Abstract
Heterogeneous populations within a tumor have varying metabolic profiles, which can muddle the interpretation of bulk tumor imaging studies of treatment response. Although methods to study tumor metabolism at the cellular level are emerging, these methods provide a single time point “snapshot” of tumor metabolism and require a significant time and animal burden while failing to capture the longitudinal metabolic response of a single tumor to treatment. Here, we investigated a novel method for longitudinal, single-cell tracking of metabolism across heterogeneous tumor cell populations using optical metabolic imaging (OMI), which measures autofluorescence of metabolic coenzymes as a report of metabolic activity. We also investigated whether in vivo cellular metabolic heterogeneity can be accurately captured using tumor-derived three-dimensional organoids in a genetically engineered mouse model of breast cancer. OMI measurements of response to paclitaxel and the phosphatidylinositol-3-kinase inhibitor XL147 in tumors and organoids taken at single cell resolution revealed parallel shifts in metaboltruic heterogeneity. Interestingly, these previously unappreciated heterogeneous metabolic responses in tumors and organoids could not be attributed to tumor cell fate or varying leukocyte content within the microenvironment, suggesting that heightened metabolic heterogeneity upon treatment is largely due to heterogeneous metabolic shifts within tumor cells. Together, these studies show that OMI revealed remarkable heterogeneity in response to treatment, which could provide a novel approach to predict the presence of potentially unresponsive tumor cell subpopulations lurking within a largely responsive bulk tumor population, which might otherwise be overlooked by traditional measurements.
Introduction
There is accumulating evidence that tumor cell populations are heterogeneous, enabling heterogeneous responses to treatments that may either enhance or inhibit treatment sensitivity [1], [2], [3], [4]. Minority populations of tumor cells with innate treatment resistance have been identified, such as CD24+ breast cancer cells, which exhibit resistance to certain chemotherapies [5], [6]. The presence of minority tumor cell subpopulations with innate resistance to treatment can ultimately result in tumor recurrence, even under circumstances when the original tumor, comprised mainly of treatment sensitive cells, responds to treatment. Clinicians lack the tools necessary to assess this heterogeneity and to recommend optimal treatment plans for each individual patient. It is also difficult to study the process by which tumors evolve to obtain variability in cellular treatment sensitivity. Current techniques to perform high-throughput in vivo drug screens and assess heterogeneity are destructive to the cells and require enormous animal burden. These limitations not only hinder our understanding of the mechanisms behind tumor heterogeneity and recurrence, but also obstruct the discovery of novel drugs and drug combinations that combat the emergence of therapy-resistant subpopulations of cells. To address these problems, a platform is needed that faithfully recapitulates and quantifies in vivo cellular heterogeneity in vitro.
Next-generation single-cell sequencing can be used to characterize genetic diversity by identifying mutant subclones [7], [8], [9], [10], [11], and the number of subclonal driver mutations has predictive value for a patient's overall survival [12]. However, the predictive ability of single cell sequencing in the clinical setting relies on genetic mutations that are already known. Therefore, the remarkable advances provided by single-cell sequencing still cannot provide a true predictive analysis of treatment-resistance tumor cell subpopulations that may lurk within a larger tumor cell population. Further, the sensitivity of a cell to a specific drug is a complex combination of both genetic and nongenetic factors, including cellular metabolism [13], [14]. To fuel aberrant proliferation rates, cancer cells reprogram their metabolic machinery to incorporate nutrients into required pathways even in the absence of growth factor signals [15], [16]. However, not all cells in a tumor alter their metabolism in the same manner. In response to unique levels of oxygen, glucose, pH, cytokines, and extracellular matrix proteins in their microenvironment, individual cells adapt appropriately to maximize their survival [14], [17]. These mechanisms of adaptation also affect cell resistance to cytotoxic drugs and immunotherapies [18], [19], resulting in subpopulations of tumor cells with differing sensitivities to treatment. This highlights the need for a functional readout of heterogeneity that is based on cell metabolism.
Given the limitations of genetic testing on predicting treatment response, alternative approaches which directly measure drug response in tumor cells are under investigation. For example, testing treatment response in mouse patient-derived xenograft models captures in vivo genetic heterogeneity and can be used to predict patient response to many therapies [20]. However, patient-derived xenografts require enormous numbers of animals for high-throughput drug screening and cannot be performed in a clinically beneficial time frame. Alternatively, in vitro cancer organoids can be used to screen drugs directly on patient cells, alleviating the burdens of time, animals, and cost [21]. Organoids maintain the genetic, histopathological, and 3-dimensional characteristics, along with the functional surface markers of the original tumor for a variety of cancer types [22], [23], [24], [25]. Additionally, organoids contain stromal cells that can facilitate therapeutic resistance [26]. Many organoids can be cultured from a single patient biopsy, supporting the feasibility of screening patient-derived tumor organoids for sensitivity to a variety of treatments.
Optical metabolic imaging (OMI) is a label-free two-photon microscopy technique that quantifies single-cell metabolic changes with treatment both in tumors in vivo [27] and in tumor-derived organoids [28]. OMI uses the endogenous fluorescent properties of metabolic coenzymes NAD(P)H and FAD. The optical redox ratio, or the ratio of the fluorescence intensity of NAD(P)H to that of FAD, reflects the redox state of the cell [29], [30], [31]. The fluorescence lifetimes of NAD(P)H and FAD are distinct for the free and protein-bound conformations of both molecules and thus reflect enzyme binding [32], [33], [34]. These OMI variables, integrated to form a composite OMI index, reflect early metabolic shifts and thus can evaluate drug response prior to changes in cell viability or tumor size [21], [35], [36]. The nondestructive nature of this technique allows heterogeneity to be tracked in organoids over time to analyze the potential for drug resistance evolution in the original tumor. This is not possible with other methods for measuring heterogeneity (e.g., flow cytometry, single-cell sequencing, or immunolabeling) because these standard methods are destructive.
OMI of cancer organoids has been validated as an accurate predictor of in vivo drug response in xenograft models generated from human breast cancer and head and neck cancer cell lines [21], [35] and a mouse model of pancreatic cancer [36], but it is unclear whether the heterogeneity measured in organoids also accurately mirrors the original tumor. Here, we investigate whether in vivo heterogeneity is reflected in vitro in organoids using OMI measurements in vivo and in organoids derived from the polyomavirus middle T (PyVmT) mouse model. The PyVmT model closely mimics the stages and progression of human breast cancer, exhibits more heterogeneity than human cell line xenografts, and can develop in a fully immunocompetent mouse [37]. This study demonstrates that OMI of in vitro tumor organoids accurately captures in vivo heterogeneous response to treatment at the single-cell level in a relevant breast cancer model.
Materials and Methods
Orthotopic PyVmT Tumors
Animal research was approved by the Institutional Animal Care and Use Committees at Vanderbilt University and the University of Wisconsin-Madison. Orthotopic tumors were initially generated by injecting 106 PyVmT cells suspended in 100 μl of a chilled 1:1 mixture of DMEM (Gibco #11965) and Matrigel (Corning #354234) into the fourth inguinal mammary fat pads of 6-week-old FVB female mice (The Jackson Laboratory #001800) using a 26-gauge needle. The PyVmT cell line was derived from tumors isolated from transgenic FVB MMTV-PyVmT mice (The Jackson Laboratory #002374). Tumors were passaged by mechanically dissociating an existing tumor, passing the tumor cell suspension through a 70 μm strainer, and injecting the cells into a new 6-week-old FVB mouse.
