Abstract
Cytotoxin is one of the important pathogenic factors, which plays a role in the virulence of Klebsiella oxytoca. The aim of this study was to investigate molecular typing of clinical isolates of the cytotoxin-producing K. oxytoca using internal transcribed spacer (ITS) PCR. A total of 75 isolates of K. oxytoca were isolated from clinical samples; they were verified as K. oxytoca by standard microbiological tests and PCR. Production of toxin determines the cytotoxic effects on HEp-2 cells. The genetic diversity of isolates of the cytotoxin-producing K. oxytoca were defined by ITS-PCR. Of all the isolates investigated, five K. oxytoca strains isolated from stool cultures, two strains from blood samples, one strain from a wound and one strain isolated from urine had cytotoxic effects on HEp-2 cells. The ITS-PCR patterns showed genetic diversity among cytotoxin-producing isolates. The ITS-PCR method had good discriminatory power; performance of this method and interpretation of the results were easy and repeatable. Five genetic diversity patterns were identified by ITS-PCR.
Keywords: Cytotoxin, genetic diversity, internal transcribed spacer PCR, Klebsiella oxytoca
Introduction
Typing of bacterial strains is an important process in diagnosis, treatment and epidemiological studies. In general, different methods of microbial agent typing are used to examine the common canons of disease, study the transmission of infection from one patient to another, investigate sporadic infections, identify the types of microbial pathogens, determine the pattern of antibiotic resistance and susceptibility, and evaluate drug therapy failure. Typing methods are classified into two groups—phenotypic methods and genotypic methods. Choosing a suitable typing method depends on the level of capability, laboratory resources and other factors. Study of genetic variation can ultimately justify and show phenotypic variability in bacteria in terms of geographical distribution, host characteristics, pathogenicity, antibiotic resistance and virulence [1], [2], [3]. Every optimal typing system must have some essential characteristics, including cost, discriminatory power, reproducibility, typability, stability, resolution, ease of use, and ease of obtaining and interpreting results [4].
Phenotypic methods such as biotyping, serotyping, bacteriocin typing and phage typing are costly and time-consuming, have limited epidemiological value and are not capable of proper differentiation [5]. There are different genotypic methods, including ribotyping, pulsed field gel electrophoresis (PFGE) [6], internal transcribed spacer (ITS) PCR [7], repetetive element sequence-based (rep-) PCR [8], random amplified polymorphic DNA (RAPD) [9], enterobacterial repetitive intergenic consensus (ERIC-) PCR [10], amplified fragment length polymorphism [11], multilocus sequence typing (MLST) [12] and multilocus variable number tandem repeat analysis (MLVA) [13]. These techniques have a high diagnostic power, but it is not possible to use them in all centres [2]. The 16S, 23S and 5S rRNA genes are found in similar genetic loci in prokaryotes and are separated by non-coding regions called the internal transcribed spacer (ITS). Because of high levels of polymorphism and a large degree of variation in strain and species, these regions are useful for identifying and subtyping bacteria. These regions are easily replicable using complementary primers designed in conserved regions of the 16S and rRNA 23S genes [14], [15], [16], [17].
The position of the ITS among the protected regions of DNA and the size of the regions are the characteristics of bacterial species that are used as the basis of species genotyping. Due to the large variability in size and sequence, these regions are suitable for identifying species and detecting different strains. There is more than one copy of ITS in the bacterial genome and the ITS sequences can vary from one species to another [18].
The ITS-PCR method is used to investigate and evaluate genetic variation and make phylogenetic descriptions of the genetic groups of Klebsiella oxytoca strains. ITS-PCR genotyping using the short intergenic regions between the 23S rRNA and 16S encoding genes is a useful, low-cost method for evaluating the genetic diversity of K. oxytoca isolates [19], [20]. After the test, the PCR product is electrophoresed on an 8% polyacrylamide gel after staining then photographed. Using software such as BioNumerics and algorithms such as unweighted pair group method using arithmetic averages (UPGMA), the genotyping patterns achieved for K. oxytoca isolates are clustered.
