, and leaf 3 obscured the view of the SAM. The bps1 mutant produced two leaf primordia that arrest growth, and no leaf primordia were observed in bps1 bps2 or bps1 bps2 bps3 double and triple mutants. (C) Cotyledons of both the wild type (Col-0) and bps1 have distinct blade and petiole, but bps1 bps2 and bps1 bps2 bps3 cotyledons blades lack petioles and are often fused along their margins. Scale bars: 1 mm in A; 100 μm in B; 500 μm in C.
The shoot apex of both bps1 bps2 and bps triple mutants showed no evidence of leaf primordia, in contrast to bps1-2, which produces two small, arrested, leaf primordia (Fig. 2). Cotyledon defects were also more pronounced in the bps1 bps2 double and bps triple mutants; cotyledons were smaller and, in most individuals, they were fused. This analysis also revealed a cotyledon defect in bps1-2 single mutants. Wild-type seedlings produce two cotyledons, each on its own separate petiole. However, about 25% of bps1 mutants in the Col-0 background (bps1-2 and bps1-5) showed cotyledon defects, such as fused cotyledons (a single wide fused cotyledon and cotyledons with lobes) or supernumerary, yet unfused, cotyledons (see supplementary material Fig. S2). By contrast, cotyledons of the bps1-1 allele (Landsberg erecta accession) were similar to the wild type. This phenotypic difference between the bps1 alleles appeared to be caused by genetic background, as bps1-5 has the same genetic lesion as bps1-1, but it is in the Col-0 accession, and its cotyledon phenotype is similar to bps1-2. We quantified the fused cotyledon phenotype in single, double and triple bps mutants (Fig. 2C, Fig. 3A; see supplementary material Table S3). Seedlings with cotyledon fusion ranged from just under 2% of the mutants for bps1-2, to 40% and 60% for bps double and triple mutants, respectively.
Fig. 3.
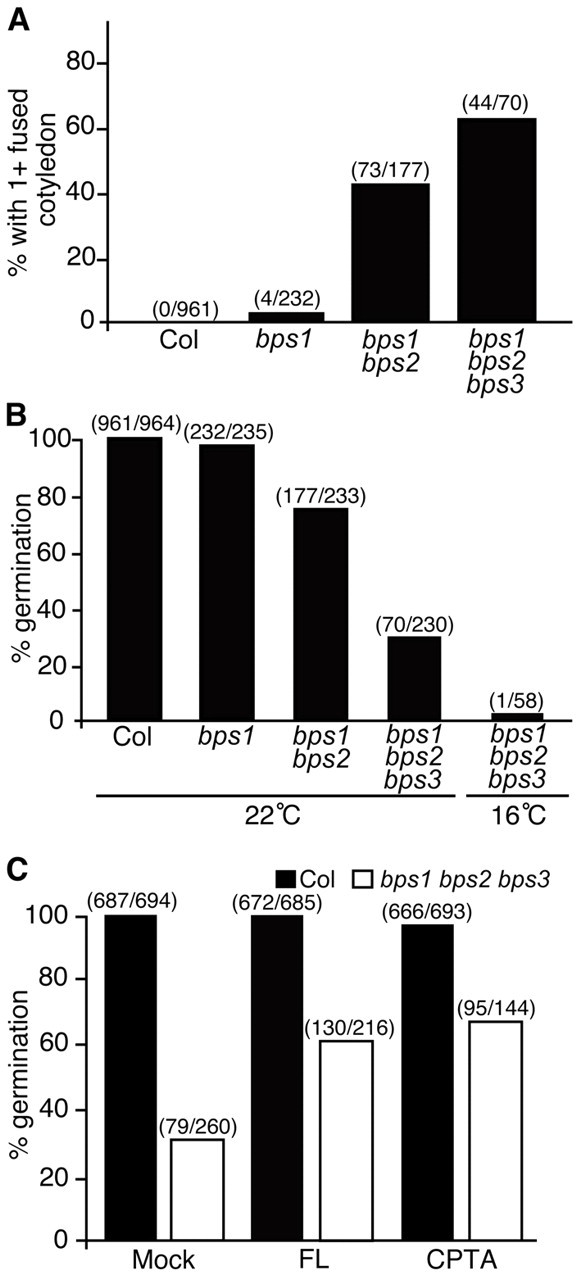
Germination and cotyledon formation are sensitive to loss of BPS activity. (A) Quantitative analysis of the cotyledon fusion phenotype revealed increased penetrance in higher-order mutants. (B,C) Quantitative analysis of germination rates in bps single and multiple mutants (B) and in wild type (Col-0) and bps triple mutants from siliques treated with the carotenoid biosynthesis inhibitors FL and CPTA (C). The partial suppression of germination defects in bps triple mutants indicated that this phenotype required carotenoid biosynthesis, and that the activity of all three BPS gene products prevent synthesis of the bps signal. These analyses are based on the number of homozygous mutants (see supplementary material Table S3). The number of observed mutants and the number of expected mutants are listed above each bar on all three graphs.
Among F2 progeny of BPS1+/bps1-2– BPS2+/bps2-2– F1 plants, the frequency of seedlings having the strong double mutant phenotype described above was close to 3/16 rather than the expected 1/16 (see supplementary material Table S5). This result suggested that, in the bps1-2 mutant background, BPS2 gene dose was limiting. We confirmed BPS2 haploinsufficiency by crossing BPS1+/bps1– bps2-2–/– and BPS1+/bps1– BPS2+/+. All F1 progeny were heterozygous for bps2-2 and the 25% bps1-2 homozygotes all showed the more severe phenotype (see supplementary material Table S5).
The severe bps triple mutant phenotype requires carotenoid biosynthesis
Because the bps1 phenotype arises from a non-cell autonomous signal (Van Norman et al., 2004; Van Norman et al., 2011b), we expected that the enhanced phenotypes of bps double and triple mutants might also arise from this same non-cell-autonomous signal. To test this possibility, we applied the same carotenoid biosynthesis inhibitors (CPTA and Fluridone) that partially rescued the bps1 phenotype (Van Norman and Sieburth, 2007) to developing siliques of BPS1+/bps1-2– bps2-2–/– bps3-2–/– plants, and assessed rescue by quantifying germination of the bps triple mutant.