FDG-PET/CT Imaging
Mice were randomized and imaged prior to treatment (when tumors grew to >200 mm3) and then imaged weekly over a 14-day time course at the University of Wisconsin Small Animal Imaging and Radiotherapy Facility by trained staff (J.J.J., J.T.S.). 10 mg/kg paclitaxel (Vanderbilt University pharmacy/University of Wisconsin pharmacy) was diluted in 150 μl PBS and injected intraperitoneally twice weekly. 50 mg/kg XL147 (Selleckchem) was suspended in 100 μl 1% DMSO and delivered by oral gavage daily. Control mice received 100 μl 1% DMSO by oral gavage daily and 150 μl PBS by intraperitoneal injection twice weekly. Prior to PET/CT imaging, mice were fasted for 12 hours. One hour prior to imaging, ~9.25 MBq of [18F]FDG was delivered by tail vein injection, and mice were immediately anesthetized under 4% isoflurane, maintained at 1.5%, and warmed with a heated bed until the end of the imaging procedure. The mice were scanned using an Inveon microPET/CT (Siemens Medical Solutions, Knoxville, TN) in the prone position. Forty-million counts per mouse were collected for the PET scan to obtain adequate signal-to-noise. PET data were histogrammed into one static frame and subsequently reconstructed using ordered-subset expectation maximization of three dimensions followed by the maximum a posteriori algorithm, and CT attenuation and scatter correction were applied based on the NEMA NU 4 image-quality parameters [38]. Inveon Research Workplace software (Siemens Medical Solution, Knoxville, TN) was used to measure tumor volume and glucose uptake. PET and CT images were co-registered, and manual regions of interest (ROIs) were drawn around each tumor. FDG uptake was measured as the decay-corrected percent injected dose normalized by the mass of the tissue (%ID/gtissue) assuming the density of the ROI tissue is equal to water. Five tumors from five mice were imaged per treatment group.
Fluorescence Lifetime Imaging
A custom multiphoton fluorescence lifetime system (Bruker Fluorescence Microscopy) was used to acquire fluorescence images. A titanium:sapphire laser (Chameleon Ultra II, Coherent) was used for excitation, while GaAsP photomultiplier tubes (H7422P-40, Hamamatsu) were used to detect emission photons. A 40x water immersion objective (Nikon, 1.15 NA) was used with an inverted microscope (Nikon, TiE). Two-photon excitation of NAD(P)H and FAD was performed with 750 nm and 890 nm light, respectively. A 440/80 nm filter was used to isolate NAD(P)H fluorescence emission, and a 550/100 nm filter was used for FAD fluorescence emission. 256×256 pixel images were acquired using a pixel dwell time of 4.8 microseconds and a 60-second collection time. Time-correlated single photon counting electronics (SPC-150, Becker & Hickl) were used to acquire fluorescence lifetime data with 256 time bins. A single Fluoresbrite YG microsphere (Polysciences) was imaged each day as a fluorescence lifetime standard. The measured lifetime (2.15 ± 0.08 ns, n = 7) was stable and consistent with previously published values [27], [33], [35], [39], [40]. For each field of view, an NAD(P)H image was acquired first followed immediately by an FAD image.
Intravital OMI
Treatment was initiated when tumors reached >200 mm3. Forty-eight hours after initial treatment, and 1 hour following final treatment, mice were anesthetized using isoflurane, and a small portion of skin was removed to expose the underlying tumor. OMI was performed on at least four fields of view and at least 450 total cells per tumor. Following imaging, mice were euthanized while under anesthesia, and tumors were extracted and fixed. Five tumors from five different mice were imaged per treatment group.
Tissue Processing and Organoid Culture
Excised tumors were rinsed in PBS and thoroughly minced in DMEM with dissecting scissors. The resulting cell macrosuspension was chilled on ice and mixed with Matrigel at a 1:2 ratio, and 100 μl of the resulting mixture was pipetted into 35 mm glass-bottom dishes (#P35G-1.5-14-C, MatTek). Gels were allowed to solidify slowly at room temperature for 30 minutes and then placed in a 37°C, 5% CO2 incubator for 1 hour before being overlaid with DMEM supplemented with 10% FBS and 1% penicillin-streptomycin (Gibco).
Organoid Imaging
Twenty-four hours prior to imaging, organoid media was replaced with fresh media containing 500 nM paclitaxel, 25 nM XL147, or 0.1% DMSO vehicle. These doses were chosen to replicate in vivo doses [21]. Organoids were imaged after 24, 48, and 72 hours of treatment. At least six organoids from each treatment group were imaged at each time point, containing at least 290 cells in total for each treatment group. The length of the longest dimension in each organoid was quantified using ImageJ [41].
OMI Endpoint Images
The optical redox ratio was computed from NAD(P)H and FAD lifetime images. First, the fluorescence intensities of NAD(P)H and FAD are determined by the total number of photons detected over the collection time. Then, the intensity of NAD(P)H is divided by the intensity of FAD in each pixel. SPCImage software was used to analyze fluorescence lifetime images (Becker & Hickl) [42]. First, fluorescence decay curves for NAD(P)H and FAD are generated for each pixel by assigning all photon events into 256 temporal bins. Detected photons from the eight neighboring pixels are also included in each curve to increase photon counts. This decay curve is deconvolved with the measured instrument response function and fit with a two-component exponential decay Eq. (1).
| (1) |
I(t) represents the fluorescence intensity measured at time t following a laser pulse; α1 and α2 represent the percentage of the overall signal made up by the short and long lifetime components, respectively; τ1 and τ2 are the short and long lifetime components, respectively; and C represents background light. The use of a two-component fit was chosen in order to differentiate the free (τ1) and bound (τ2) states of NAD(P)H and the free (τ2) and bound (τ1) states of FAD [32], [43]. The mean lifetime (τm) of NAD(P)H and FAD for each pixel is the weighted average of the free and bound lifetimes Eq. (2).
| (2) |
A CellProfiler routine was written to automatically distinguish between individual cell cytoplasms and the nuclei they encircle [44], [45]. For each cytoplasm detected, values for the redox ratio and all NAD(P)H and FAD lifetime variables were calculated by averaging the incorporated pixels within the cytoplasm.
OMI Index
The redox ratio, NAD(P)H τm, and FAD τm were normalized to average values in control cells for the OMI index calculation. In vivo, values are normalized to averages of all cells imaged in all control mice on the same day. In vitro, values are normalized to averages of all cells from all control organoids imaged on the same day. In both cases, the OMI index is calculated for each cell using a linear combination of the control-normalized redox ratio, NAD(P)H τm, and FAD τm with coefficients of (1, 1, −1), respectively. A decrease in OMI index correlates with drug sensitivity [21].
Heterogeneity Analysis
Histograms were generated for all cells within a group using individual cell OMI indices. The histogram is fit to Gaussian mixture distribution models containing one, two, or three components using an iterative expectation maximization algorithm. Each component represents a distinct subpopulation of cells. Goodness of fit for each model was evaluated using the Akaike information criterion [46], and only the model with best fit was implemented. For each group, this process is repeated 1000 times. For visual comparison, example distributions are normalized to have equal areas under the curve. Similar numbers of cells were imaged in each tumor and in each organoid treatment group to provide similar power to identify subpopulations in each distribution [47]. The heterogeneity index is a validated metric for quantifying heterogeneity in a population [27], [48]. Here we present a modified version, the weighted heterogeneity index (wH-index), which also takes into account the standard deviations of all subpopulations.
| (3) |
Here, i represents each subpopulation, d represents the distance between the median of each subpopulation and the median of the entire distribution, p represents the proportion of all cells belonging to that subpopulation, and σ is the standard deviation.