Klebsiella oxytoca, as an opportunistic pathogen, produces cytotoxin that results in peritoneal colon destruction. Cytotoxin is one of the pathogenic factors of this bacterium and one of the important factors involved in its virulence [21], [22] and pathogenicity. This bacterium is the aetiological factor in haemorrhagic colitis, which is associated with the use of antibiotics in adults and the elderly [23]. It mainly cause nosocomial infections, especially in immunocompromised patients or those in need of intensive care. People with immune deficiency and weakness, including organ transplant recipients, neonates, the elderly, individuals undergoing dialysis, individuals who are HIV positive, people with cancer, and recipients of immunosuppressive drugs are among the groups at high risk of K. oxytoca infection in hospital wards. Outbreak of K. oxytoca infections in hospitals is often associated with the contamination of natural reservoirs [24], [25]. The aim of this study was to investigate the molecular typing of cytotoxin-producer K. oxytoca clinical isolates, using ITS-PCR.
Materials and methods
Bacterial isolates
A total of 75 K. oxytoca strains were collected from several hospitals in Tehran between 2015 and 2016. Clinical strains were isolated from stool, blood, urine, sputum and wounds.
Verification of K. oxytoca isolates
All bacterial strains that underwent standard microbiological tests in microbiology laboratories and were detected as K. oxytoca strains were also detected and verified by PCR through amplification of polygalacturonase-specific gene (pehX). The ITS-PCR and cytotoxicity assay of K. oxytoca isolates were performed according to the previously described methods [17], [26].
Cytotoxin production assay
The cytotoxicity assay of K. oxytoca strains was performed using previously described methods [27], [28]. In brief, an HEP-2 cell line (American Type Culture Collection, Manassas, VA, USA; CCL-23) was used for screening this cytotoxin. A 1:1 dilution with phosphate-buffered saline of the filtered supernatant from the cultures of K. oxytoca strains was added to each well of a 96-well plate seeded with 1 × 105 HEp-2 cells, followed by incubation in 5% carbon dioxide at 37°C for 72 h. A positive cytotoxic effect was recorded as cell rounding under light microscopy. The positive control—the cytotoxin-producing K. oxytoca MH43-1—was a gift from Dr Christoph Hoegenauer, Department of Internal Medicine, Medical University of Graz, Austria. The K. oxytoca strain ATCC 13182 served as a negative control.
ITS-PCR
To amplify the ITS region, we used primers forward: 5′-GAAGTCGTAACAAGG-3′ and reverse: 5′-CAAGGCATCCACCGT-3′ as previously described [17]. The PCR was performed with 20-μL reaction volumes comprising 10 μL Master mix (Ampliqon, Odense, Denmark), 0.5 μL forward primer 10 pmol (Bioneer, Daejeon, Korea), 0.5 μL reverse primer 10 pmol (Bioneer, Daejeon, Korea), 8.5 μL distilled water and 50 ng bacterial DNA.
The PCR was performed in a thermocycler (PEQLAB, Erlangen, Germany) with an initial denaturation at 95°C for 5 min; and 25 cycles, including denaturation steps at 95°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min, and final extension at 72°C for 5 min. Electrophoresis of the PCR product was performed in an 8% polyacrylamide gel. The gel was stained using 1% silver nitrate and detection used Gel Doc (GVM20 model syngene, Cambridge, UK). The digital image was stored electronically as a TIFF image and analysed with GelCompar software (Applied Maths, Sint-Martens-Latem, Belgium) by using the Dice correlation coefficient and the UPGMA method.
Results
A total of 75 isolates of K. oxytoca were collected from clinical samples of stool, blood, urine, sputum and wound. The samples were collected from individuals admitted to four hospitals in Tehran between 2015 and 2016. At the early stage of the study, all strains with the galacturonase-specific gene (pehX) were detected and verified as K. oxytoca. Of these 75 isolates, 11 (14.7%) were recovered from blood, 2 (2.7%) were recovered from wounds, 4 (5.3%) were recovered from respiratory cultures, 51 (68%) were recovered from urine cultures and 7 (9.3%) were recovered from stool cultures.
All the K. oxytoca strains isolated from the stool cultures of patients with diarrhoea, and from urine, blood, wounds and sputum cultures were evaluated in terms of their production of cytotoxin by HEp-2 cell culture. Of all the isolates, five strains isolated from stool cultures, two strains isolated from blood cultures, one strain isolated from a wound culture, and one strain isolated from a urine culture had cytotoxic effects on HEp-2 cells. None of the strains isolated from sputum cultures had a cytotoxic effect on HEp-2 cells.