Treated siliques became white, indicating plastid photo-oxidation. Seeds from both CPTA- and FL-treated wild-type siliques showed normal germination frequency, and the CPTA- and FL-treated BPS1+/bps1– bps2–/– bps3–/– siliques produced triple mutants with much higher germination frequencies than controls (Fig. 3C). Together with the enhanced phenotypes of higher order mutants, these observations indicate that BPS gene products function redundantly during embryogenesis, and suggest that they negatively regulate a common signaling molecule whose synthesis requires carotenoid biosynthesis.
To further assess whether the severe bps triple mutant phenotype was caused by the bps signal, we compared germination of seedlings that had undergone embryogenesis at 16°C. The bps1 phenotype is enhanced at low temperature (Van Norman et al., 2004), and germination defects of bps triple mutants were also enhanced by low temperature (Fig. 3B; see supplementary material Table S3). This result further links the increased severity of the multiple bps mutants to the non-cell-autonomous signal characterized in the bps1 single mutant. This, together with partial phenotypic suppression by carotenoid biosynthesis inhibitors, indicates that all three BPS genes contribute to preventing synthesis of the bps signal, and that it affects both embryonic and post-embryonic stages of development.
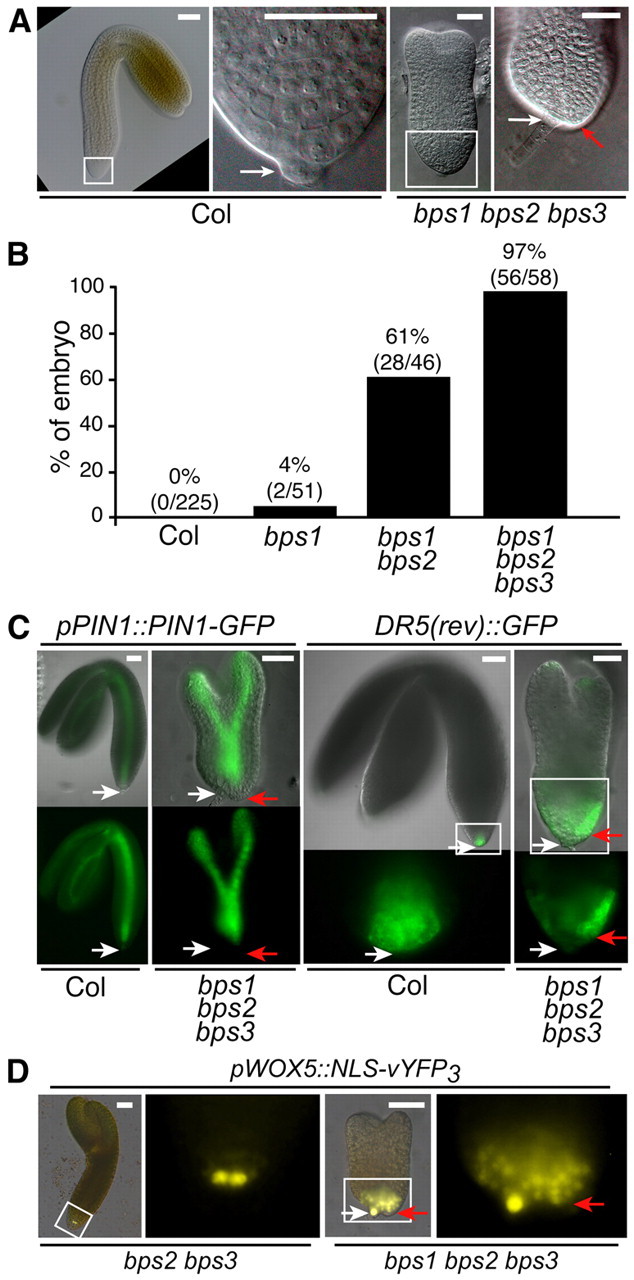
Embryogenesis defects in bps triple mutants
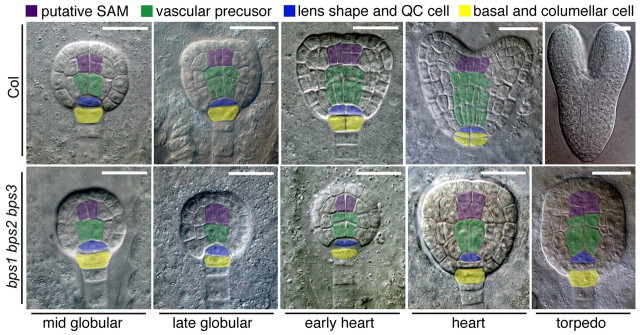
Many of the phenotypes present in bps triple mutant seedlings, such as cotyledon and meristem formation, represent defects in embryonic processes. Moreover, seedling analyses only allowed us to examine those uncommon triple mutants that had germinated. We therefore characterized embryogenesis in the bps triple mutant. Major defects were apparent in walking-stick and mature embryos, whereas defects in heart-stage embryos were modest (see supplementary material Fig. S3). We characterized more than 1200 embryos dissected from siliques of BPS1+/bps1-2– bps2-2–/– bps3-2–/– plants (see supplementary material Table S6). Using the phenotypically normal bps2 bps3 double mutants as internal staging controls, the bps triple mutant embryos had a pronounced developmental delay starting at the late globular stage. This coincided with the first appearance of morphological defects, a cell division defect in the inner-most cells. In the wild type, these internal cells differentiate into procambium, which is reflected by a stereotyped division resulting in narrow elongated cells aligned along the embryo axis (Scheres et al., 1994), but the bps triple mutants failed to undergo this stereotypical division (Fig. 4; supplementary material Table S6).
Fig. 4.
Embryonic cell division defects in vascular and root meristem precursors of bps triple mutants. Top row: wild-type (Col-0) embryos. Bottom row: bps1-2 bps2-2 bps3-2 triple mutant embryos. Embryo staging was based on phenotypically wild-type siblings (bps2 bps3), and is indicated below the images. DIC images were false colored to facilitate comparisons of cell types between the wild type and the bps triple mutant. The major defects identified in this analysis were provascular/procambium development and cell divisions at the root apex. Scale bars: 25 μm in all images except the wild-type torpedo embryo (50 μm).