Histological Analysis
Immediately following in vivo tumor imaging at 48 hours of treatment, tumors were collected and fixed in 10% neutral buffered formalin for 48 hours. Tumors were oriented and paraffin embedded such that 5 μm slices were taken from the imaged portion of the tumor. Also immediately following imaging at 72 hours of treatment, Matrigel droplets containing organoids were detached from glass coverslips and encased in agarose. These agarose gels were placed in 10% neutral buffered formalin for 48 hours, paraffin embedded, and sliced into 5 μm sections starting from the center. Organoid histology experiments were duplicated and results were combined. Tumors and organoid sections were stained using immunohistochemistry (IHC) for CD45 (ab10558, Abcam, 32 minutes), vimentin (ab92547, Abcam, 32 minutes), alpha smooth muscle actin (αSMA) (ab5694, Abcam, 32 minutes), wide-spectrum cytokeratin (ab9377, Abcam, 28 minutes), polyoma virus middle T antigen (sc-53,481, SantaCruz, 60 minutes), Ki67 (ab15580, Abcam, 28 minutes), cleaved caspase 3 (9661, Cell Signaling, 28 minutes), phospho-histone H3 (9701, Cell Signaling, 16 minutes), and phospho-PRAS40 (2997, Cell Signaling, 12 minutes). Positive staining was visualized with diaminobenzidine (DAB) and counterstained with hematoxylin.
Histology Imaging and Quantification
Multispectral image cubes were acquired using a Nuance multispectral imaging microscope and software (Perkin Elmer) [49]. Images of transmitted light were taken from 420 nm to 720 nm with 20-nm steps and converted to optical densities using a blank reference image. Image cubes were unmixed using a spectral library to isolate individual stains and exclude background signals. Individual cells (stained with DAB and counterstained with hematoxylin) in tumor and organoid images were automatically segmented using inForm Cell Analysis software (Perkin Elmer). In organoid images, machine learning was used to train a feature-recognition algorithm to automatically recognize and distinguish between individual cellular organoids, extracellular matrix, and background regions. For each stain, a random selection of 10% of all images was used as a training set. Organoid, background, and extracellular matrix regions were manually defined in this set, which was then used to train a segmentation algorithm until it had >92% accuracy. Finally, this algorithm was used to segment all images. For tumors, algorithms were similarly trained to distinguish between tumor and stromal compartments. Next, individual cells were automatically segmented by thresholding the intensity of the nuclear hematoxylin counterstain. Only cells in tumor or organoid compartments were quantified. Individual cells were scored as positive or negative for a particular target antibody by thresholding for the mean pixel intensity of DAB within each cell. At least four random fields of view were acquired per stain per tumor, for a total of at least 23 images per stain per treatment group. Organoids containing less than five cells were rejected from analysis, and at least 35 organoids were quantified per stain per treatment group.
Statistical Analysis
Normalized tumor volumes and glucose uptake values were compared using a Student t‐test with Bonferroni correction for multiple comparisons. Significant differences in OMI variables, wH-indices, IHC percentages, and organoid longest dimensions between treatment groups were tested using a Wilcoxon rank-sum test. Treatment effect size was calculated with Glass's Δ [50], with directionality determined by the response of HER2+ cell line xenografts to trastuzumab [21]. Preliminary data from our group show that an OMI index treatment effect size threshold of 0.75 in tumor-derived organoids accurately classified pancreatic cancer patients based on their recurrence-free survival time during adjuvant therapy [51]. This is similar to a previously suggested cutoff of 0.8 for large effect sizes [52].
Results
PET/CT of In Vivo Treatment Response
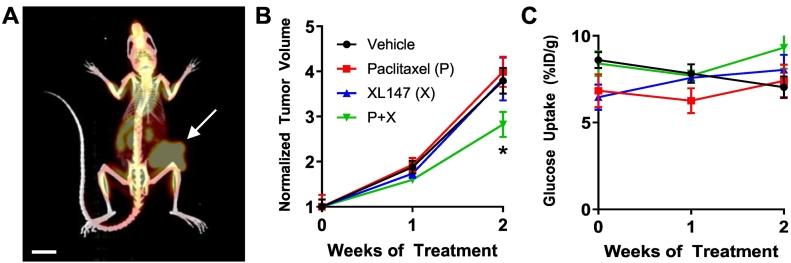
Response to treatment with paclitaxel and XL147 was measured in orthotopic PyVmT allograft tumors and tumor organoids. Paclitaxel is an FDA-approved chemotherapy for breast cancer that has been shown to reduce tumor growth rates in orthotopic PyVmT tumors [53]. XL147, an inhibitor of the phosphatidylinositol-3 kinase family (PI3K), was also chosen due to the dependence of the PyVmT model on PI3K activity [54], [55], [56] and for its use in clinical trials for breast cancer. To verify response to treatment, FDG-PET measurements were taken of tumors treated with paclitaxel, XL147, and combination (P + X) over 14 days (Figure 1). Subsequent CT measurements were also taken in order to better visualize the tumor in the mammary fat pad and distinguish tumor-associated signal from background (Figure 1A). While XL147 alone and paclitaxel alone did not affect tumor growth after 2 weeks, the combination of both drugs significantly decreased tumor growth vs. vehicle (P < .05, Figure 1B). No differences in FDG uptake were observed with treatment (Figure 1C). A reduction in pPRAS40 staining in XL147-treated tumors and organoids confirms successful PI3K inhibition (Supplementary Figure S1).
Figure 1.
FDG-PET/CT of PyVmT in vivo treatment response. (A) Example PET/CT image showing glucose uptake in an orthotopic PyVmT tumor (arrow) growing on the right mammary fat pad. Scale bar = 1 cm. (B) The effect of drug treatment on tumor growth. All volumes are normalized to pretreatment volume. N = 5 tumors per treatment group. * P < .05 vs. vehicle. (C) Glucose uptake did not change with drug treatment.
OMI of Cellular Metabolic Heterogeneity In Vivo
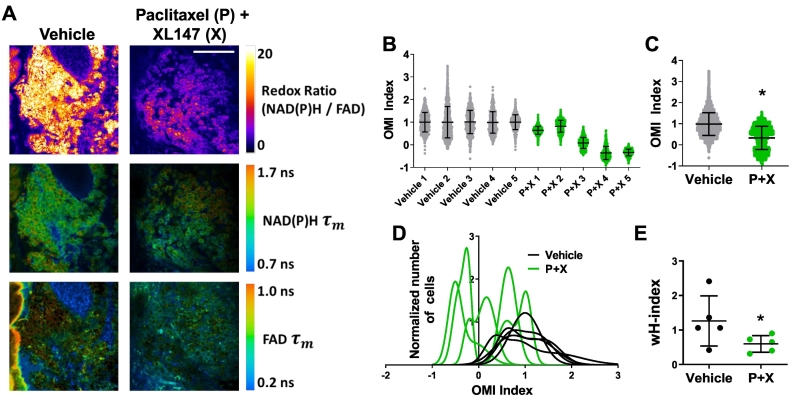
Intravital OMI was performed to determine how treatment affects tumor metabolic heterogeneity in vivo at a cellular level. Representative images demonstrate that OMI can visualize single-cell metabolic properties in these tumors (Figure 2A). Response was quantified using the OMI index after 48 hours of treatment with either vehicle or P + X in five mice per group (Figure 2B). Across all cells from all tumors, P + X treated cells had a significantly lower OMI index compared with vehicle controls (P < .0001, Figure 2C). Histograms of cell OMI indices were fit to Gaussian mixture distribution models (Figure 2D, plotted separately in Supplementary Figure S2). Vehicle-treated tumors exhibited higher degrees of variance in their single-cell OMI index distributions compared to P + X treated tumors. The smallest subpopulation detected in tumors represented 29.0% of the overall population (vehicle). The degree of heterogeneity in each tumor is quantified using a modified form of the heterogeneity index [27], [48] with an additional term added for the standard deviation of each individual subpopulation (wH-index). P + X treatment was found to significantly decrease the in vivo wH-index vs. vehicle (P < .05, Figure 2E).