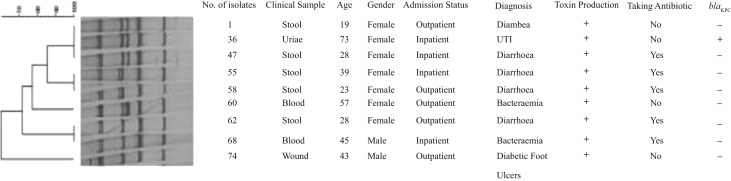
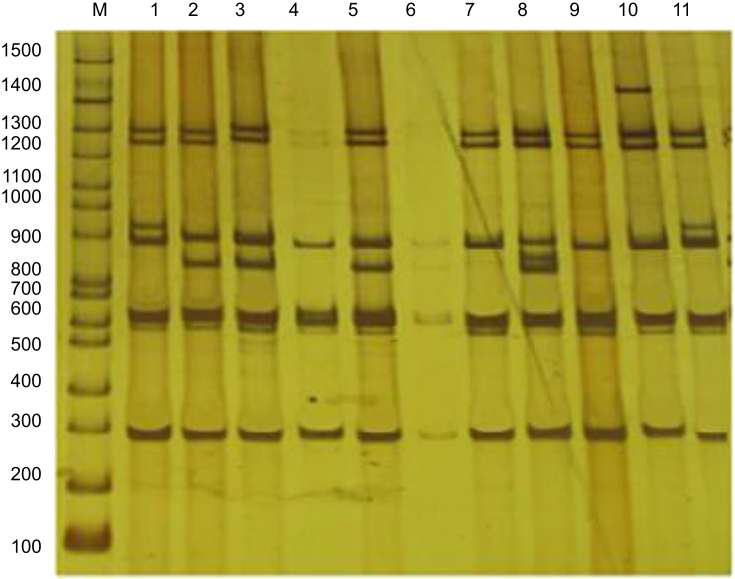
The patterns obtained were clustered from I to V. Two isolates were placed in cluster I, three in cluster II, two in cluster III, one in cluster IV and one in cluster V (Fig. 2). Cluster I contained cytotoxin-producing isolates that produced ITS-PCR with sizes of 295, 620, 890, 948, 1280 and 1300 bp. Cluster II included ITS-PCR products with sizes of 290, 620, 847, 890, 1280 and 1300 bp. Cluster III included ITS-PCR products with sizes of 290, 620, 890, 1280 and 1300 bp. Cluster IV included ITS-PCR products with sizes of 290, 620, 847, 871, 890, 1280 and 1300 bp. Cluster V included ITS-PCR products with sizes of 290, 620, 890, 1280, 1300 and 1400 bp (Fig. 1).
Fig. 2.
Dendrogram of cytotoxin-producing Klebsiella oxytoca isolates based on internal transcribed spacer PCR method by using the Dice correlation coefficient and the UPGMA method.
Fig. 1.
Electrophoresis of PCR products amplified from internal transcribed spacer region in 8% polyacrylamide gel. Lanes 1, 2, 3, 5, 7, 8, 9, 10, 11: cytotoxin-producing Klebsiella oxytoca isolates; lane M, 100-bp ladder.
The two isolates in cluster I were obtained from stool cultures of an inpatient and an outpatient. Of the three isolates in cluster II, one isolate was obtained from an inpatient's urine culture and two isolates were obtained from stool cultures of an inpatient and an outpatient. The two isolates in cluster III were obtained from an outpatient's stool culture and an inpatient's blood culture. The isolate in cluster IV was obtained from an inpatient's blood culture. The isolate in cluster V was obtained from an outpatient's wound. A toxin-producing K. oxytoca isolate (isolate 36) was obtained from a urine specimen of a 73-year-old woman hospitalized in a gynaecology ward in one of the hospitals in the study.
The patterns obtained using ITS-PCR were clustered from I to V. Two isolates were placed in cluster I, three isolates in cluster II, two isolates in cluster III, one isolate in cluster IV and one isolate in cluster V.
Discussion
The drawn dendrogram showed that the toxin-producing isolates had different genetic patterns. In this study, most of the toxin-producing isolates were isolated from stool samples. Four toxin-producing isolates were isolated from inpatients and five toxin-producing isolates were isolated from outpatients. The two isolates that were obtained from the two inpatients hospitalized in the women's surgery ward and the blood and oncology ward with a similar genetic pattern placed in a cluster [1] were isolated from patients hospitalized in the same hospital. Because of the similarity of the genetic patterns and the admission of two patients in a hospital, it can be concluded that these isolates may have been transmitted from physicians, hospital staff, medical equipment, or the pollution of environmental reservoirs. The two isolates (nos 36 and 47) have the same genetic pattern and isolate 36 not only was able to produce toxin but also had the blaKPC gene; the identification of these isolates can be an alarm signal for a sudden outbreak of isolated blaKPC genes among hospitalized patients. Therefore, it is important to identify and prevent the release of these isolates [29].