As wild-type embryos progress to transition stage, stereotypical cell divisions in the apical subepidermal cells give rise to the presumptive SAM (Barton and Poethig, 1993) and at the basal end, the lenticular and basal cells both undergo anticlinal divisions to give rise to the quiescent center and columella precursor cells, respectively (Fig. 4) (Scheres et al., 1994). The age-matched bps triple mutants failed to undergo both these sets of cell divisions, and instead continued to resemble mid-globular stage embryos.
The wild-type heart-stage embryos have distinct cotyledon lobes, an increased number of vascular precursor cells and two layers of cells in the columella lineage (Fig. 4). The age-matched bps triple mutant embryos lacked distinct cotyledon lobes, although some ground cells underwent periclinal cell divisions. The vascular precursors appeared to increase in size, but there was no evidence of cell division or their elongation into typical procambial cells. Cells in the SAM precursor position were enlarged relative to those in wild-type embryos, and the increased number of SAM precursors indicated some cell division activity in that region. Finally, at the root pole, the bps triple mutant displayed an abnormal three-tiered structure consisting of broad cells, which appeared to have arisen from an abnormal periclinal division of the lenticular cell.
The wild-type torpedo embryos are much larger than the earlier stages, especially in cotyledon and hypocotyl length. By contrast, age-matched bps triple mutants showed remarkably little change. They resembled a slightly elongated globular embryo, with isodiametric cells in place of procambium, an abnormal root pole and no cotyledon lobes (Fig. 4). These observations reflect an acute requirement for BPS activity for progression through the globular stage, and revealed a particularly severe impact on the vascular, root and shoot stem cell populations.
bps triple mutant embryo defects are accompanied by altered gene expression
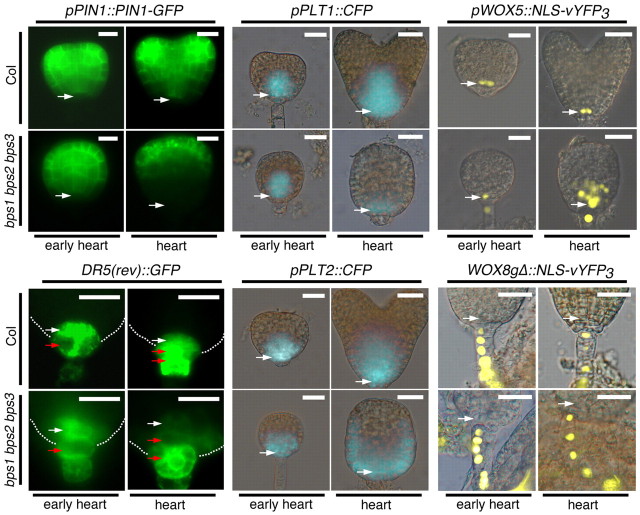
To assess the interaction between the bps signal and known pathways regulating embryogenesis, we analyzed expression patterns of six reporter genes (Fig. 5). Possible links to processes requiring auxin transport and transcriptional response were assessed using pPIN1::PIN1-GFP and DR5(rev)::GFP. Patterns of pPIN1::PIN1-GFP have been well documented in wild-type embryos (Steinmann et al., 1999; Benková et al., 2003; Friml et al., 2003). It is expressed throughout wild-type globular-stage embryos, and as the embryo progresses to heart stage, it is increasingly localized to cotyledon primordia and the procambial cells (Fig. 5). We observed no defect in pPIN1::PIN1-GFP expression in bps triple mutants until mid-heart stage, when PIN1-GFP failed to localize within the procambial tissue, and instead expression was concentrated in the upper tier of cells. In addition, expression failed to become concentrated in the incipient cotyledons as is seen in the wild type (supplementary material Fig. S4). By the torpedo stage, bps triple mutants showed some PIN1-GFP localization in the procambial region, albeit in a broad, short and diffuse pattern (see supplementary material Fig. S4). In late torpedo bps triple embryos, PIN1-GFP had resolved into distinct expression through the center of small cotyledon lobes, and into a broad region that corresponds to the procambial area of the presumptive hypocotyl.
Fig. 5.
Embryogenesis defects in bps triple mutants are accompanied by altered expression of developmental regulators. Fluorescent protein marker expression in wild type (Col) and bps triple mutant embryos at early and mid-heart stages. Expression patterns from six markers are shown: pPIN1::PIN1-GFP; DR5(rev)::GFP; pPLT1::CFP; pPLT2::CFP; pWOX5::NLS-vYFP3; and WOX8gΔ::NLS-vYFP3. In general, the bps triple mutant establishes normal expression patterns of these markers during the early heart stage, but shows altered expression by mid-heart stage, except WOX8, which shows normal expression in the suspensors of the triple mutant embryo. White arrows indicate lens-shape or quiescent center cells; red arrows indicate basal or columella cells. Scale bars: 50 μm.
DR5(rev)::GFP expression in the developing root apex of wild-type globular and heart-stage embryos is primarily in the lenticular/QC and basal/columella cells (Friml et al., 2003). Again, we observed no defect in DR5(rev)::GFP expression in bps triple mutants until mid-heart stage, when DR5(rev)::GFP expression was reduced overall, and nearly undetectable in the QC. Instead, DR5(rev)::GFP was largely restricted to the lower-most columella cells, with very faint signal in the flanking lateral root cap cells. Altered DR5(rev):GFP expression in the developing root of bps triple mutants is consistent with the pattern of PIN1-GFP expression; loss of PIN1-GFP in the central procambial region suggests that auxin is not properly transported to the basal pole at heart stage.
Another important pathway for root specification uses the PLETHORA (PLT) transcription factors (Aida et al., 2004; Blilou et al., 2005; Galinha et al., 2007; Smith and Long, 2010). We analyzed expression from PLT1 and PLT2 transcriptional fusions in bps triple mutants (Fig. 5). pPLT1:CFP is expressed in central regions of the lower pole of the early heart-stage embryo, and in later heart stages, expression expands to include the provascular tissues and a broad region of the presumptive root apex. The bps triple mutants showed normal pPLT1:CFP expression through the early heart-stage, but by mid-heart stage, expression was diminished and was slightly less concentrated at the root pole. Wild-type expression of PLT2 is similar to PLT1, and in bps triple mutants, pPLT2::CFP showed normal expression until mid-heart stage, when it was diminished in the center of the embryo, yet had expanded to a large U-shaped domain that encompassed the peripheral cells of the lower half of the embryo.