Figure 2.
OMI of drug response heterogeneity in vivo. (A) Representative images of the redox ratio, NAD(P)H mean lifetime (τm), and FAD τm in vivo in PyVmT tumors treated with vehicle or paclitaxel + XL147 (P + X) for 48 hours. Scale bar = 100 μm. (B) OMI Index quantified at the single-cell level 48 hours after treatment in vivo for each tumor. Each dot represents one cell. A decrease in the OMI index indicates drug response. Error bars indicate mean ± SD. N > 450 cells/tumor. (C) Decrease in average OMI Index with treatment across all cells from all tumors. Error bars indicate mean ± SD. * P < .0001 vs. vehicle. N > 3300 cells/group. (D) Population density distributions of drug response at the single-cell level for each tumor. (E) The effect of treatment on the degree of OMI Index heterogeneity per tumor quantified by the wH-index. Error bars indicate mean ± SD. N = 1000 fits/tumor. Each dot represents one tumor. *P < .05 vs. vehicle.
OMI of Cellular Metabolic Heterogeneity in Organoids
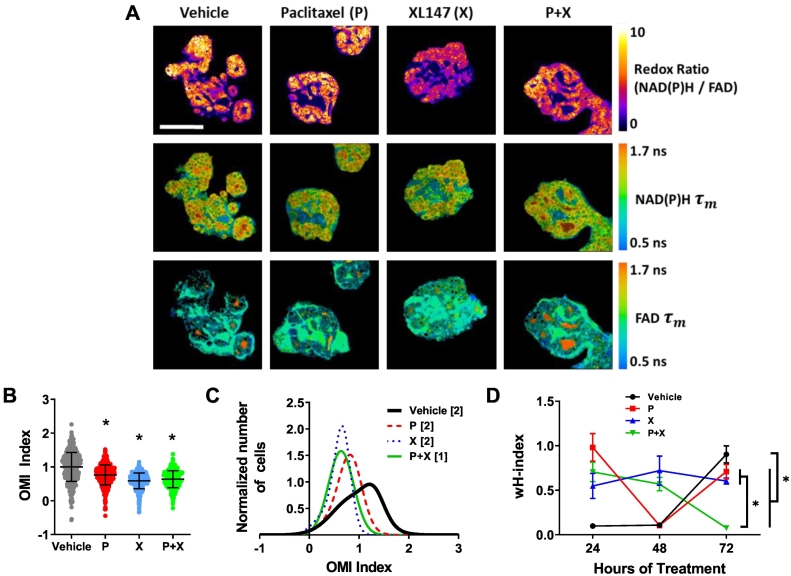
Next, responses to the same therapies were measured in PyVmT tumor-derived organoids to determine if organoid heterogeneity is similar to PyVmT tumors in vivo. Representative images demonstrate the size and morphology of the organoids and the OMI endpoint spatial distributions (Figure 3A). Single therapies were tested in parallel to P + X combined therapy, and OMI index was calculated at the single-cell level after 24, 48, and 72 hours of treatment. Organoids treated with P + X had a lower mean OMI index compared to control organoids (P < .0001, Figure 3B and Supplementary Figure S3, A and B), in agreement with the in vivo results. Examples of Gaussian mixture modeling showed multiple subpopulations in the control organoids and only a single population with smaller overall variance in the P + X treated organoids (Figure 3C). The degree of heterogeneity was significantly decreased in P + X treated organoids vs. control organoids at 72 hours (P < .0001, Figure 3D), just as it was in vivo at 48 hours. The mean wH-index values were higher at 48 hours in vivo (1.26 and 0.60 for vehicle and P + X treated, respectively) compared to 72 hours in vitro (0.90 and 0.08 for vehicle and P + X treated, respectively). Similar wH-indices were found between treatment groups in organoids at 24 and 48 hours of treatment (Figure 3D, example distributions in Supplementary Figure S3, C and D). The smallest subpopulation detected in organoids represented 12.2% of the overall population (paclitaxel, 24 hours). The optical redox ratio, NAD(P)H τm, and FAD τm also changed significantly with P + X treatment in both organoids and tumors (Supplementary Figure S4). Coefficients of variation (CV) for NAD(P)H and FAD intensities and lifetimes showed similar trends within organoids and in vivo tumors (Supplementary Figure S5). Notably, CV values for FAD intensity are large compared to other variables in both tumors and organoids. This may be because FAD is localized to mitochondria, leading to a high level of spatial variance compared to NAD(P)H, which is present throughout the mitochondria and cytoplasm. Thus, FAD intensity values for individual cells can vary depending on how many mitochondria are present in the focal plane. This supports the complimentary use of FAD lifetime imaging, as lifetime signals are independent of fluorophore concentration. This is evident in the lower CV values for FAD lifetime variables.
Figure 3.
OMI of drug response heterogeneity in PyVmT organoids. (A) Representative images of the redox ratio, NAD(P)H τm, and FAD τm in organoids at 72 hours of treatment. Scale bar = 100 μm. (B) OMI index quantified at the single-cell level. N = 6 organoids/group. N > 300 cells/group. Each dot represents one cell. Error bars indicate mean ± SD. * P < .0001 vs. vehicle. (C) Example subpopulation analysis of the OMI index in all organoid cells. Brackets in legend indicate number of subpopulations for each group. (D) The effect of treatment on the degree of OMI index heterogeneity per tumor quantified by the wH-index. Error bars indicate mean ± SD. N = 1000 fits/group for all 6 organoids/group. * P < .0001 at 72 hours.
Comparison of Drug Treatment Effect Sizes on OMI Variables
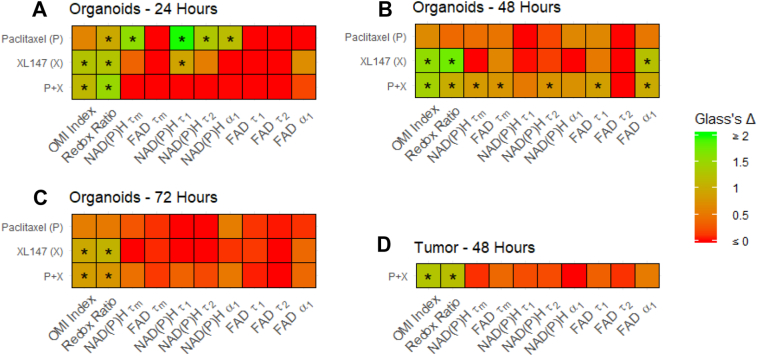
The effect size of drug treatment on individual OMI variables was calculated for all cells in tumors and organoids using Glass's Δ [50] (Figure 4). P + X treatment had large effects (Δ > 0.75) on the redox ratio and OMI index at 24, 48, and 72 hours in organoids (Figure 4, A-C) and in tumors at 48 hours (Figure 4D). P + X did not have a large effect on individual lifetime variables, except at 48 hours in tumor-derived organoids. XL147 alone also had a large effect on the OMI index and redox ratio at all time points in organoids. Paclitaxel alone had a large effect on the redox ratio at only 24 hours in organoids, and did not have a large effect on OMI index at any time point. The correlation between Glass's Δ values with P + X treatment in vivo at 48 hours versus organoids at 24, 48, and 72 hours was significant at all time points (P < .05, Pearson's correlation), indicating that changes in OMI variables with P + X treatment in organoids faithfully mirror in vivo changes.