In a study by Stojowska et al. in the Netherlands in 2009 [16], the researchers investigated and analysed the genetic variation of 209 isolates of K. oxytoca obtained in a period of 50 years (from 1954 to 2007) using the two methods of ITS-PCR and PCR melting profile (PCR-MP). In the ITS-PCR method, the isolates were classified into KoX and KoY clusters on the basis of the results of analysis of patterns, dendrogram and electropherogram of the strains. Accordingly, 30 strains were placed in the KoY cluster, 28 strains in the KoY1 cluster and 2 strains in the KoY2 cluster. In addition, 170 strains were placed in the KoX1 cluster, 4 strains in the KoX2 cluster, 4 strains in the KoX3 cluster and 1 strain in the KoX4 cluster [16].
In the study by Krawczyk and colleagues in the Netherlands, in 2009 [17], the researchers investigated the genotyping analysis of 14 isolated K. oxytoca strains obtained from the neonatal intensive care unit over 15 years (from 1992 to 2007) using the two methods ITS-PCR and PCR-MP. Based on the results of this study, using the ITS-PCR method, electrophoretic patterns of K. oxytoca strains were grouped into the two main KoX1 and KoY1 clusters. In addition, there was genetic diversity between 14 isolates of K. oxytoca isolates (a similarity rate of >75%) [17].
Ryberg et al., in Sweden in 2011, used GTG (5-PCR) and ITS-PCR methods for molecular typing of Klebsiella isolates. The dendrogram of 19 Klebsiella pneumoniae isolates and 35 K. oxytoca isolates in GTG (5-PCR) indicated that K. pneumoniae strains were placed in clusters III and I whereas K. oxytoca strains were placed in clusters III and II and sub-clusters A-B-C, D. The dendrogram of 19 K. pneumoniae isolates and 35 K. oxytoca isolates in the ITS-PCR method indicated that K. pneumoniae strains were placed in clusters III and I, and K. oxytoca strains were placed in cluster II [8].
In Austria in 2010, Grisold et al. investigated automated rep-PCR and PFGE methods for molecular typing of extended spectrum β-lactamase-producing K. oxytoca isolates. The results of the automated rep-PCR method, compared with PFGE, were more accurate for the study of the outbreak of extended spectrum β-lactamase-producing K. oxytoca isolates [30].
In Joainnig et al.’s study in Austria in 2010, the results of the pulsotype of 70 K. oxytoca isolates via PFGE showed no clonal relationship between the K. oxytoca stains isolated from individuals with antibiotic-associated haemorrhagic colitis (AAHC). In five isolates obtained from the stool of a patient in the acute stage of antibiotic-associated haemorrhagic colitis, the results of pulsotype and cytotoxin production were evaluated. The macrorestriction profile showed that there were three different genetic variants among the five isolates. One of the strains had cytotoxin and two other strains lacked cytotoxin [31].
Tsakris et al., in Greece in 2011, used the two methods of PFGE and ERIC-PCR for molecular typing of nine K. oxytoca isolates. The results of ERIC-PCR showed that all isolates were basically from a single genotype. Based on the results of PFGE, five isolates were categorized in a single type clone (Ia) and four other isolates in different type clones (Ib to Ie) [32].
In contrast to our study, Stojowska et al.’s study classified the K. oxytoca isolates into KoX and KoY clusters, 60 strains were placed in the KoY cluster and 179 strains were placed in the KoX cluster. In our study, the patterns obtained using ITS-PCR were clustered from I to V. Two isolates were placed in cluster I, three isolates in cluster II, two isolates in cluster III, one isolate in cluster IV, and one isolate in cluster V. Also, In Krawczyk et al.’s study, 14 K. oxytoca strains isolated from neonatal intensive care were grouped into the KoX1 and KoY1 clusters. In contrast to our results, Ryberg et al. placed 35 K. oxytoca isolates in cluster II using ITS-PCR.