Another class of developmental regulators is the WUSCHEL-RELATED HOMEOBOX (WOX) genes (Haecker et al., 2004; Sarkar et al., 2007; Breuninger et al., 2008). We examined expression of WOX5 (pWOX5::NLS-vYFP3) and WOX8 (WOX8gΔ::NLS-vYFP3) transcriptional reporters in bps triple mutants. In the wild type, WOX5 is expressed in the hypophysis and lens-shaped cell during globular stages, and then is restricted to QC cells in heart and later stages. The bps triple mutant showed a normal pWOX5::NLS-vYFP3 expression pattern until the mid-heart stage, when ectopic expression was found though much of the basal pole of the embryo (Fig. 5). Expression of WOX8gΔ::NLS-vYFP3 was the same in wild type and bps triple mutants, where its expression was restricted to the suspensor. Together, these expression analyses suggest a normal establishment of embryonic patterning, but that broad developmental defects arose during the heart stage.
Ectopic root pole development in bps triple mutants
The root pole normally forms in alignment with the suspensor; however, we frequently observed a putative ectopic root pole extending from the side of bps mutant embryos (Fig. 6A). Quantitative analysis of embryonic root pole-suspensor alignment revealed this phenotype to be a general feature of bps mutants, and with higher penetrance in the higher order mutants (Fig. 6B). Because auxin is strongly linked to root development (Steinmann et al., 1999; Benková et al., 2003; Friml et al., 2003), we analyzed pPIN1::PIN1-GFP and DR5(rev)::GFP expression in bent-cotyledon stage embryos (Fig. 6C). In the bps triple mutant hypocotyl, pPIN1::PIN1-GFP expression was broader than that observed in the wild type, and at the basal pole of the embryo, asymmetric expression extended towards the putative ectopic root pole, instead of adjacent towards the suspensor (Fig. 6C). In wild type, DR5(rev)::GFP is prominent in the quiescent center (QC) and columella cells (Friml et al., 2003; Blilou et al., 2005). However, in bps triple mutants, DR5(rev)::GFP expression appeared broad and asymmetrical at the root apex, and with the strongest expression aligned with the putative ectopic root pole.
Fig. 6.
Root pole formation defects in bps triple mutants. (A) DIC images of bent-cotyledon stage embryos of wild type (Col) and bps1-2 bps2-2 bps3-2 triple mutants. Each pair of images shows the embryo (left), and a higher magnification of the boxed area (right). (B) Frequency of root pole-suspensor misalignment; the graph depicts the number of mutants with misaligned poles relative to the total number of mutants examined. (C) Expression of pPIN1::PIN1-GFP and DR5(rev)::GFP in bent-cotyledon stage embryos of wild-type and bps1-2 bps2-2 bps3-2 triple mutants. The asymmetry of both pPIN1::PIN1-GFP and DR5(rev)::GFP in the bps triple mutants is consistent with auxin-directed recruitment of an ectopic root pole. (D) Expression of pWOX5::NLS-vYFP3 in walking-stick stage embryos of wild type and bps1-2 bps2-2 bps3-2. Ectopic expression in the bps triple mutants is consistent with altered root specification at the basal pole of the bps triple mutant. White arrows indicate suspensor; red arrows indicate the ectopic root pole. Scale bars: 50 μm.
The PIN1 and DR5 expression patterns suggested the recruitment of a new root pole in the bps triple mutant embryo. To assess this possibility, we followed expression of pWOX5::NLS-vYFP (Haecker et al., 2004). The phenotypically wild-type bps2-2 bps3-2 walking-stick stage embryos showed normal pWOX5::NLS-vYFP expression in the QC, whereas stage-matched bps triple mutants expressed pWOX5::NLS-vYFP in a broader region of the developing root (Fig. 6D). Expression was typically tilted away from alignment with the suspensor, similar to DR5(rev)::GFP, and was stronger at the morphological root pole. Ectopic pWOX5::NLS-YFP expression at the top of the suspensor suggests that this cell type might also be mis-specified in bps triple mutants. Together with pPIN1::PIN1-GFP and DR5(rev)::GFP expression patterns, these observations suggest that bps triple mutants show auxin-mediated recruitment of a second root pole.
Auxin responses in bps1 leaves
Analysis of bps triple mutant embryos revealed changes in expression of both auxin-related markers (PIN1 and DR5) and developmentally important transcription factors (PLT1, PLT2 and WOX5). The growth, patterning and cell differentiation phenotypes of bps triple mutant embryos could be consistent with altered auxin response; however, because changes in marker expression lagged behind the morphological defects, whether or not the primary response to the bps signal was altered auxin signaling was unclear.
Previously, we found that DR5::GUS expression in bps1 leaves was recalcitrant to exogenous auxin activation (Van Norman et al., 2004). We revisited this experiment, testing whether auxin responsiveness of DR5::GUS was restored if the source of the bps1 signal (the root) was removed. Five hours after root excision, the bps1 leaf primordia were able to induce DR5::GUS expression in response to auxin (Fig. 7A). This result linked DR5:GUS auxin responsiveness to the bps signal.
Fig. 7.