Figure 4.
Effect sizes of treatment on OMI variables in PyVmT tumors and organoids. (A-C) Glass’s Δ effect size of treatment on OMI variables across all PyVmT tumor-derived or ganoid cells at (A) 24, (B) 48, and (C) 72 hours. * Δ > 0.75. (D) Glass’s Δ effect size of treatment on OMI variables across all PyVmT tumor cell.
Characterization of Cell Types in PyVmT Tumors and Organoids
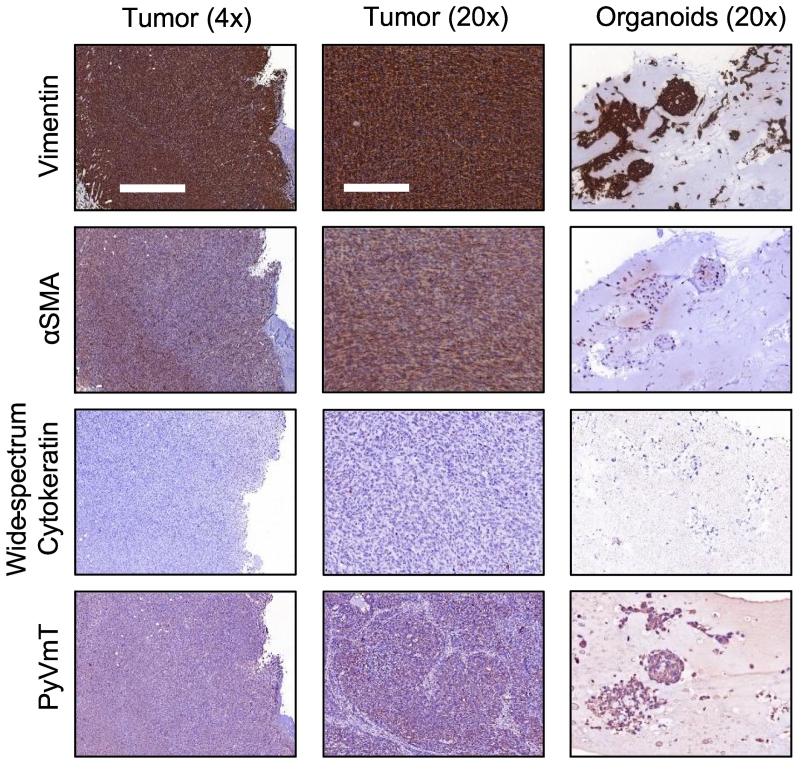
Histology of PyVmT tumors and organoids was performed to determine if the distinct cell subpopulations measured in Figure 2, Figure 3 were comprised of different cell types, cells undergoing apoptosis or proliferation, or cells simply employing unique metabolic strategies. Expression of vimentin and αSMA and loss of wide spectrum cytokeratin expression in all tumors and organoids indicate that cells had undergone epithelial to mesenchymal transition prior to imaging (Figure 5). Expression of PyVmT antigen in both tumors and organoids (Figure 5) confirms that cells matched the tumors of origin.
Figure 5.
Characterization of tumors and organoid cells. Example images demonstrate that tumor and organoid cells express vimentin and α-smooth muscle actin and do not express cytokeratins. Cells are confirmed positive for PyVmT antigen. 4x scale bar = 1 mm. 20x scale bar = 200 μm.
Quantification of Potential Sources of Heterogeneity in PyVmT Tumors and Organoids
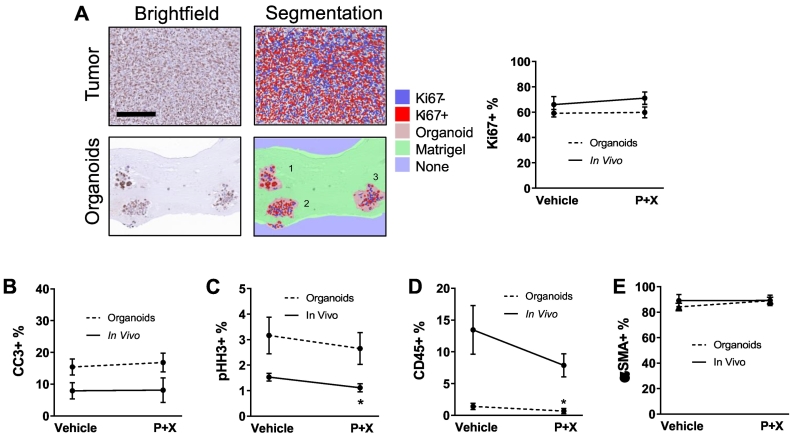
Machine learning algorithms were trained using inForm software to distinguish between individual cellular organoids in histology images, and to identify cells and quantify expression both in organoids and in vivo. The expression of Ki67 and phospho–histone H3 (pHH3), two proliferation markers, was quantified along with cleaved caspase 3 (CC3), a marker of apoptosis. Ki67 is expressed in the S, G2, and M phases of the cell cycle, while pHH3 is specific to cells in M phase [57]. The percentage of cells expressing Ki67 was similar between tumors and organoids and did not change after 48 hours of treatment in either setting (Figure 6A). Likewise, the percentage of cells in apoptosis was similar between tumors and organoids (both <20%) and did not change after 48 hours of treatment in either setting (Figure 6B and Supplementary Figure S6A). pHH3 was expressed in significantly fewer cells in tumors and organoids compared to Ki67 (Figure 6C and Supplementary Figure S6B). Significantly fewer of these cells in M-phase were found in P + X treated tumors compared to vehicle controls (P < .05), but treatment did not significantly change the number of M-phase cells in organoids. Small subpopulations of CD45+ leukocytes were present in both tumors and organoids (Figure 6D and Supplementary Figure S6C). A smaller percentage of CD45+ leukocytes were present in organoids compared to the original tumors, and fewer were present in P + X treated organoids compared to vehicle (P < .05). A majority of cells in organoids and tumors were positive for αSMA (Figure 6E and Supplementary Figure S6D). The percentage of αSMA+ cells was not significantly different between organoids and tumors and did not significantly change with treatment in either setting. The variance of positive staining of all markers across all fields of view (FOVs) and organoids was not affected by treatment. Overall, these cell fates and cell types are unlikely to be responsible for the decrease in OMI index and its heterogeneity with P + X treatment because none of the positive percentages of these markers significantly changed with treatment in both tumors and organoids.
Figure 6.