Epidemiological studies and typing methods provide molecular tools for identifying the genetic diversity between bacterial isolates and population structure of the bacterium. Typing plays an important role in understanding the epidemiology of the studied bacterium [33]. Different typing methods have been used such as phage typing, serotyping, antibiogram typing, ITS-PCR, rep-PCR, RAPD-PCR, ERIC-PCR, PCR-restriction fragment length polymorphism, PCR-MP, ribotyping, PFGE, MLVA and MLST. Every optimal typing system must have a set of essential features such as discriminatory power, reproducibility, typability, stability, resolution, ease of performance, and ease of interpreting the results [4]. Due to the inadequacy of using a single method alone, the supervisory and complementary studies must investigate strains based on a combination of results from different typing methods. As one method alone does not have all the features and since each type of typing method alone provides different results, some studies have used different typing methods such as rep-PCR, ITS-PCR or PFGE for typing K. oxytoca. The ITS-PCR method is suitable for differentiation, especially for closely related strains; it is easy to perform and to repeat, and results are easily interpreted.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by Vice-Chancellor for Research grant (no. 27720) of Tehran University of Medical Sciences, Tehran, Iran.
References
- 1.Wenjun Li, Raoult D., Fournier P.E. Bacterial strain typing in the genomic era. FEMS Microbiol Rev. 2009;33:892–916. doi: 10.1111/j.1574-6976.2009.00182.x. [DOI] [PubMed] [Google Scholar]
- 2.Ranjbar R., Karami A., Farshad S., Giammanco G.M., Mammina C. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. N Microbiol. 2014;37:1–15. [PubMed] [Google Scholar]
- 3.Yıldırım1 İ.H., Yıldırım S.C., Koçak N. Molecular methods for bacterial genotyping and analyzed gene regions. J Microb Infect Dis. 2011;1:42–46. [Google Scholar]
- 4.Murchan S., Kaufmann M.E., Deplano A., de Ryck R., Struelens M., Zinn C.E. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J Clin Microbiol. 2003;41:1574–1585. doi: 10.1128/JCM.41.4.1574-1585.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisse S., Grimont F., Grimont P. The genus Klebsiella. Prokaryotes. 2006;6:159–196. [Google Scholar]
- 6.Davis M.A., Hancock D.D., Besser T.E., Call D.R. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J Clin Microbiol. 2003;41:1843–1849. doi: 10.1128/JCM.41.5.1843-1849.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M., Cao B., Yu Q., Liu L., Gao Q., Wang L. Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species. J Clin Microbiol. 2008;46:3555. doi: 10.1128/JCM.00927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryberg A., Olsson C., Ahrné S., Monstein H.J. Comparison of (GTG)5-oligonucleotide and ribosomal intergenic transcribed spacer (ITS)- PCR for molecular typing of Klebsiella isolates. J Microbiol Methods. 2011;84(2):183–188. doi: 10.1016/j.mimet.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Lin A.W., Usera M.A., Barret T.J., Golds R.A. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J Clin Microbiol. 1996;34:870–876. doi: 10.1128/jcm.34.4.870-876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Versalovic J., Koeuth T., Lupski J.R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vos P., Hogers R., Bleeker M., Reijans M., Van De Lee T., Hornes M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sullivan C.B., Diggle M.A., Clarke S.C. Multilocus sequence typing. Mol Biotechnol. 2005;25:245–253. doi: 10.1385/MB:29:3:245. [DOI] [PubMed] [Google Scholar]
- 13.van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA) FEMS Immunol Med Microbiol. 2007;49:22–27. doi: 10.1111/j.1574-695X.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 14.Versalovic J., Lupski J.R. Molecular detection and genotypingof pathogens: more accurate and rapid answers. Trends Microbiol. 2002;10(Suppl. 10):S15–S21. doi: 10.1016/s0966-842x(02)02438-1. [DOI] [PubMed] [Google Scholar]
- 15.Fournier P.E., Drancourt M., Raoult D. Bacterial genome sequencing and its use in infectious diseases. Lancet Infect Dis. 2007;7:711–723. doi: 10.1016/S1473-3099(07)70260-8. [DOI] [PubMed] [Google Scholar]
- 16.Stojowska K., Krawczyk B., Kałuzewski S., Kur J. Retrospective analysis of the genetic diversity of Klebsiella oxytoca isolated in Poland over a 50-year period. Eur J Clin Microbiol Infect Dis. 2009;28:1263–1266. doi: 10.1007/s10096-009-0768-7. [DOI] [PubMed] [Google Scholar]