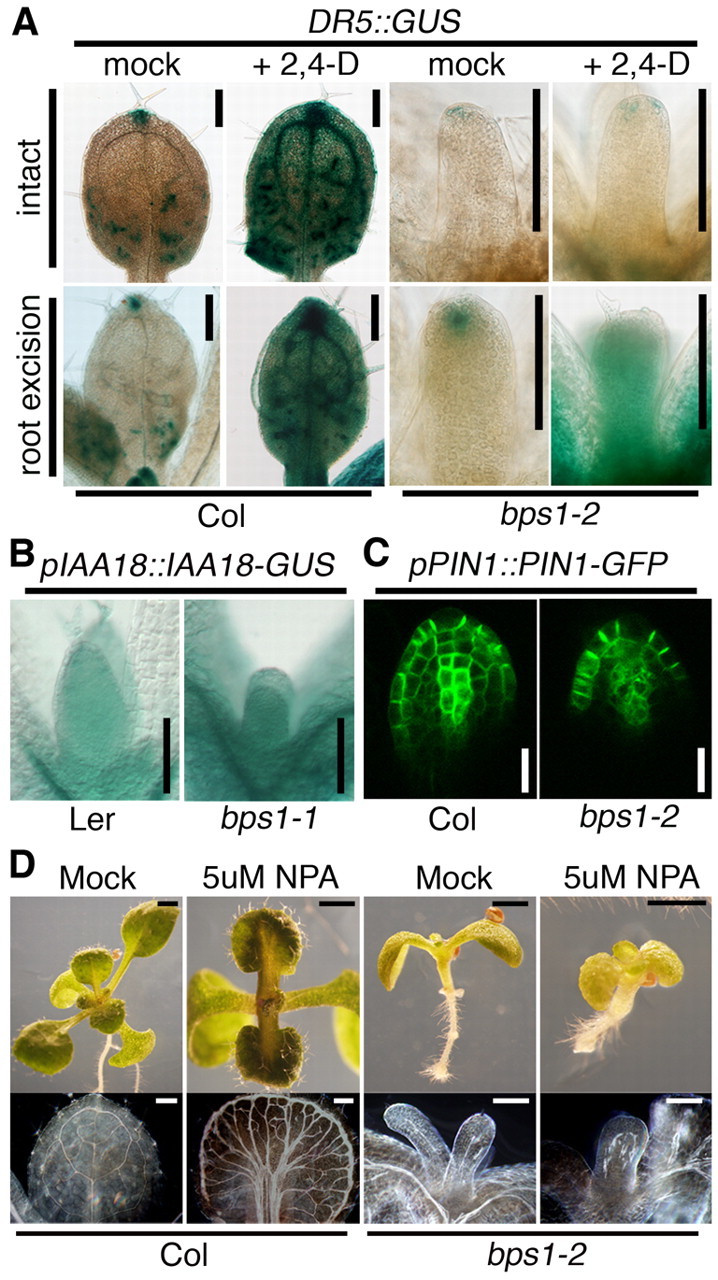
Leaf development and auxin responses in bps1 mutants. (A) Seven-day-old seedlings were either mock treated or supplied with 10 μM 2,4-D for 6 hours, and GUS stained for 12 hours, either with intact seedlings (top row) or with seedlings that had undergone root excision (bottom row). Auxin-induced DR5::GUS expression was observed only in bps1 leaf primordia following root excision. (B) GUS-stained leaf primordia of 5-day-old wild type and bps1 carrying pIAA18::IAA18-GUS. (C) PIN1-GFP patterns in leaf primordia of 3-day-old wild type and bps1 mutants. (D) Growth on NPA-supplemented media. NPA-treated bps1 leaf shows partial vein formation at the periphery of the leaf. Scale bars: 1 mm in the top row of D; 200 μm in A and the bottom row of D; 20 μm in C. in B 100 μm.
Next, we examined whether the bps signal inhibited auxin-induced proteolysis using plants carrying a pIAA18::IAA18-GUS reporter gene. Transcription of this reporter is auxin independent, but its protein product is destabilized by the motif II auxin receptor recognition sequence (Ploense et al., 2009). We reasoned that if the bps signal induced stabilization of this class of proteins, then bps1 seedlings would show much stronger pIAA18::IAA18-GUS expression than the wild type (Plonse et al., 2009). However, wild-type and bps1 leaf primordia showed very similar expression patterns (Fig. 7B), indicating normal IAA18 proteolysis in the bps1 leaves, and suggesting that the bps signal is not a general inhibitor of auxin-induced proteolysis of the AUX/IAA transcriptional repressors.
Another possible mechanism for bps signal interference with auxin processes would be to inhibit establishment of normal auxin transport patterns. To assess whether auxin transport patterns were established normally, we compared pPIN1::PIN1-GFP localization in three-day leaf primordia. This is the stage when leaf development begins to deviate in the bps1 mutant. However, PIN1-GFP expression pattern and localization appeared the same in the two genotypes (Fig. 7C), indicating that the bps signal does not directly interfere with PIN1 expression or localization.
We also examined how bps1 leaf primordia respond to an altered auxin distribution by growing seedlings on media supplemented with the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA). In wild-type leaves, NPA inhibits differentiation of the primary vein, and results in an increased number of secondary veins around the leaf periphery (Mattsson et al., 1999; Sieburth, 1999). The leaves of NPA-grown bps1 lacked the single primary vein that is typically produced, and instead produced veins at the periphery of the leaf (Fig. 7D). These similar developmental responses to NPA suggest that the bps signal does not interfere with leaf developmental responses to altered auxin transport.
Of these four assays to analyze leaf-specific auxin processes in bps1 mutants, three suggested that auxin processes are normal. The sole exception was auxin inducibility of DR5::GUS. DR5::GUS is a very useful reporter; however, it is also responsive to brassinolide, and can be inconsistent with auxin levels (Nakamura et al., 2003; de Reuille et al., 2006).
Auxin responses in bps1 seedlings
To further investigate the relationship between the bps signal and auxin, we also carried out whole-seedling analyses. Double mutants that combined auxin resistant 1 (axr1-3) (Leyser et al., 1993) and bps1-2 were nearly indistinguishable from bps1 single mutants (Fig. 8A). However, they produced a novel phenotype, a bulbous expansion at the base of the cotyledon petioles. Although the developmental significance of this phenotype is unclear, it revealed that, in bps1 single mutants, AXR1 is functional at the hypocotyl-cotyledon junction, a position predicted to experience a high level of root-derived bps signal. Intact auxin response was also suggested by the phenotypic response of bps1 to excess auxin. rooty/superroot (rty-1) mutants produce excess auxin endogenously, which results in epinastic cotyledons and excess lateral roots (Boerjan et al., 1995; Mikkelsen et al., 2004). The bps1 rty double mutants showed an additive phenotype: arresting as small plants with bps1-like roots and small epinastic cotyledons (Fig. 8B). Similarly, bps1 mutants grown on auxin-supplemented media showed similar reduced growth, thickened hypocotyl and altered cotyledons (Fig. 8C).
Fig. 8.
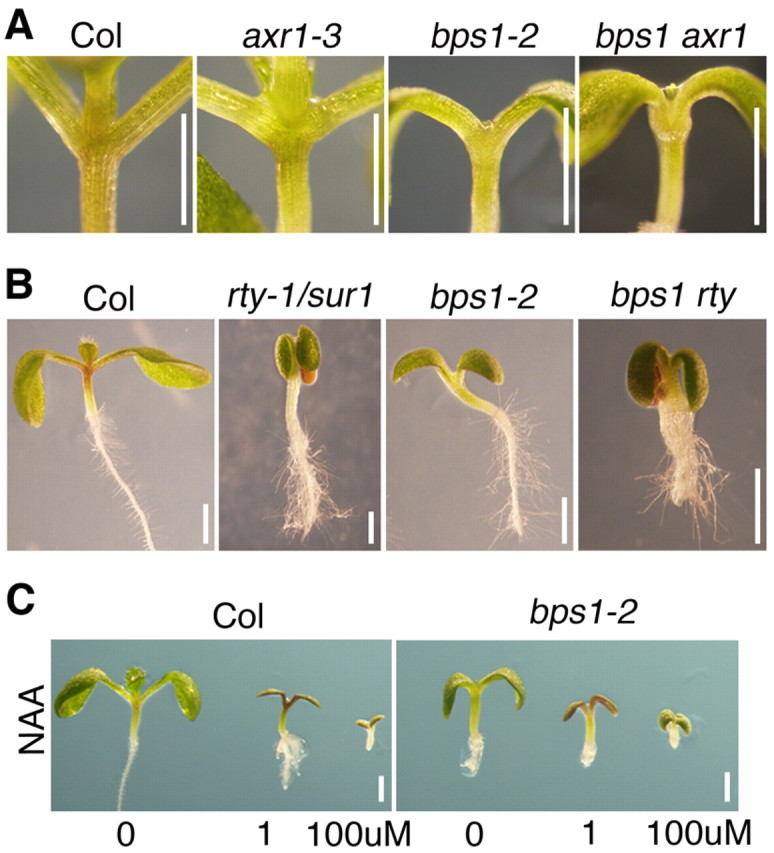
Auxin responses in bps1 and wild-type seedlings. (A) Double mutant analysis reveals a novel phenotype in bps1-2 axr1-3 double mutants. (B) Double mutant analysis reveals an additive phenotype between bps1-2 and rty-1. (C) Growth on auxin-supplemented medium causes similar responses in the wild type (Col) and bps1-2. Scale bars: 1 mm
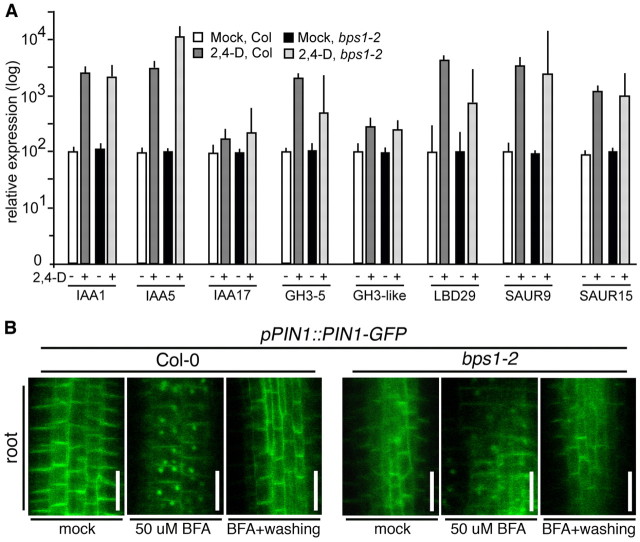
To further address whether auxin-induced transcriptional responses are intact in bps1 mutants, we used real-time RT-PCR to compare auxin-induced expression of primary auxin response genes (Paponov et al., 2008). Wild-type and bps1 showed a similar strong increase in transcript levels in response to exogenous auxin (Fig. 9A), supporting intact auxin responses in bps1 mutants.
Fig. 9.
Auxin-responsive gene expression and PIN-GFP cycling are intact in bps triple mutant. (A) Real-time RT-PCR analysis of transcript levels in 4-day-old seedlings. Relative expression, graphed on a log scale, shows a similar response to auxin in the wild type (Col) and bps1-2. Error bars show s.d. (B) Patterns of pPIN1::PIN1-GFP in 3-day-old wild type and bps1-2 roots. PIN1-GFP expression was diminished in bps1-2, and the cells are less organized; however, the PIN1-GFP shows normal localization and normal responses to BFA treatment. Scale bars: 20 μm
PIN polar localization and cycling is intact in bps1 mutants
Finally, it was possible that the bps signal affected PIN protein trafficking. PIN proteins show highly stereotyped polar membrane localization patterns, and also traffic between the plasma membrane and the endosome (Steinmann et al., 1999; Geldner et al., 2001; Geldner et al., 2003). We analyzed PIN1-GFP localization in wild-type and bps1 roots of 3-day-old seedlings. Similar plasma membrane localization of PIN1-GFP was observed, although its expression was strongly reduced in bps1 (Fig. 9B). In addition, both genotypes showed a similar response to brefeldin A (BFA), which inhibits membrane trafficking and leads to PIN1 accumulation in an endosome-like compartment. These observations suggest that PIN1 trafficking is not the target of the bps signal.
DISCUSSION
The Arabidopsis BYPASS1 gene has previously been shown to prevent root tissues from producing a mobile compound (the bps signal) that is transported to the shoot, where it arrests development (Van Norman et al., 2004). This pathway remains largely uncharacterized, and major questions include the identity of the bps signal, biochemical functions of the BPS1 protein, and the mechanism by which the bps signal affects plant development. In this study, we extended the functional analysis to include the other two Arabidopsis homologs, BPS2 and BPS3, and analyzed the developmental responses to bps signal-induced growth arrest in embryos and bps1 single mutants. We found that the three BPS genes have partially overlapping roles in preventing excess synthesis of the bps signal. How the bps signal affected development was further analyzed using developmental markers and through analysis of auxin responses. Our results indicate that the bps signal affects development by an auxin-independent mechanism.
Functional redundancy and haploinsufficiency among BPS gene family
The Arabidopsis BPS1 gene is both the highest expressed of the gene family, and it is the only one for which we observed a single mutant phenotype, indicating that this gene plays the most prominent role. However, BPS2 and BPS3 carry out related functions, because we found progressively more severe phenotypes in bps1 bps2 and bps1 bps2 bps3 mutants. Phenotypes of the higher order mutants originated during embryogenesis, which is consistent with their expression profiles. A common function for these three genes was revealed by experiments using carotenoid biosynthesis inhibitors. Like bps1 single mutants, the inhibitors partially suppressed the embryonic phenotype of bps triple mutants; thus, all three genes appear to regulate the synthesis of the same mobile compound.