Quantification of heterogeneity and treatment response in PyVmT tumors and organoids with histology. (A) Automatic cell segmentation and quantification of Ki67 expression in 20x images of PyVmT tumor and organoids. Scale bar = 200 μm. N > 4 FOVs per tumor, 5 tumors per group. N > 39 organoids per group. (B) CC3 expression in tumors and organoids. N = 5 FOVs per tumor, 5 tumors per group. N > 44 organoids per group. (C) pHH3 expression in tumors and organoids. N = 5 FOVs per tumor, 5 tumors per group. N > 60 organoids per group. (D) CD45 expression in tumors and organoids. N > 4 FOVs per tumor, 5 tumors per group. N > 45 organoids per group. (E) αSMA expression in tumors and organoids. N > 4 FOVs per tumor, N > 5 tumors per group. N > 20 organoids per group. Error bars indicate mean ± SD. * P < .05 vs. control. All tumors and organoids were fixed after 48 hours and 72 hours of treatment, respectively.
Discussion
To optimize a treatment plan for an individual cancer patient, a clinician must understand how various drug options will affect all the cells in a patient's uniquely heterogeneous tumor. While recent technological advances have improved our fundamental understanding of intratumoral heterogeneity, we still lack tools for cancer treatment planning that incorporate distinct metabolic subpopulations. Cancer organoids allow for rapid, high-throughput drug screening directly on tumor cells in a relevant three-dimensional microenvironment [21], [23], [35], [36], [58]. Current methods for evaluating drug response in organoids either damage the sample, overlook cellular heterogeneity, or do not directly screen drugs on patient-derived cells. To address these limitations, we have developed OMI to noninvasively quantify metabolic heterogeneity within living organoids using the fluorescent properties of metabolic coenzymes NAD(P)H and FAD [21], [35]. OMI detects minority metabolic subpopulations of drug-resistant cells within organoids, and this information can predict long-term tumor drug response. These capabilities have been confirmed in breast [21], [47] and head and neck [35] cell line xenografts and in a pancreatic cancer mouse model [36]. However, cell line tumors in immunocompromised mice do not capture the cellular heterogeneity of primary human tumors, and it remains unclear whether metabolic heterogeneity in primary tumor-derived organoids mirrors the heterogeneity of the original tumor. In this study, OMI was used to compare subpopulations of cell metabolism in tumors and tumor-derived organoids of the immunocompetent PyVmT breast cancer model. We confirm that organoids accurately capture in vivo metabolic heterogeneity, demonstrating the feasibility of using organoids to study tumor response to treatment.
CT images confirmed that P + X treatment reduced PyVmT tumor volume in vivo over 14 days in immunocompetent allografts (Figure 1). Tumor volume did not change over the first week of treatment, and FDG uptake did not change with therapy over 2 weeks. However, OMI was sensitive to metabolic treatment response in only 48 hours (Figure 2). This highlights the enhanced sensitivity of OMI to early metabolic changes compared to traditional methods of measuring treatment response, in agreement with previous reports in immunocompromised xenografts [27].
Intravital OMI was performed in mice treated with vehicle or P + X for 48 hours (Figure 2). Variation in mean OMI index between P + X treated tumors may be due to slight variations in tumor sizes and vascularization, and/or variations in drug absorption, especially for the final dose that was given 1 hour prior to imaging. The degree of heterogeneity within each tumor was characterized by the wH-index. P + X treated tumors exhibited a significantly lower degree of heterogeneity compared to control tumors (P < .05), further suggesting that this treatment combination could be beneficial. OMI was also used to measure response to P + X in PyVmT tumor-derived organoids (Figure 3). Paclitaxel and XL147 single therapies were also screened with low additional effort and cost, underlining the high-throughput nature of organoid versus animal drug screens. Additionally, due to the noninvasive nature of OMI, measurement of response was performed at 24, 48, and 72 hours in the same set of organoids.
OMI measurements at 72 hours in organoids, of all the time points acquired, most accurately replicated in vivo heterogeneity. This agrees with previous reports that early metabolic drug response at 72 hours in organoids predicts long-term in vivo tumor drug response [21]. The wH-index is low for control organoids at 24 and 48 hours and P + X treated organoids at 72 hours because the populations of cells at these time points were best fit by only one Gaussian component, giving d = 0 in Eq. (3). The wH-index gives significant weight to heterogeneous distributions that are best fit by multiple Gaussians, and thus, these single-component populations have low wH-indices. A potential explanation for the increase in heterogeneity in vehicle-treated organoids from 48 to 72 hours is a significant increase in organoid size between time points (Supplementary Figure S7). These larger organoids may have developed metabolic heterogeneity due to gradients of drug delivery, fuel sources, and metabolic waste. The higher degree of heterogeneity in organoids at 72 hours posttreatment for single-agent treatments compared to P + X treatment (Figure 3) may explain why single therapies alone did not reduce overall tumor growth (Figure 1) despite a decrease in mean OMI index. It is possible that only a subpopulation of the cells was sensitive, which led to disease progression. In contrast, P + X treated organoids exhibited a lower wH-index at 72 hours compared to controls and single therapies (Figure 3D), which suggests that this drug combination worked synergistically to overcome the resistance to single-agent treatments. The higher fold change in wH-index with P + X treatment in organoids compared to in vivo tumors may be due to differences in efficiency of drug delivery. While drug gradients caused by drug diffusion do exist in large organoids, this may not completely capture the irregularity of drug diffusion throughout solid tumors. Additionally, while drug doses in vivo and in vitro were chosen to be as analogous as possible, differences in effective dose may also explain why the decrease in weighted heterogeneity in organoids at 72 hours of treatment required less time to occur in tumors (48 hours).
Evaluating the degree of drug response heterogeneity in organoids using the wH-index was a better predictor of long-term tumor growth than the average response across all cells. P + X treatment also had similar effects on individual OMI variables in tumors and organoids, including a large effect on the optical redox ratio, and small effects on FAD lifetime variables (Figure 4). These parallels suggest that cellular-level drug response in tumor-derived organoids can be used to analyze heterogeneous responses to treatment in the tumor of origin.
We next assessed potential sources of the shifts in heterogeneity found in tumors after 48 hours of treatment and in organoids after 72 hours of treatment using IHC (Figure 5). These results indicate that PyVmT tumors in this study progressed to a mesenchymal phenotype at the time of imaging, closely resembling basal-like human breast cancer [59]. Organoid cultures successfully captured these mesenchymal properties of the original tumor (Figure 5).
Traditional IHC measures of therapeutic response (CC3, Ki67, pHH3) were also evaluated as potential sources of metabolic heterogeneity (Figure 6) because metabolic activities are linked to apoptosis and proliferation [60], [61]. PyVmT tumor allografts incorporate the host's acquired immune system, which is an advantage over cell line xenografts grown in athymic nude mice. Immune cells and tumor cells have distinct metabolic properties [19], [62] and thus could also contribute to OMI heterogeneity. A small subset of CD45+ leukocytes was found in both tumors and organoids using IHC, and P + X treatment significantly reduced the percentage of leukocytes in organoids but not in vivo. This may be because organoid culture conditions were not optimized to maintain mouse leukocytes. Finally, mesenchymal transition can also affect cellular drug response and metabolism [63], but our IHC results indicate that this mesenchymal phenotype is nearly homogenous and consistent with treatment, and thus not the cause of OMI index heterogeneity.