- 17.Stojowska K., Kałuzewski S., Krawczyk B. Usefulness of PCR melting profile method for genotyping analysis of Klebsiella oxytoca isolates from patients of a single hospital unit. Pol J Microbiol. 2009;58:247–253. [PubMed] [Google Scholar]
- 18.Jensen M.A., Webster J.A., Stratus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphism. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.González N., Romero J., Espejo R.T. Comprehensive detection of bacterial populations by PCR amplification of the 16S–23S rRNA spacer region. J Microbiol Methods. 2003;55:91–97. doi: 10.1016/s0167-7012(03)00110-6. [DOI] [PubMed] [Google Scholar]
- 20.Martínez G., Bescos J., Rodriguez-Sala J.J., Valera R.F. RISSC: a novel database for ribosomal 16S–23S RNA genes spacer regions. Nucleic Acids Res. 2001;29:178–180. doi: 10.1093/nar/29.1.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneditz G., Rentner J., Roier S. Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc Natl Acad Sci USA. 2014;111:13181–13186. doi: 10.1073/pnas.1403274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darby A., Lertpiriyapong K., Sarkar U. Cytotoxic and pathogenic properties of Klebsiella oxytoca isolated from laboratory animals. PLoS One. 2014;9:e100542. doi: 10.1371/journal.pone.0100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J., Cachay E.R., Hunt G.C. Klebsiella oxytoca: a rare cause of severe infectious colitis: first North American case report. Gastrointest Endosc. 2004;60:142–145. doi: 10.1016/s0016-5107(04)01537-8. [DOI] [PubMed] [Google Scholar]
- 24.Kola A., Holst M., Chaberny I.F., Ziesing S., Suerbaum S., Gastmeier P. Surveillance of extended-spectrum β-lactamase-producing bacteria and routine use of contact isolation: experience from a three-year period. J Hosp Infect. 2007;66:46–51. doi: 10.1016/j.jhin.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Lowe C., Willey B., O'Shaughnessy A., Lee W., Lum M., Pike K.M. Outbreak of extended-spectrum β-lactamase producing Klebsiella oxytoca infections associated with contaminated handwashing sinks. Emerg Infect Dis. 2012;18:1242–1247. doi: 10.3201/eid1808.111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovtunovych G., Lytvynenko T., Negrutska V., Lar O., Brisse S., Kozyrovska N. Identification of Klebsiella oxytoca using a specific PCR assay targeting the polygalacturonase pehX gene. Res Microbiol. 2003;154:587–592. doi: 10.1016/S0923-2508(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 27.Cheng V.C., Yam W.C., Tsang L.L., Yau M.C., Siu K.H., Wong C.Y. Epidemiology of Klebsiella oxytoca-associated diarrhea detected by Simmons citrate agar supplemented with inositol, tryptophan and bile salts. J Clin Microbiol. 2012;50:1571–1579. doi: 10.1128/JCM.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Validi M., Soltan-Dallal M.M., Douraghi M., Fallah-Mehrabadi J., Rahimi-Foroushani A., Frohesh-Tehrani H. Identification of cytotoxin-producing Klebsiella oxytoca strains isolated from clinical samples with cell culture assays. Microb Pathog. 2017;113:1–4. doi: 10.1016/j.micpath.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 29.Validi M., Soltan Dallal M.M., Douraghi M., Fallah Mehrabadi J., Rahimi Foroushani A. Identification of Klebsiella pneumoniae carbapenemase-producing Klebsiella oxytoca in clinical isolates in Tehran hospitals, Iran by chromogenic medium and molecular method. Osong Pub Health Res Perspect. 2016;7:301–306. doi: 10.1016/j.phrp.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grisold A.J., Zarfel G., Strenger V., Feierl G., Leitner E., Masoud L. Use of automated repetitive-sequence-based PCR for rapid laboratory confirmation of nosocomial outbreaks. J Infect. 2010;60:44e51. doi: 10.1016/j.jinf.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 31.Joainig M.M., Gorkiewicz G., Leitner E., Weberhofer P., Zollner-Schwetz I., Lippe I. Cytotoxic effects of Klebsiella oxytoca strains isolated from patients with antibiotic-associated hemorrhagic colitis or other diseases caused by infections and from healthy subjects. J Clin Microbiol. 2010;48:817–824. doi: 10.1128/JCM.01741-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsakris A., Poulou A., Markou F., Pitiriga V., Piperaki E.T., Kristo I. Dissemination of clinical isolates of Klebsiella oxytoca harboring CMY-31, VIM-1 and a New OXY-2-type variant in the community. Antimicrob Agents Chemother. 2011;55:3164–3168. doi: 10.1128/AAC.00102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanc D.S. The use of molecular typing for epidemiological surveillance and investigation of endemic nosocomial infections. Infect Genet Evol. 2004;4:193–197. doi: 10.1016/j.meegid.2004.01.010. [DOI] [PubMed] [Google Scholar]