We also found that BPS2 behaves in a dose-dependent manner, as BPS2 is haploinsufficient in a bps1 homozygous mutant background. Other dose-dependent regulators include miR165/166, where dosage in root tissues arises due to miRNA mobility, and leads to fine-scale patterning of root vascular cell types (Carlsbecker et al., 2010; Miyashima et al., 2011), stomagen, which regulates stomatal density based on the amount produced in mesophyll cells (Sugano et al., 2010), and the PLT genes, the products of which appear in gradients that confer spatial patterning to the root meristem (Galinha et al., 2007). Dose dependence of BPS2 suggests that plants can tightly regulate the amount of bps signal produced, a feature consistent with a developmentally important molecule.
The bps signal modulates development in parallel with auxin
Identification of the molecular target of the bps signal is crucial for understanding this enigmatic pathway. To address this question, we traced the earliest morphological defect to procambium specification in the late globular embryo. Procambial identity has been linked to auxin (Reinhardt, 2003; Scarpella et al., 2004; Scarpella et al., 2006). However, two auxin-related embryonic markers, pPIN1::PIN1-GFP and DR5(rev)::GFP, showed normal expression through to mid-heart stage, suggesting that auxin transport and responses were intact when the procambial defect arose. Nevertheless, auxin signaling was an attractive candidate for the target of the bps signal, as expression of the DR5::GUS auxin reporter in bps1 leaf primordia is only auxin inducible if the bps signal source, the root, is removed, and because many of the bps triple mutant phenotypes are similar to those reported for mutants with auxin-related defects (Bennett et al., 1995; Hadfi et al., 1998; Mravec et al., 2008). However, we found no independent support for defects in auxin signaling in bps1 leaf primordia or whole plants.
Instead, the bps signal appears to reveal a novel developmental pathway with functions during embryogenesis and in vegetative growth. The strongest effect of the bps signal appears to be on stem cells, e.g. the root and shoot meristems in bps1 seedlings, and the procambial stem cells of the bps triple mutant embryo. An attractive molecular target of the bps signal might be something shared by all three of these cell types. Indeed, parallel signaling pathways using a small peptide, a receptor and a WOX transcription factor that play crucial roles in regulation of all three stem cell types have been described (Sablowski, 2010; Stahl and Simon, 2010). Consistent with this possibility, the pattern of PIN1-GFP distribution during bps triple mutant embryogenesis is similar to that of mutants with defects in two protein phosphatase 2A genes, POLTERGEIST (POL) and POLTERGEIST-LIKE (PLL), which function downstream from CLV1 (Song et al., 2008). Recently, the CLE41/TDIF-PXY/TDR-WOX4 module has been shown to be required for vascular development and procambium auxin responsiveness (Hirakawa et al., 2010; Ji et al., 2010; Suer et al., 2011), and is a possible candidate for the developmental target of the bps signal. Alternative developmental regulators that might be attractive candidates for the bps signal include KANADI and the Class II HD-ZIP transcription factors (Ito et al., 2006; Hirakawa et al., 2010; Ilegems et al., 2010), which also function upstream of auxin, and in vascular patterning pathways.
Targeting of developmental pathways by the bps signal appears to be supported by the pPLT1::CFP and pPLT2::CFP marker expression patterns; in bps triple mutants, we found broad ectopic expression of pPLT2::CFP in mid-heart-stage embryos, but reduced, and only slightly ectopic, expression of PLT1. Although ectopic PLT expression has been observed in pin mutants, and has been attributed to the failure to concentrate auxin at the root meristem (Blilou et al., 2005), PLT1 and PLT2 tend to show similar expression patterns. Thus, these data suggest a novel developmental mechanism of the bps signal. Determining how the bps signal modulates development, including PLT expression, is an important goal of our future studies.
Acknowledgments
We thank Arabidopsis Biological Resource Center for providing bps2, bps3, axr1-3 and rty mutant alleles; Jiri Friml for DR5(rev)::GFP; Elliot Meyerowitz for pPIN1::PIN1-GFP; Thomas Laux for pWOX5::NLS-vYFP3 and WOX8gΔ::NLS-vYFP3; and Ben Scheres for pPLT1::CFP and pPLT2::CFP. We are also grateful to A. M. Hornsby for general laboratory assistance and to Hui Tian for helpful discussions.
Footnotes
Funding
This research was supported by grants from the National Science Foundation [IOS-0922288] and the National Institute of Food and Agriculture [2008-35304-04488] to L.E.S.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.077313/-/DC1
References
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.-S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Barton M., Poethig R. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Development 119, 823–831. [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443–465. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett S., Alvarez J., Bossinger G., Smyth D. (1995). Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J. 8, 505–520. [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. [DOI] [PubMed] [Google Scholar]
- Boerjan W., Cervera M. T., Delarue M., Beeckman T., Dewitte W., Bellini C., Caboche M., Van Onckelen H., Van Montagu M., Inzé D. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuninger H., Rikirsch E., Hermann M., Ueda M., Laux T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867–876. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A., Lee J.-Y., Roberts C. J., Dettmer J., Lehesranta S., Zhou J., Lindgren O., Moreno-Risueno M. A., Vatén A., Thitamadee S., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Dai X., Zhao Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S., Shinohara N., Sieberer T., Williamson L., George G., Hepworth J., Muller D., Domagalska M. A., Leyser O. (2010). Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137, 2905–2913. [DOI] [PubMed] [Google Scholar]
- de Reuille P. B., Bohn-Courseau I., Ljung K., Morin H., Carraro N., Godin C., Traas J. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc. Natl. Acad. Sci. USA 103, 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska M. A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell. Biol. 12, 211–221. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T., Offringa R., Jürgens G. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y. D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 212–230. [DOI] [PubMed] [Google Scholar]
- Hadfi K., Speth V., Neuhaus G. (1998). Auxin-induced developmental patterns in Brassica juncea embryos. Development 125, 879–887. [DOI] [PubMed] [Google Scholar]
- Haecker A., Groß-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131, 657–668. [DOI] [PubMed] [Google Scholar]
- Hardtke C. S., Berleth T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., Long J. A., Meyerowitz E. M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hejátko J., Blilou I., Brewer P. B., Friml J., Scheres B., Benková E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1, 1939–1946. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Till B. J., Comai L. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22, 2618–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilegems M., Douet V., Meylan-Bettex M., Uyttewaal M., Brand L., Bowman J. L., Stieger P. A. (2010). Interplay of auxin, KANADI and Class III HD-ZIP transcription factors in vascular tissue formation. Development 137, 975–984. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. [DOI] [PubMed] [Google Scholar]
- Jenik P. D., Jurkuta R. E. J., Barton M. K. (2005). Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. Plant Cell 17, 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Strable J., Shimizu R., Koenig D., Sinha N., Scanlon M. J. (2010). WOX4 promotes procambial development. Plant Physiol. 152, 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser H. M., Lincoln C. A., Timpte C., Lammer D., Turner J., Estelle M. (1993). Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364, 161–164. [DOI] [PubMed] [Google Scholar]
- Li Y., Rosso M. G., Viehoever P., Weisshaar B. (2007). GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 35, D874–D878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. A., Ohno C., Smith Z. R., Meyerowitz E. M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523. [DOI] [PubMed] [Google Scholar]
- Marhavy P., Bielach A., Abas L., Abuzeineh A., Duclercq J., Tanaka H., Parezova M., Petrasek J., Friml J., Kleine-Vehn J., et al. (2011). Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev. Cell 21, 796–804. [DOI] [PubMed] [Google Scholar]
- Mattsson J., Sung Z. R., Berleth T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- McCallum C. M., Comai L., Greene E. A., Henikoff S. (2000a). Targeted screening for induced mutations. Nat. Biotechnol. 18, 455–457. [DOI] [PubMed] [Google Scholar]
- McCallum C. M., Comai L., Greene E. A., Henikoff S. (2000b). Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M. D., Naur P., Halkier B. A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. [DOI] [PubMed] [Google Scholar]
- Miyashima S., Koi S., Hashimoto T., Nakajima K. (2011). Non-cell-autonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313. [DOI] [PubMed] [Google Scholar]
- Mravec J., Kubes M., Bielach A., Gaykova V., Petrasek J., Skupa P., Chand S., Benkova E., Zazimalova E., Friml J. (2008). Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135, 3345–3354. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. (2008). Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Higuchi K., Goda H., Fujiwara M. T., Sawa S., Koshiba T., Shimada Y., Yoshida S. (2003). Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol. 133, 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T., Lukowitz W., Bayer M. (2008). Talk global, act local-patterning the Arabidopsis embryo. Curr. Opin. Plant Biol. 11, 28–33. [DOI] [PubMed] [Google Scholar]
- Paponov I. A., Paponov M., Teale W., Menges M., Chakrabortee S., Murray J. A. H., Palme K. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1, 321–337. [DOI] [PubMed] [Google Scholar]
- Ploense S. E., Wu M. F., Nagpal P., Reed J. W. (2009). A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D. (2003). Vascular patterning: more than just auxin?. Curr. Biol. 13, R485–R487. [DOI] [PubMed] [Google Scholar]
- Sablowski R. (2010). Plant stem cell niches: from signalling to execution. Curr. Opin. Plant Biol. 14, 4–9. [DOI] [PubMed] [Google Scholar]
- Sarkar A. K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Francis P., Berleth T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131, 3445–3455. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Marcos D., Friml J., Berleth T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B., Wolkenfelt H., Willemsen V., Terlouw M., Lawson E., Dean C., Weisbeek P. (1994). Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120, 2475–2487. [Google Scholar]
- Sieburth L. (1999). Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 121, 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L. E., Muday G. K., King E. J., Benton G., Kim S., Metcalf K. E., Meyers L., Seamen E., Van Norman J. M. (2006). SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18, 1396–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z., Long J. (2010). Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature 464, 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.-K., Hofhuis H., Lee M. M., Clark S. E. (2008). Key divisions in the early arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev. Cell 15, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R. (2010). Plant primary meristems: shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 13, 53–58. [DOI] [PubMed] [Google Scholar]
- Steinmann T., Geldner N., Grebe M., Mangold S., Jackson C. L., Paris S., Gälweiler L., Palme K., Jürgens G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286, 316–318. [DOI] [PubMed] [Google Scholar]
- Suer S., Agusti J., Sanchez P., Schwarz M., Greb T. (2011). WOX4 imparts auxin responsiveness to cambium cells in Arabidopsis. Plant Cell 23, 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S. S., Shimada T., Imai Y., Okawa K., Tamai A., Mori M., Hara-Nishimura I. (2010). Stomagen positively regulates stomatal density in Arabidopsis. Nature 463, 241–244. [DOI] [PubMed] [Google Scholar]
- Till B. J., Reynolds S. H., Greene E. A., Codomo C. A., Enns L. C., Johnson J. E., Burtner C., Odden A. R., Young K., Taylor N. E., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman J. M., Sieburth L. E. (2007). Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J. 49, 619–628. [DOI] [PubMed] [Google Scholar]
- Van Norman J. M., Frederick R. L., Sieburth L. E. (2004). BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr. Biol. 14, 1739–1746. [DOI] [PubMed] [Google Scholar]
- Van Norman J. M., Breakfield N. W., Benfey P. N. (2011a). Intercellular communication during plant development. Plant Cell 23, 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman J. M., Murphy C., Sieburth L. E. (2011b). BYPASS1: synthesis of the mobile root-derived signal requires active root growth and arrests early leaf development. BMC Plant Biol. 11, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Sauer M., Meurette O., Friml J., Ljung K., Sandberg G., Hooykaas P., Offringa R. (2005). Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 17, 2517–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Schlereth A., Ehrismann J. S., Schwank G., Kientz M., Jürgens G. (2006). Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265–270. [DOI] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev Genet. 10, 305–317. [DOI] [PubMed] [Google Scholar]
- Zhang W., Murphy C., Sieburth L. E. (2010). Conserved RNaseII domain protein functions in cytoplasmic mRNA decay and suppresses Arabidopsis decapping mutant phenotypes. Proc. Natl. Acad. Sci. USA 107, 15981–15985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., To J. P., Cheng C. Y., Eric Schaller G., Kieber J. J. (2011). Type-A response regulators are required for proper root apical meristem function through post-transcriptional regulation of PIN auxin efflux carriers. Plant J. 68, 1–10. [DOI] [PubMed] [Google Scholar]