None of the markers tested changed significantly in both tumors and organoids, indicating that apoptosis, proliferation, and the presence of immune cells and αSMA+ cells are unlikely to be responsible for the decreases in OMI index and its heterogeneity with P + X treatment. These markers are likely not associated with the high OMI index subpopulations in control organoids at 72 hours and control tumors, which were affected by P + X treatment. Overall, we did not identify a marker of cell type or cell fate that accounted for the metabolic heterogeneity detected with OMI. These results suggest that the subpopulations identified with OMI are likely due to metabolic differences between cells. Future studies could confirm this finding using emerging single-cell mass spectrometry on dissociated tumors and organoids, but these techniques are still in development [64], [65]. Single-cell RNA analysis of dissociated tumors and organoids could also support our findings but does not offer a comparable measure to the more downstream metabolic activity of cells that OMI provides. Similarly, fluorescent reporters can quantify metabolic properties within single live cells that are correlated with the optical metabolic variables reported here (e.g., glucose uptake [66], pH [67], membrane potential [68], hypoxia [69], NADH/NAD+ redox state [70]). Overall, comparison of OMI with IHC highlights that OMI provides an early measure of tumor response compared to traditional markers, consistent with previous results [35].
This study validates OMI of primary tumor-derived organoids as a unique, powerful tool to study in vivo tumor metabolic heterogeneity. This tool could also improve clinical treatment decisions because minority subpopulations of treatment-resistant cells within an otherwise responsive tumor can initiate patient recurrence [2], [3], [21]. These results are the first to show that in vitro organoids capture the early metabolic changes in heterogeneity that occur in vivo with treatment in an immunocompetent tumor. OMI of organoids quantifies single-cell response to many drugs in a relevant three-dimensional microenvironment, using a single tissue sample, and can be performed longitudinally to track the evolution of heterogeneity over time. This technology could provide a personalized medicine platform to perform high-throughput screening of drugs directly on patient cells, and detect metabolic heterogeneity. This would allow clinicians to quickly analyze cellular heterogeneity in response to numerous treatment options for an individual patient to more robustly inform on treatment decisions. Additionally, this technique could reduce the animals and time required to develop new therapeutic strategies that overcome tumor heterogeneity and achieve better outcomes in patients.
Acknowledgements
The authors thank the UW Translational Research Initiatives in Pathology laboratory and UW Small Animal Imaging and Radiotherapy Facility (both supported by the UWCCC grant P30 CA014520) for use of their facilities and services. Thank you to Sophie Mancha for assistance with organoid culture. Thank you to Dr. Dustin Deming and Dr. Suzanne Ponik for helpful conversations about organoids and cancer cell heterogeneity. The Skala laboratory is supported by an NSF Graduate Research Fellowship (DGE-1445197; J.T.S.), and grants from the NSF (CBET-1642287), Stand Up to Cancer, United States (SU2C-AACR-IG-08-16, SU2C-AACR-PS-18), and the NIH (R01 CA185747, R01 CA205101, R01 CA211082, R21 CA224280, U01 TR002383).
Footnotes
Conflicts of interest: The authors declare no potential conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.04.004.
Appendix A. Supplementary Data
Supplementary Figures
References
- 1.Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5(8):806–820. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805(1):105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108(3):479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103(8):1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, Gonzalez-Farre X, Munoz M, Russnes HG, Helland A. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep. 2014;6(3):514–527. doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X, Apple S, Zhao H, Song J, Lee M, Luo W, Wu X, Chung D, Pietras RJ, Chang HR. CD24 Expression and differential resistance to chemotherapy in triple-negative breast cancer. Oncotarget. 2017;8(24):38294–38308. doi: 10.18632/oncotarget.16203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki Y, Ng SB, Chua C, Leow WQ, Chng J, Liu SY, Ramnarayanan K, Gan A, Ho DL, Ten R. Multiregion ultra-deep sequencing reveals early intermixing and variable levels of intratumoral heterogeneity in colorectal cancer. Mol Oncol. 2017;11(2):124–139. doi: 10.1002/1878-0261.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, Mitchell TJ, Grossmann S, Lightfoot H, Egan DA. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556(7702):457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 9.Sievers CK, Zou LS, Pickhardt PJ, Matkowskyj KA, Albrecht DM, Clipson L, Bacher JW, Pooler BD, Moawad FJ, Cash BD. Subclonal diversity arises early even in small colorectal tumours and contributes to differential growth fates. Gut. 2017;66(12):2132–2140. doi: 10.1136/gutjnl-2016-312232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, Navin NE. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell. 2018;173(4):879–893. doi: 10.1016/j.cell.2018.03.041. (e813) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleppe M, Levine RL. Tumor heterogeneity confounds and illuminates: assessing the implications. Nat Med. 2014;20(4):342–344. doi: 10.1038/nm.3522. [DOI] [PubMed] [Google Scholar]
- 13.Caiado F, Silva-Santos B, Norell H. Intra-tumour heterogeneity - going beyond genetics. FEBS J. 2016;283(12):2245–2258. doi: 10.1111/febs.13705. [DOI] [PubMed] [Google Scholar]
- 14.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2(10):881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 18.Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys Acta Rev Cancer. 2017;1868(1):1–6. doi: 10.1016/j.bbcan.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Renner K, Singer K, Koehl GE, Geissler EK, Peter K, Siska PJ, Kreutz M. Metabolic Hallmarks of Tumor and Immune Cells in the Tumor Microenvironment. Front Immunol. 2017;8:248. doi: 10.3389/fimmu.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, Pogrebniak K, Sandoval J, Cassidy JW, Tufegdzic-Vidakovic A. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016;167(1):260–274. doi: 10.1016/j.cell.2016.08.041. (e222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh AJ, Cook RS, Sanders ME, Aurisicchio L, Ciliberto G, Arteaga CL, Skala MC. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 2014;74(18):5184–5194. doi: 10.1158/0008-5472.CAN-14-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, MS Spector Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1-2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, Balgobind AV, Wind K, Gracanin A, Begthel H. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell. 2018;172(1-2):373–386. doi: 10.1016/j.cell.2017.11.010. (e310) [DOI] [PubMed] [Google Scholar]
- 24.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359(6378):920–926. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majety M, Pradel LP, Gies M, Ries CH. Fibroblasts Influence Survival and Therapeutic Response in a 3D Co-Culture Model. PLoS One. 2015;10(6) doi: 10.1371/journal.pone.0127948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah AT, Diggins KE, Walsh AJ, Irish JM, Skala MC. In Vivo Autofluorescence Imaging of Tumor Heterogeneity in Response to Treatment. Neoplasia. 2015;17(12):862–870. doi: 10.1016/j.neo.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh AJ, Cook RS, Skala MC. Functional Optical Imaging of Primary Human Tumor Organoids: Development of a Personalized Drug Screen. J Nucl Med. 2017;58(9):1367–1372. doi: 10.2967/jnumed.117.192534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgakoudi I, Quinn KP. Optical Imaging Using Endogenous Contrast to Assess Metabolic State. Annu Rev Biomed Eng. 2012;14:351–367. doi: 10.1146/annurev-bioeng-071811-150108. [DOI] [PubMed] [Google Scholar]
- 30.Chance B, Schoener B, Oshino R, Itshak F, Nakase Y. Oxidation-reduction ratio studies of mitochondria in freeze-trapped samples. NADH and flavoprotein fluorescence signals. J Biol Chem. 1979;254(11):4764–4771. [PubMed] [Google Scholar]
- 31.Walsh A, Cook RS, Rexer B, Arteaga CL, Skala MC. Optical imaging of metabolism in HER2 overexpressing breast cancer cells. Biomed Opt Express. 2012;3(1):75–85. doi: 10.1364/BOE.3.000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakowicz JR. 2nd ed. Kluwer Academic/Plenum; New York: 1999. Principles of Fluorescence Spectroscopy; p. 698. xxiii. [Google Scholar]
- 33.Bird DK, Yan L, Vrotsos KM, Eliceiri KW, Vaughan EM, Keely PJ, White JG, Ramanujam N. Metabolic mapping of MCF10A human breast cells via multiphoton fluorescence lifetime imaging of the coenzyme NADH. Cancer Res. 2005;65(19):8766–8773. doi: 10.1158/0008-5472.CAN-04-3922. [DOI] [PubMed] [Google Scholar]
- 34.Sharick JT, Favreau PF, Gillette AA, Sdao SM, Merrins MJ, Skala MC. Protein-bound NAD(P)H Lifetime is Sensitive to Multiple Fates of Glucose Carbon. Sci Rep. 2018;8(1):5456. doi: 10.1038/s41598-018-23691-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shah AT, Heaster TM, Skala MC. Metabolic Imaging of Head and Neck Cancer Organoids. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsh AJ, Castellanos JA, Nagathihalli NS, Merchant NB, Skala MC. Optical Imaging of Drug-Induced Metabolism Changes in Murine and Human Pancreatic Cancer Organoids Reveals Heterogeneous Drug Response. Pancreas. 2016;45(6):863–869. doi: 10.1097/MPA.0000000000000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Disselhorst JA, Brom M, Laverman P, Slump CH, Boerman OC, Oyen WJ, Gotthardt M, Visser EP. Image-quality assessment for several positron emitters using the NEMA NU 4-2008 standards in the Siemens Inveon small-animal PET scanner. J Nucl Med. 2010;51(4):610–617. doi: 10.2967/jnumed.109.068858. [DOI] [PubMed] [Google Scholar]
- 39.Skala MC, Riching KM, Gendron-Fitzpatrick A, Eickhoff J, Eliceiri KW, White JG, Ramanujam N. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc Natl Acad Sci U S A. 2007;104(49):19494–19499. doi: 10.1073/pnas.0708425104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh AJ, Cook RS, Manning HC, Hicks DJ, Lafontant A, Arteaga CL, Skala MC. Optical metabolic imaging identifies glycolytic levels, subtypes, and early-treatment response in breast cancer. Cancer Res. 2013;73(20):6164–6174. doi: 10.1158/0008-5472.CAN-13-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergmann A. Becker & Hickl GmbH; 2003. SPCImage: data analysis software for fluorescence lifetime imaging microscopy. available on www becker-hickl com. [Google Scholar]
- 43.Nakashima N, Yoshihara K, Tanaka F, Yagi K. Picosecond fluorescence lifetime of the coenzyme of D-amino acid oxidase. J Biol Chem. 1980;255(11):5261–5263. [PubMed] [Google Scholar]
- 44.Walsh AJ, Skala MC. An automated image processing routine for segmentation of cell cytoplasms in high-resolution autofluorescence images. SPIE Proc. 2014;8948 [Google Scholar]
- 45.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19(6):716–723. [Google Scholar]
- 47.Walsh AJ, Skala MC. Optical metabolic imaging quantifies heterogeneous cell populations. Biomed Opt Express. 2015;6(2):559–573. doi: 10.1364/BOE.6.000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almendro V, Kim HJ, Cheng YK, Gonen M, Itzkovitz S, Argani P, van Oudenaarden A, Sukumar S, Michor F, Polyak K. Genetic and phenotypic diversity in breast tumor metastases. Cancer Res. 2014;74(5):1338–1348. doi: 10.1158/0008-5472.CAN-13-2357-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mansfield JR, Hoyt C, Levenson RM. Visualization of microscopy-based spectral imaging data from multi-label tissue sections. Curr Protoc Mol Biol. 2008;14:19. doi: 10.1002/0471142727.mb1419s84. Chapter 14. (Unit) [DOI] [PubMed] [Google Scholar]
- 50.Glass GV. Primary, secondary, and meta-analysis of research. Educ Res. 1976;5(10):3–8. [Google Scholar]
- 51.Sharick JT, Walsh CM, Sprackling CM, Pasch CA, Parikh AA, Matkowskyj KA, Deming DA, Skala MC. Optical Metabolic Imaging of Heterogeneous Drug Response in Pancreatic Cancer Patient Organoids. bioRxiv. 2019;542167 [Google Scholar]
- 52.Sawilowsky SS. New effect size rules of thumb. J Mod Appl Stat Methods. 2009;8(2):597–599. [Google Scholar]
- 53.DeNardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, Gallagher WM, Wadhwani N, Keil SD, Junaid SA. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sai J, Owens P, Novitskiy SV, Hawkins OE, Vilgelm AE, Yang J, Sobolik T, Lavender N, Johnson AC, McClain C. PI3K Inhibition Reduces Mammary Tumor Growth and Facilitates Antitumor Immunity and Anti-PD1 Responses. Clin Cancer Res. 2017;23(13):3371–3384. doi: 10.1158/1078-0432.CCR-16-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cook RS, Garrett JT, Sanchez V, Stanford JC, Young C, Chakrabarty A, Rinehart C, Y Zhang Y Wu, Greenberger L. ErbB3 ablation impairs PI3K/Akt-dependent mammary tumorigenesis. Cancer Res. 2011;71(11):3941–3951. doi: 10.1158/0008-5472.CAN-10-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webster MA, Hutchinson JN, Rauh MJ, Muthuswamy SK, Anton M, Tortorice CG, Cardiff RD, Graham FL, Hassell JA, Muller WJ. Requirement for both Shc and phosphatidylinositol 3' kinase signaling pathways in polyomavirus middle T-mediated mammary tumorigenesis. Mol Cell Biol. 1998;18(4):2344–2359. doi: 10.1128/mcb.18.4.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee LH, Yang H, Bigras G. Current breast cancer proliferative markers correlate variably based on decoupled duration of cell cycle phases. Sci Rep. 2014;4:5122. doi: 10.1038/srep05122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15(12):747–756. doi: 10.1038/nrc4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 60.Andersen JL, Kornbluth S. The tangled circuitry of metabolism and apoptosis. Mol Cell. 2013;49(3):399–410. doi: 10.1016/j.molcel.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vander Heiden MG, Lunt SY, Dayton TL, Fiske BP, Israelsen WJ, Mattaini KR, Vokes NI, Stephanopoulos G, Cantley LC, Metallo CM. Metabolic pathway alterations that support cell proliferation. Cold Spring Harb Symp Quant Biol. 2011;76:325–334. doi: 10.1101/sqb.2012.76.010900. [DOI] [PubMed] [Google Scholar]
- 62.Gentric G, Mieulet V, Mechta-Grigoriou F. Heterogeneity in Cancer Metabolism: New Concepts in an Old Field. Antioxid Redox Signal. 2017;26(9):462–485. doi: 10.1089/ars.2016.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spitzer MH, Nolan GP. Mass Cytometry: Single Cells, Many Features. Cell. 2016;165(4):780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2018;144(3):782–793. doi: 10.1039/c8an01581c. [DOI] [PubMed] [Google Scholar]
- 66.O'Neil RG, Wu L, Mullani N. Uptake of a fluorescent deoxyglucose analog (2-NBDG) in tumor cells. Mol Imaging Biol. 2005;7(6):388–392. doi: 10.1007/s11307-005-0011-6. [DOI] [PubMed] [Google Scholar]
- 67.Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133(26):10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poburko D, Santo-Domingo J, Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem. 2011;286(13):11672–11684. doi: 10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howard SS, Straub A, Horton N, Kobat D, Xu C. Frequency Multiplexed In Vivo Multiphoton Phosphorescence Lifetime Microscopy. Nat Photonics. 2013;7(1):33–37. doi: 10.1038/nphoton.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14(4):545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures