Abstract
Background
In developed countries, global developmental disorders are encountered in approximately 1% of all children. The causes are manifold, and no exogenous cause can be identified in about half of the affected children. The parallel investigation of the coding sequences of all genes of the affected individual (whole exome sequencing, WES) has developed into a successful diagnostic method for identifying the cause of the problem. It is not yet clear, however, when WES should best be used in routine clinical practice in order to exploit the potential of this method to the fullest.
Methods
In an interdisciplinary study, we carried out standardized clinical phenotyping and a systematic genetic analysis (WES of the index patient and his or her parents, so-called trio WES) in 50 children with developmental disturbances of unclear etiology and with nonspecific neurological manifestations.
Results
In 21 children (42% of the collective), we were able to identify the cause of the disorder by demonstrating a mutation in a gene known to be associated with disease. Three of these children subsequently underwent specific treatment. In 22 other children (44%), we detected possibly etiological changes in candidate genes not currently known to be associated with human disease.
Conclusion
Our detection rate of at least 42% is high in comparison with the results obtained in other studies from Germany and other countries to date and implies that WES can be used to good effect as a differential diagnostic tool in pediatric neurology. WES should be carried out in both the index patient and his or her parents (trio-WES) and accompanied by close interdisciplinary collaboration of human geneticists and pediatricians, by comprehensive and targeted phenotyping (also after the diagnosis is established), and by the meticulous evaluation of all gene variants.
Divergence from the expected psychomotor development (global developmental delay) is observed in around 1% of children in the industrialized countries (1, 2). Global developmental delay thus accounts for a considerable proportion of all cases of neuropediatric illness, although there are no reliable statistical data on the prevalence of neurological diseases in childhood. The symptoms of global developmental disorders are often unspecific, so that in many cases no precise diagnosis is possible. Establishment of the diagnosis is, however, a necessary precondition for initiating any disease-specific treatment that may be available, drawing up an individualized support and prevention program, assessment of the developmental prognosis, and accurate estimation of the risk of similar disorders in the patient’s siblings or other family members.
There are multiple different factors that may be responsible for developmental delay, and in around half of the children affected no causative exogenous factor is identified (3). Probably most of these cases are of genetic origin (3– 9). Genetic techniques therefore play a crucial role in identifying the cause and pinpointing the diagnosis in this group of patients. In the past, molecular genetic tests were extremely laborious, usually proceeding “gene by gene.” However, the recently introduced “next-generation” sequencing (NGS) enables analysis of large numbers of genes (right up to the whole human genome) in a short amount of time at reasonable cost (10– 12) (table 1).
Table 1. Comparison of sequencing techniques.
| Next-generation sequencing (NGS) | Single-gene sequencing | |||
|
Whole-genome sequencing (WGS) |
Whole-exome sequencing (WES) |
Panel diagnostics | Sanger sequencing | |
|
Portion of genetic material analyzed |
Entire genome | All known exons, individual regulatory sequences |
Exons of selected genes | Exons of a single selected gene |
| Scope of analysis | ˜3 billion base pairs = 3000 Mb |
˜1% of the genome + regulatory sequences + miRNA = ˜50 Mb |
Depending on the genes/ exons selected, several Kb to a few Mb |
Depending on the gene/exon selected, a few Kb |
|
Number of variants identified |
> 3 000 000 | 15 000–40 000 | Few or none | Few or none |
| Detection | Point mutations, indels, numerical and structural chromosome variants |
Point mutations, indels | Variants in the selected genes | Variants in the selected gene/ exon |
| Incidental findings | Incidental findings | No incidental findings | No incidental findings | |
| New disease genes | New disease genes | No new disease genes | No new disease genes | |
Kb, Kilobase (1 Kb = 1000 base pairs); Mb, megabase (1 Mb = 1 000 000 base pairs); miRNA, microRNA; indels, insertions/deletions
The ethical aspects of such all-embracing genetic analysis techniques have been and continue to be widely discussed (13– 16). Particularly intensively debated topics are the explanation of and consent to genetic analysis, storage of and access to genetic data, and how to deal with incidental findings. The latter are genetic variants that happen to be identified in the course of NGS-based analyses. They are not related to the index patient’s illness, but mean there is an increased likelihood of a second, independent disease in the patient (and possibly his/her relatives). Country-specific and international guidelines and regulations vary widely in their recommendations on how to deal with incidental observations, and the debate is probably far from over. There is unanimity, however, on the need actively to include patients and guardians in the discussion of these questions.
Despite this, NGS methods have become established in routine genetic diagnosis. The technique generally used in Germany is “panel diagnostics,” in which a defined number of genes are investigated depending on the disease of interest. In highly heterogeneous illnesses (e.g., unspecific childhood developmental disorders) with hundreds of associated genes it does not make sense to limit the amount of genes to be analyzed. Patients affected by such diseases should be offered analysis of their entire genetic coding material (i.e., all genomic regions that are translated into proteins = all exons of the circa 20 000 human genes), known as whole-exome sequencing (WES).
Numerous studies from various countries have shown that the causes of developmental disorders and neurological illnesses in childhood can often be uncovered by means of WES (8, 17– 25). The success rate of WES has been reported as 25 to 68% and is particularly high when:
The exome sequencing is extended to parents (trio exome sequencing) and/or other family members (family exome sequencing) (17, 26)
The analysis of the exome data embraces all known genes, not only those associated with a human illness at the given point in time (27, 28).
It has not yet been adequately investigated how exome sequencing can be effectively integrated into concrete clinical routine (5).
Many questions remain unanswered. These include what patients should be offered WES, to what extent it should be extended to the patient’s relatives, and in what clinical and analytical context it should take place.
To address these questions, we carried out a single-center pilot study of 50 children with undiagnosed developmental delay and neurological illness presumed to be of genetic origin. Thorough clinical examination, biochemical analyses, electroencephalography, and diagnostic imaging were accompanied by trio WES.
The aim of this study was to establish a practical procedure for phenotyping and genotyping that would achieve a high rate of diagnosis for unspecific neurological illnesses of childhood and could be accommodated into the clinical routine.
Method
Study design
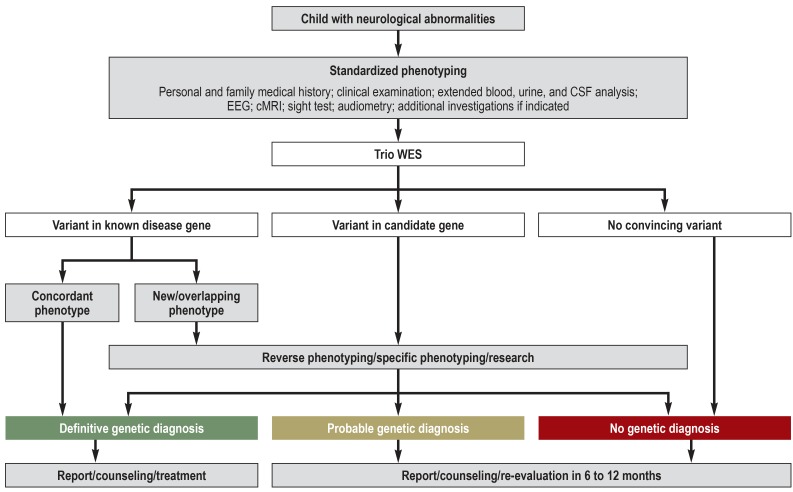
The study was a joint project of the Department of Pediatrics and the Institute of Human Genetics at the University Medical Center Hamburg-Eppendorf, Germany. Over a period of 17 months, a total of 50 consecutive children with undiagnosed neurological illnesses underwent a standardized assessment program in the context of a complex neuropediatric diagnostic work-up (German surgical procedure code [OPS] 1–942) (Figure 1, eMethods). The inclusion criteria were as follows:
Figure 1.
Study protocol including evaluation algorithm for genetic variants
cMRI, Cranial magnetic resonance imaging; CSF, cerebrospinal fluid; EEG, electroencephalography; trio WES, whole-exome sequencing of index patient and parents
Neurological symptoms (e.g., global developmental delay, ataxia, seizures)
Suspected genetic etiology (i.e., no sign of serious perinatal complications, infection, injury, other exogenic factors)
No specific provisional diagnosis
Signed consent from the parents, following appropriate explanation, for exhaustive investigations including trio WES
The study was approved by the ethics committee of the Hamburg Medical Association (project number PV3802).
Phenotype documentation
The comprehensive assessment program always included detailed questioning about the medical history of the patient and their family, extensive clinical evaluation by a neuropediatrician and a clinical geneticist, wide-ranging investigation of blood, urine, and cerebrospinal fluid parameters, diagnostic imaging (1.5-T or 3-T cranial magnetic resonance imaging (cMRI), and electroencephalography (EEG). Depending on the clinical findings, other tests were added in individual patients, for example further diagnostic imaging procedures (e.g., MR spectroscopy), audiometry, echocardiography, or ophthalmological examination. We documented the reported medical history and the clinical data systematically using the Phenomizer software (29), which is based on Human Phenotype Ontology (HPO) (30). The degree of cognitive and/or physical impairment was classified according to Zhang et al. (31) (etable 1).
eTable 1. Classification of mental retardation (MR) according to Zhang et al. (e5).
| Level |
Degree of disability(IQ) |
Adults | Children |
| 0 | None | No impairment | Development appropriate for age |
| 1 | Borderline (<70) | Attendance of regular schools for several years; needs little support | Milestones reached at normal ages; slight developmental delay apparent in first years at school |
| 2 | Very mild (<65) | Attendance of regular schools for a few years; needs a lot of support; simple skills in reading, writing, and numeracy | Milestones reached at normal ages for the first few years; slight developmental delay apparent from age of 2 to 3 years |
| 3 | Mild (<50) | Good language comprehension, even long sentences are understood; limited skills in reading, writing, and numeracy | Achievement of milestones delayed by a few months; apparent from the age of 1 to 2 years |
| 4 | Moderate (<35) | Good language comprehension; speaks short sentences, makes a lot of gestures | Achievement of milestones delayed by several months; apparent from the 2nd year of life |
| 5 | Severe (<20) | Understands simple common sentences; speaks two-to three word sentences, makes a lot of gestures; can walk | Achievement of milestones delayed by several months to a year; apparent from the 1st year of life |
| 6 | Very severe (<10) | Understands single words; speaks single words or not at all; can walk unsteadily/with assistance | Developmental delay by several years; becomes apparent in first 6 months of life |
| 7 | Profound | No or only slight reaction; can sit and stand with assistance, walks rarely or not at all | No unassisted sitting at age of 5 years |
IQ, Intelligence quotient
Genotype documentation
The genetic data were obtained from WES of EDTA blood from each index patient and their biological parents (trio WES). The WES procedure is described in the eMethods. Closer attention was paid to genetic variants which were very rarely identified in the population (minor alleles frequency [MAF]; occurrence of the rarer allele in the population <0.01%) and which, according to several different bioinformatic prediction algorithms, impact negatively on gene function (functionally relevant variants). The variants filtered out in this way were mostly nonsynonymous point mutations (variants that changed the amino acid sequence in the coded protein), losses/gains of one or several base pairs (indels) or losses/gains of submicroscopic chromosome regions (microdeletions/microduplications; copy number variations [CNVs]). These affected a known disease gene or a candidate gene. More detailed information on the WES procedure and the classification of variants and genes can be found in the eMethods.
Interdisciplinary interpretation of findings and reverse phenotyping
In each individual case, the members of a multidisciplinary team consisting at least of pediatricians and human geneticists discussed the findings and assessed the potential relevance of the genetic variants identified in the context of the medical history and the patient’s symptoms. Whenever necessary, other experts (e.g., neuroradiologists) were added to the panel. In many cases the team ordered additional specific clinical investigations (reverse phenotyping) to enable more detailed assessment of a genetic variant. The results of reverse phenotyping were then interpreted by the assembled team.
Statistical analysis
Descriptive statistical analyses were carried out using the Statistical Package for the Social Sciences (SPSS, version 23).
Results
Study participants
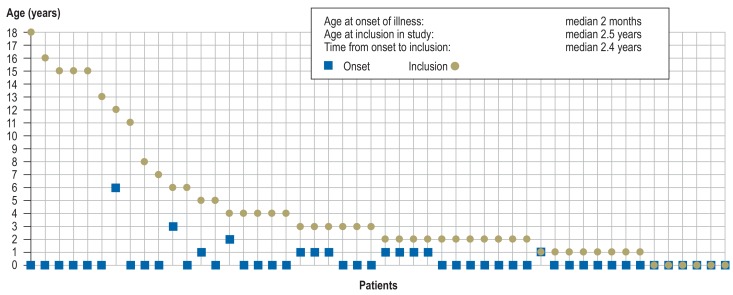
The median age of the 50 unrelated children (19 girls [38%], 31 boys [62%]) at study inclusion was 2.5 years (range 4 days to 18 years; Figure 2). Thirty-three (66%) of them had already undergone genetic testing (chromosome analysis, array-based comparative genomic hybridization [array CGH]), and/or single-gene sequencing. In no case had the results been abnormal (for details see the eMethods).
Figure 2.
Age at onset of illness (square) and age at inclusion in study (circle) for each of the 50 patients
Results of phenotyping
The first symptoms were observed at a median age of 2 months (0 days to 6 years; Figure 2). Sixteen patients (32%) had delayed development of motor functions and/or speech, six (12%) showed abdominal symptoms (e.g., omphalocele, esophageal stenosis/atresia, feeding difficulties), and five (10%) evinced ophthalmological abnormalities (e.g., congenital cataract, nystagmus, strabismus). A median 2.4 years (4 days to 18 years; Figure 2) elapsed between occurrence of the first symptoms and inclusion in the study.
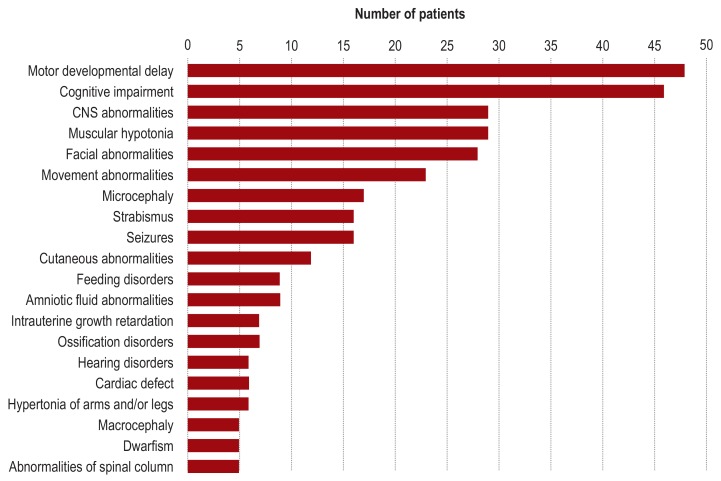
In line with the inclusion criteria, all 50 patients had neurological symptoms at the time of entry into the study. These comprised global developmental delay (88%), cognitive impairment without motor symptoms (4%), or motor symptoms without cognitive impairment (8%). As classified according to Zhang et al. (31), the developmental disorder was mild in 10% of patients, moderate in 32%, severe in 56%, and profound in one patient. Psychomotor development was categorized in 68% of patients as “gradually progressive,” in 6% as “stagnating,” and in 26% as “regressive.” At the time of clinical examination, body weight, body length, and head circumference were abnormal (?- 2 standard deviations [SD] or ? + 2 SD according to Kromeyer-Hauschild et al. [32]) in 13 (26%), 12 (24%), and 21 (42%) of patients, respectively. Thirty-six patients (72%) showed abnormalities of the head and neck (e.g., facial dysmorphism), 34 (68%) had abnormalities of the musculature (e.g., muscular hypotonia), and 28 (56%) exhibited ocular abnormalities (e.g., strabismus) (figure 3). Further details of the phenotyping findings are documented in the eMethods and in eTable 2.
Figure 3.
Clinical characterization of the study group according to the HPO system (HPO, Human Phenotype Ontology)
eTable 2. Findings of instrument-based diagnostic methods.
| Method | Number of patients with abnormalities |
| EEG | 20 of 47 (42.6%) |
| cMRI | 29 of 50 (58.0%) |
| Audiometry | 8 of 16 (50.0%) |
| Sight test | 23 of 37 (62.2%) |
cMRI, Cranial magnetic resonance imaging; EEG, electroencephalography
Results of genotyping
In 21 patients (42%) we found a pathogenic variant (mutation) in a disease gene known at the time of analysis or a known disease-related microdeletion (Table 2, eTable 3). Twelve (57.1%) of these 21 mutations were de novo events, i.e., they had arisen in the index patient. In nine index patients (42.9%) the mutations were biallelic (on both alleles of the gene affected), so that the inheritance was autosomal recessive. Strikingly, during the course of our study variants in three genes (CHAMP1, SSR4, and SON) initially classified as candidate genes were convincingly shown by newly published data to be mutations in new disease genes.
Table 2. Findings of whole-exome sequencing (WES).
| Number of patients | |
| Findings | 50 (100%) |
| – Mutation in a known disease gene | 21 (42.0%) |
| – Variant in a candidate gene | 22 (44.0%) |
| – No convincing variant | 7 (14.0%) |
| Characterization of the mutations in known disease genes | 21 (100%) |
| – Inheritance | |
| – De novo | 12 (57.1%) |
| – Autosomal recessive | 9 (42.9%) |
| – X-chromosomal | 0 (0%) |
| – Mutation type | |
| – Missense | 11 (52.4%) |
| – Nonsense | 3 (14.3%) |
| – Frameshift | 4 (19.0%) |
| – CNV | 3 (14.3%) |
CNV, Copy number variations
eTable 3. Patients with mutations in known disease genes or CNVs known to cause disease.
| Patient | Consanguinity of parents | Other family members with similar symptoms |
Phenotype (cardinal symptoms) |
OMIM disease gene | Variant | Mutation nucleotide | Mutation protein | Inheritance | Associated disease | OMIM disease number |
| a) Mutations in known disease genes with concordant phenotype | ||||||||||
| 1 | No | No | Severe global developmental delay, regression, dysmorphism | del 15q11q13 | CNV | chr.15: 23,684,690– 28,544,611x1 |
De novo | Angelman syndrome | # 105830 | |
| 2 | No | No | Severe global developmental delay, congenital cataract, microcephaly | COL4A1 | Missense | c.1973C>A | p.Gly658Val | De novo | BSVD | # 607595 |
| 3 | No | No | Mild global developmental delay, dystonia, gait disorder | SGCE | Nonsense | c.771_772delAT | p.Cys258Stop | De novo | DYT11 | # 159900 |
| 4 | No | No | Moderate global developmental delay, short stature, facial dysmorphism | ANKRD11 | Nonsense | c.4886G>C | p.Ser1629Stop | De novo | KBG syndrome | # 148050 |
| 5 | No | No | Severe global developmental delay, short stature, dysmorphism | KAT6A | Nonsense | c.1096G>A | p.Arg366Stop | De novo | MRD32 | # 616268 |
| 6 | No | No | Severe global developmental delay, spastic tetraplegic cerebral palsy, microcephaly | RNASEH2B | Missense | c.529G>A; c.529G>A |
p.Ala177Thr; p.Ala177Thr |
Autosomal recessive | AGS2 | # 610181 |
| 7 | Yes | No | Severe global developmental delay, regression, movement disorder, myelinization disorder | RNASEH2C | Missense | c.205G>A; c.205G>A |
p.Arg69Trp; p.Arg69Trp |
Autosomal recessive | AGS3 | # 610329 |
| 8 | No | No | Moderate global developmental delay, ataxia, stereotypes | MYT1L | Nonsense | c.223G>A | p.Arg75Stop | De novo | MRD39 | # 616521 |
| 9 | No | Yes | Severe global developmental delay, leukodystrophy, cerebellar hypoplasia | TREX1 | Missense | c.290G>A; c.290G>A |
p.Arg97His; p.Arg97His |
Autosomal recessive | AGS1 | # 225750 |
| 10 | No | Yes | Mild global developmental delay, regression, hypotonia | TH | Missense | c.1273C>T; c.1096G>T |
p.Glu425Lys; p.Leu366Met |
Autosomal recessive | Segawa syndrome | # 605407 |
| 11 | No | No | Severe global developmental delay, microcephaly, autism, stereotypes | MBD5 | CNV | chr.2: 149,130,689–149, 227,038 |
De novo | MRD1 | # 156200 | |
| 12 | No | No | Mild global developmental delay, neurodegenerative disorder | FA2H | Missense | c.1T>G | p.Met1Leu | Autosomal/ recessive | SPG35 | # 612319 |
| 13 | Yes | No | Moderate global developmental delay, regression, neuropathy, pyloric stenosis | ARSA | Missense | c.679G>C | p.Arg227Gly | Autosomal/ recessive | MLD | # 250100 |
| b) Mutations in known disease genes with extension of phenotype | ||||||||||
| 14 | No | No | Severe global developmental delay, severe encephalopathy, dystonia | GLE1 | Missense | c.1706G>A; c.1750C>T |
p.Arg569His; p.Arg584Trp |
Autosomal/ recessive | LCCS1, LAAHD | # 253310, # 611890 |
| 15 | Yes | Yes | Severe global developmental delay, tapetoretinal degeneration, coordination disorder | RPIA | Missense | c.627G>C; c.627G>C |
p.Trp209Cys; p.Trp209Cys |
Autosomal/ recessive | RPIA deficiency | # 608611 |
| 16 | No | No | Moderate delay in motor development, connective tissue weakness | IGHMBP2 | Missense | c.745G>A; c.61C>T |
p.Asp249Asn; p.Arg21Cys |
Autosomal/ recessive | DSMA1, CMT2S | # 604320, # 616155 |
| 17 | No | No | Severe global developmental delay, hypo‧tonia, hearing and vision deficiencies | CHD2 | Missense | c.1854A>T | p.Glu618Asp | De novo | EEOC | # 615369 |
| c) Mutations initially categorized as variants in candidate genes but reclassified as mutations in new disease genes in the course of the study | ||||||||||
| 18 | No | No | Severe global developmental delay, microcephaly, muscular hypotonia | CHAMP1 | Frameshift | c.1865_1866delAC | p.Asp622 Glufs*7 |
De novo | MRD40 | # 616579 |
| 19 | No | No | Moderate global developmental delay, short stature, deafness | del Xq28 (SSR4, PLXNB3, SRPK3, IDH3G) | CNV | chr.X: 153,034,617– 153,060,208 |
De novo | CDG1Y | # 300934 | |
| 20 | Yes | No | Severe global developmental delay, cardiac abnormalities, muscular hypotonia, facial dysmorphism | SON | Frameshift | c.268delC | p.Ser90Valfs*59 | De novo | ZTTK syndrome | # 617140 |
| 21 | No | No | Severe global developmental delay, cardiac abnormalities, epilepsy, renal cyst | SON | Frameshift | c.4055delC | p.Pro1352Glnfs*14 | De novo | ZTTK syndrome | # 617140 |
AGS2, Aicardi–Goutières syndrome 2; AGS3, Aicardi–Goutières syndrome 3; BSVD, brain small vessel disease with or without ocular anomalies; CDG1Y, ?congenital disorder of glycosylation type 1y; CMT2S, Charcot–Marie–Tooth disease, axonal, type 2S; CNV, copy number variations; DSMA1, spinal muscular atrophy, distal, autosomal recessive 1; DYT11, dystonia 11; EEOC, epileptic encephalopathy childhood onset; KBG syndrome, designation composed of the initials of the surnames of the first three patients described; LAAHD, lethal arthrogryposis with anterior horn cell disease; LCCS1, lethal congenital contracture syndrome 1; MLD, metachromatic leukodystrophy; MRD1, mental retardation, autosomal dominant 1; MRD32, mental retardation, autosomal dominant 32; MRD39, mental retardation, autosomal dominant 39; MRD40, mental retardation, autosomal dominant 40; OMIM, Online Mendelian Inheritance in Man; SPG35, spastic paraplegia, autosomal recessive 35; ZTTK syndrome, Zhu–Tokita–Takenouchi–Kim syndrome
In an additional 22 patients (44%) we identified a variant in candidate genes that probably caused illness. The disease association of these candidate genes has been or is currently being investigated in international cooperation projects. Since the conclusion of our work, two of these 22 candidate genes have been confirmed as disease genes and the research to date supports a disease association for some of the others.In only seven patients there (14%) were no abnormal genetic findings.
Consequences for clinical care
In 17 (81%) of the 21 patients with a mutation in a known disease gene, diagnosis resulted in recommendations to change/modify the clinical management. In 14 children (66.7%) the recommendations affected the prevention and monitoring program (e.g., the institution of new follow-up investigations or discontinuation of existing investigations), and in 13 patients (62%) the individual support measures (e.g., use of sign language or a talker, planning of future education) were involved. Moreover, three patients (3/21 = 14%; 3/50 = 6%) were offered specific treatment: In an 11-month-old girl with developmental regression, severe muscular hypotonia, and eye movement disorders, treatment with L-Dopa was initiated immediately after the detection of compound heterozygous mutations in the tyrosine hydroxylase gene (TH-Segawa syndrome, OMIM #605407). This led to swift regression of the neurological symptoms and to catch-up development over the course of time. In a 4-year-old boy with global developmental delay, dystonia, and gait abnormality, treatment with trihexyphenidyl was started after identification of a de novo mutation in SGCE (dystonia type 11, OMIM #159900). Follow-up showed decreased severity of the dystonic movement disorder and improved gait. Detection of a homozygous mutation in ARSA (metachromatic leukodystrophy, OMIM #250100) in a 3-year-old boy with peripheral neuropathy and leukodystrophy led to his inclusion in a treatment study (eTable 4 eTable 5).
eTable 4. Recommended management of treatment in the 21 patients with a genetic diagnosis.
| Patient |
Phenotype (cardinal symptoms) |
OMIM disease gene | Associated disease | Individually recommended measures |
| 1 | Severe global developmental delay, regression, dysmorphism | del 15q11q13 | Angelman syndrome | Preventive (EEG monitoring initiated), symptomatic (speech therapy modified) |
| 2 | Severe global developmental delay, congenital cataract, microcephaly | COL4A1 | BSVD | Preventive (ophthalmological monitoring modified, nephrological monitoring initiated) |
| 3 | Mild global developmental delay, dystonia, gait disorder | SGCE | DYT11 | Curative (trihexyphenidyl), symptomatic (physiotherapy modified) |
| 4 | Moderate global developmental delay, short stature, facial dysmorphism | ANKRD11 | KBG syndrome | None |
| 5 | Severe global developmental delay, short stature, dysmorphism | KAT6A | MRD32 | Preventive (EEG, cardiological, ophthalmological, and orthopedic monitoring initiated), symptomatic (ergotherapy and speech ‧therapy modified) |
| 6 | Severe global developmental delay, spastic tetraplegic cerebral palsy, microcephaly | RNASEH2B | AGS2 | Preventive (cardiological monitoring discontinued, ‧orthopedic and ophthalmological monitoring initiated), symptomatic ‧(physiotherapy ‧modified) |
| 7 | Severe global developmental delay, regression, movement disorder, myelinization disorder | RNASEH2C | AGS3 | Preventive (neurological and ophthalmological monitoring ‧initiated), symptomatic (physiotherapy adjusted) |
| 8 | Moderate global developmental delay, ataxia, stereotypes | MYT1L | MRD39 | None |
| 9 | Severe global developmental delay, leukodystrophy, cerebellar hypotrophy | TREX1 | AGS1 | None |
| 10 | Mild global developmental delay, regression, hypotonia | TH | Segawa syndrome | Curative (L-Dopa), preventive (EEG monitoring initiated), symptomatic (physiotherapy adjusted, ergotherapy initiated) |
| 11 | Severe global developmental delay, microcephaly, autism, stereotypes | MBD5 | MRD1 | Preventive (EEG monitoring initiated) |
| 12 | Mild global developmental delay, neurodegenerative disorder | FA2H | SPG35 | Preventive (EEG-, urological, ophthalmological, and orthopedic monitoring initiated), symptomatic (physiotherapy modified, baclofen/Botox discussed) |
| 13 | Moderate global developmental delay, regression, neuropathy, pyloric stenosis | ARSA | MLD | Curative (inclusion in an MLD study), preventive (referral for specialist consultation) |
| 14 | Severe global developmental delay, severe encephalopathy, dystonia | GLE1 | LCCS1, LAAHD |
None |
| 15 | Severe global developmental delay, tapetoretinal degeneration, coordination disorder | RPIA | RPIA defiziency | Preventive (EEG monitoring initiated), symptomatic (physiotherapy modified) |
| 16 | Moderate delay in motor development, connective tissue weakness | IGHMBP2 | DSMA1, CMT2S |
Symptomatic (physiotherapy modified) |
| 17 | Severe global developmental delay, hypotonia, hearing and vision deficiencies | CHD2 | EEOC | Preventive (EEG monitoring initiated) |
| 18 | Severe global developmental delay, microcephaly, muscular hypotonia | CHAMP1 | MRD40 | Symptomatic (speech therapy modified) |
| 19 | Moderate global developmental delay, short stature, deafness |
del Xq28 (SSR4, PLXNB3, SRPK3, IDH3G) |
CDG1Y | Preventive (coagulation and EEG monitoring, gastroenterological referral initiated), symptomatic (physiotherapy modified) |
| 20 | Severe global developmental delay, cardiac abnormalities, muscular hypotonia, facial dysmorphism | SON | ZTTK syndrome | Preventive (nephrological and dental monitoring, hearing test initiated; avoidance of radiography), symptomatic (speech therapy modified) |
| 21 | Severe global developmental delay, cardiac abnormalities, epilepsy, renal cyst | SON | ZTTK syndrome | Preventive (nephrological and dental monitoring, hearing test initiated; avoidance of radiography), symptomatic (ergotherapy adjusted, speech therapy initiated) |
AGS2, Aicardi–Goutières syndrome 2; AGS3, Aicardi–Goutières syndrome 3; BSVD, brain small vessel disease with or without ocular anomalies; CDG1Y, congenital disorder of glycosylation type 1y; CMT2S, Charcot–Marie–Tooth disease, axonal, type 2S; DSMA1, spinal muscular atrophy, distal, autosomal recessive 1; DYT11, dystonia 11; EEOC, epileptic encephalopathy childhood onset; KBG syndrome, designation composed of the initials of the surnames of the first three patients described; LAAHD, lethal arthrogryposis with anterior horn cell disease; LCCS1, lethal congenital contracture syndrome 1; MLD, metachromatic leukodystrophy; MRD1, mental retardation, autosomal dominant 1; MRD32, mental retardation, autosomal dominant 32; MRD39, mental retardation, autosomal dominant 39; MRD40, mental retardation, autosomal dominant 40; OMIM, Online Mendelian Inheritance in Man; SPG35, spastic paraplegia, autosomal recessive 35; ZTTK syndrome, Zhu–Tokita–Takenouchi–Kim syndrome
eTable 5. Overview of recommendations for management of treatment in the 21 patients with a genetic diagnosis.
| Number of patients | |
| Change in management | 17 (81.0%) |
| Preventive measures: | 14 (66.7%) |
| − Initiation of specific preventive measures recommended (e.g., monitoring [EEG, cardiological, ophthalmological, orthopedic], avoidance of radiography) |
14 (66.7%) |
| − Initiation of specific preventive measures recommended (ophthalmological monitoring) |
1 (4.8%) |
| − Discontinuation of specific preventive measures recommended (cardiological monitoring) |
1 (4.8%) |
| Symptomatic measures: | 13 (61.9%) |
| − Initiation of specific symptomatic measures recommended (ergotherapy, speech therapy) |
2 (9.5%) |
| − Modification of specific symptomatic measures recommended (speech therapy, ergotherapy, physiotherapy) |
13 (61.9%) |
| Curative measures: | 3 (14.3%) |
| − Initiation of specific curative measures recommended (trihexyphenidyl, L-Dopa, inclusion in MLD study) |
3 (14.3%) |
| No change in management | 4 (19.0%) |
EEG, Electroencephalography; L-Dopa, L-3,4-dihydroxyphenylalanine;
MLD, metachromatic leukodystrophy
Systematic documentation and comparative statistical evaluation of the implementation of the treatment recommendations and their success were not among the aims of our study and were in any case not compatible with the selected study design.
Discussion
Our standardized search for the cause of undiagnosed developmental delay and neurological illnesses in a pediatric collective revealed mutations in known disease genes compatible with the phenotype in 21 (42%) of 50 patients. This involved wide-ranging clinical, biochemical, and instrument-based investigations together with trio WES. Our rate of identification of the cause of developmental disorders in a general pediatric cohort is towards the high end of the spectrum of results from international research: comparable studies from, among other countries, Israel, the UK, and the USA report diagnosis rates of 25 to 49% (8, 20, 22, 24). Our findings emphasize that pediatric patients with unspecific neurological symptoms can often be helped by means of the powerful genetic diagnostic technique of exome sequencing.
Individual out-of-hospital exome sequencing is covered by German health insurance under EBM fee schedule code number 11514 (remuneration currently circa €3300). However, the necessary individual approval by the health insurance provider is rarely forthcoming. In the hospital setting, genetic diagnosis, including WES, can be carried out under OPS code 1-942.
The mutations affected 20 different disease genes in the 21 confidently diagnosed patients, reflecting the extreme heterogeneity of childhood developmental disorders and neuropediatric disorders and underlining the necessity of systematic genetic analyses (WES) in this group. Our experience leads us to recommend trio WES, because confident and effective identification of de novo (newly arising) mutations is facilitated by comparing the genetic variants in the index patient with those found in the patient’s parents. In fact, de novo mutations comprise a high proportion of the genetic causes of disease: in our study 57% of the mutations arose de novo, and in the DDD study the figure was as high as 65% (9).
Analysis of the genetic data was not limited to known disease genes, and indeed a high number of probably disease-relevant variants were found in various candidate genes. Even before the conclusion of the study, three genes we had originally categorized as candidate genes (CHAMP1, SSR4, SON) were reclassified as “new disease genes” (33, 34). After completion of the study there were 22 index patients (44%) with a probably disease-relevant variant in a candidate gene. We are convinced, on the basis of the trio WES data, bioinformatic predictions, published research, and data from cooperating study groups, that in the course of time most of these candidate genes will be confirmed as disease relevant and reclassified as new disease genes. As of November 2018, two of them (DHX30, ANO3) had already been reclassified, increasing our rate of diagnosis to 46% (23 of the 50 index patients).
Conclusion
This pilot study has confirmed the high diagnostic potential of exome sequencing in a heterogeneous cohort of neuropediatric patients whose unspecific symptoms gave no clues to the diagnosis. The high rate of successful diagnosis was achieved by means of a interdisciplinary clinical–genetic approach, with:
Detailed phenotyping, followed, whenever necessary, by specific investigations (reverse phenotyping)
Exome sequencing of the index patient and their parents (trio WES)
Comprehensive evaluation of the entire data obtained by exome sequencing
Interdisciplinary interpretation of the clinical and genetic data
Close cooperation and data sharing with physicians and study groups in Germany and other countries
Under these conditions exome sequencing is a highly powerful genetic investigation technique that can be used in patients with disease of probable genetic origin in whom no specific diagnosis is suspected. In pinpointing the diagnosis, it opens the door to the specific treatments that are increasingly becoming available for genetic diseases.
Limitations
One limitation of our study is the relatively small number of patients. However, data from independent international studies seem to show that the central results can be reproduced in a larger cohort of patients. It must be borne in mind that the detection rates relate to the data available at the time the study was carried out.
Supplementary Material
eMETHODS
Method
Study design
The 50 index patients were admitted consecutively to the Department of Pediatrics, University Medical Center Hamburg-Eppendorf for complex neuropediatric diagnostic work-up (German surgical procedure code OPS 1–942). All patients underwent the following standardized program of investigations:
The diagnostic procedures, including trio WES, were billed as “complex neuropediatric diagnostic work-up with extended genetic work-up” (OPS 1–942.2) or “complex neuropediatric diagnostic work-up with neurometabolic laboratory testing and/or infectiological/autoimmune inflammatory laboratory testing with extended genetic work-up” (OPS 1–942.3).
Documentation of genotype—WES technique
The exome sequencing was performed at the Institute of Human Genetics, Helmholtz Center Munich. All known coding DNA fragments from a patient and their biological parents were enriched with the SureSelect Human All Exon 50Mb V5 Kit (Agilent, Santa Clara, CA, USA) and sequenced using the HiSeq2500 system (Illumina, San Diego, CA, USA). The reads were assigned to the reference genome “human genome assembly hg19” (UCSC Genome Browser) with the aid of the Burrows-Wheeler Aligner (BWA, v.0.5.87.5). The detection of genetic variants (deviations from the norm) was achieved by means of SAMtools (v0.1.18), PINDEL (v 0.2.4t), and ExomeDepth (v1.0.0). In this way 95 to 99% of the exome sequences were covered at least 20-fold.
Documentation of genotype—classification of variants and genes
The genetic data generated by exome sequencing were analyzed at the Institute of Human Genetics, University Medical Center Hamburg-Eppendorf.
The disease relevance of a given identified variant was evaluated with the aid of a number of variables: MAF, assessment by bioinformatic prediction programs, comparison with databases (e.g., Database of Exome Aggregation Consortium, ExAC [e1]; Online Mendelian Inheritance of Man, OMIM [e2]), and current knowledge of the coded protein and its function.
A variant in a gene was classified as causing disease (i.e., as a mutation) if:
We defined known disease genes as those whose association with disease had been demonstrated by published clinical–genetic and/or functional data.
A variant in a gene was classified as probably causing disease if:
A gene was classified as a candidate gene if:
Phenotyping results
Sociodemographic data
For 49 patients (98%) data were available on the consanguinity of the parents. The parents’ ethnicity was documented for 45 patients (90%). The parents came from 12 different countries, including Germany (23 [46%]), Turkey (5 [10%]), Iran, Afghanistan, and Kosovo (each 3 [6%]). Nine patients (18%) had consanguineous parents. The consanguineous couples were from Germany (1 [2%]), Turkey, Iran, Afghanistan (each 2 [4%]), Egypt, and Pakistan (each 1 [2%]).
Perinatal abnormalities
Abnormal features of gestation or delivery had been reported for 18 patients (36%): an abnormal amount of amniotic fluid in nine cases (18%), vaginal bleeding in three (6%), intrauterine growth retardation (IUGR) in seven (14%), and premature delivery in five (10%). Other complications (e.g., placental insufficiency, gestational diabetes) were documented in individual cases. Nine patients showed abnormal measurements at birth: One patient was small for gestational age (SGA) (birth weight, length, and head circumference ?-2 standard deviations [SD] according to Voigt et al. [e4]), and isolated microcephaly was reported for two patients. In five patients (10%) birth weight and/or body length were ?-2 SD, and in one patient birth weight and head circumference were >+ 2 SD.
Imaging, functional analysis, and metabolic analysis
The EEG results were abnormal in 20 (43%) of 47 patients, for example “pathological waking EEG with generalized susceptibility to seizures and activation by hyperventilation and photostimulation” or “pathological waking EEG with inconstant bilateral temporal foci and left temporal susceptibility to seizures.” The findings on cMRI were abnormal in 29 (58%) of 50 patients, for instance “midline deformity with hypoplasia of the corpus callosum and absent septum pellucidum. Prominent cerebellar tonsils, but currently no Chiari I. Residual hemorrhages” or “suspicion of small right frontal subdural hematoma with no appreciable space-occupying effect. Increasing diffuse leptomeningeal and subarachnoid contrast-enhancing substrate over time, primarily compatible with a progressive angiomatous process with leptomeningeal accentuation.” Ophthalmological examination revealed abnormalities in 23 (62%) of 37 children, e.g., “left: optic nerve hypoplasia,” “right/left: mild tortuosity of veins,” or “right/left: hyperopia, astigmatism (etable 2). Blood gas analysis (BGA), performed in 45 index patients, revealed elevated lactate concentrations in 13 cases (29%) and striking base excess in nine patients (20%) (decreased levels in two children, elevated levels in seven). Of the 49 children who underwent cerebrospinal fluid analysis, one (2%) showed an elevated glucose concentration, 12 (25%) had abnormal lactate levels (low in two children, high in 10), and three children (6%) displayed elevated concentrations of protein. The majority of patients underwent extended metabolic screening, including analysis of amino acids in plasma and cerebrospinal fluid together with determination of acylcarnitines in dried blood spots and measurement of organic acids in urine. In no patient was there a constellation of biochemical findings pointing to a specific disease.
Genetic analysis had been carried out previously in 33 children (66%). Four had undergone chromosome analysis; five, array-CGH analysis; 10, chromosome analysis plus array-CGH analysis; five, individual or panel genetic analysis; and nine, individual or panel genetic analysis plus chromosome analysis or array-CGH analysis. None of these earlier analyses had revealed any abnormalities (e.g., chromosome analysis; normal male karyotype 46,XY; array-CGH analysis: no abnormal findings; ARX genetic analysis: no abnormal findings).
At least detailed questioning regarding the medical history of the index patient and family members
Extended clinical assessment
Wide-ranging examinations of blood, urine, and cerebrospinal fluid parameters
Cranial magnetic resonance imaging (cMRI) at 1.5 T or 3 T
Electroencephalography (EEG)
Trio WES
It affected a known disease gene
The very same variant had previously been described as causing disease, or the variant was comparable in type with known disease-causing variants in the same gene
There was overlap in phenotype between our patient and the published patients with causative mutations in the same gene
It affected a candidate gene (see below)
More than one prediction program (Polyphen 2, SIFT and CADD) classified it as pathogenic
In the case of a de novo mutation, it was not listed in the ExAC database
Based on the trio WES results it was the only one of the patient’s genes in which a rare variant was found, or based on the trio WES results and bioinformatic prediction algorithms it was classified as probably causing disease
Previously published data (e.g., functional in-vivo or in-vitro studies) pointed to disease association in humans or there was, via Genematcher (3), contact to other study groups who had also detected variants in the same gene in patients with overlapping symptoms
Key Messages.
The achieved diagnosis rate of at least 42% underlines the efficiency of exome sequencing as a method for detection of the genetic causes of developmental delay and neurological illness in childhood.
Exploitation of the potential of exome analysis to the full involves comprehensive phenotyping of the patient, extension of exome analysis to the patient’s parents, and detailed analysis of the genetic data.
Exome analysis can, in the absence of a concrete provisional diagnosis, be used as the first step of the genetic diagnostic work-up in neuropediatric patients. In no way does it replace explicit phenotyping of the child affected.
Timely use of exome analysis enables early diagnosis and thus avoidance or termination of a diagnostic odyssey for the patient and their family, and also hastens the use of any specific treatment that may be available in the individual case.
The complex nature of this detailed genetic analysis, including the ethical aspects, must be explicitly discussed in advance with the patients or their guardians.
Acknowledgments
Translated from the original German by David Roseveare Acknowledgments
We are grateful to the patients who took part in this study and their families. We also thank all the clinicians who contributed data as well as the molecular geneticists, bioinformaticians, and laboratory technicians who lent us their assistance.
Compliance with ethical standards
We obtained signed consent from each study participant or, if applicable, their parents. All procedures in studies with human participants complied with the ethical standards of the institution and country concerned and adhered to the tenets of the Helsinki Declaration of 1964 and its subsequent revisions or to comparable ethical standards.
Footnotes
Conflict of interest statement
Prof. Meitinger is the director of a molecular genetics laboratory authorized to issue private invoices at Rechts der Isar Hospital, TUM, Munich.
Dr. Hempel and Prof. Kubisch also work at the Martin Zeitz Center for Rare Diseases in Hamburg. This center is a project partner of the health care project TRANSLATE NAMSE.
Prof. Muntau is a project partner of the health care project TRANSLATE NAMSE.
The remaining authors declare that no conflict of interest exists.
References
- 1.Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 2.McKenzie K, Milton M, Smith G, Ouellette-Kuntz H. Systematic review of the prevalence and incidence of intellectual disabilities: Current trends and issues. Curr Dev Disord Rep. 2016;3:104–115. [Google Scholar]
- 3.McLaren J, Bryson SE. Review of recent epidemiological studies of mental retardation: prevalence, associated disorders, and etiology. Am J Ment Retard. 1987;92:243–254. [PubMed] [Google Scholar]
- 4.Karam SM, Barros AJ, Matijasevich A, et al. Intellectual disability in a birth cohort: prevalence, etiology, and determinants at the age of 4 years. Public Health Genomics. 2016;19:290–297. doi: 10.1159/000448912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shashi V, McConkie-Rosell A, Rosell B, et al. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 6.Vasudevan P, Suri M. A clinical approach to developmental delay and intellectual disability. Clin Med (Lond) 2017;17:558–561. doi: 10.7861/clinmedicine.17-6-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rauch A, Hoyer J, Guth S, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140:2063–2074. doi: 10.1002/ajmg.a.31416. [DOI] [PubMed] [Google Scholar]
- 8.Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flore LA, Milunsky JM. Updates in the genetic evaluation of the child with global developmental delay or intellectual disability. Semin Pediatr Neurol. 2012;19:173–180. doi: 10.1016/j.spen.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 11.van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;371 doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 13.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claustres M, Kozich V, Dequeker E, et al. Recommendations for reporting results of diagnostic genetic testing (biochemical, cytogenetic and molecular genetic) Eur J Hum Genet. 2014;22:160–170. doi: 10.1038/ejhg.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthijs G, Souche E, Alders M, et al. Guidelines for diagnostic next-generation sequencing. Eur J Hum Genet. 2016;24:2–5. doi: 10.1038/ejhg.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauer P, Wildhardt G, Gläser D, et al. S1 Leitlinie: Molekulargenetische Diagnostik mit Hochdurchsatzverfahren, beispielsweise mit Next-Generation Sequencing 2017. www.gfhev.de/de/leitlinien/LL_und_Stellungnahmen/2017_09_15_GfH-S1-LL_NGS-Diagnostik_final.pdf (last accessed on 8 February 2019) [Google Scholar]
- 17.Tarailo-Graovac M, Shyr C, Ross CJ, et al. Exome sequencing and the management of neurometabolic disorders. N Engl J Med. 2016;374:2246–2255. doi: 10.1056/NEJMoa1515792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thevenon J, Duffourd Y, Masurel-Paulet A, et al. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet. 2016;89:700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 19.Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–1096. doi: 10.1038/gim.2016.1. [DOI] [PubMed] [Google Scholar]
- 20.Kuperberg M, Lev D, Blumkin L, et al. Utility of whole exome sequencing for genetic diagnosis of previously undiagnosed pediatric neurology patients. J Child Neurol. 2016;31:1534–1539. doi: 10.1177/0883073816664836. [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Deignan JL, Dorrani N, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivastava S, Cohen JS, Vernon H, et al. Clinical whole exome sequencing in child neurology practice. Ann Neurol. 2014;76:473–483. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 24.Farwell KD, Shahmirzadi L, El-Khechen D, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 25.Sawyer SL, Hartley T, Dyment DA, et al. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin Genet. 2016;89:275–284. doi: 10.1111/cge.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eldomery MK, Coban-Akdemir Z, Harel T, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9 doi: 10.1186/s13073-017-0412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gambin T, Yuan B, Bi W, et al. Identification of novel candidate disease genes from de novo exonic copy number variants. Genome Med. 2017;9 doi: 10.1186/s13073-017-0472-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartley T, Wagner JD, Warman-Chardon J, et al. Whole-exome sequencing is a valuable diagnostic tool for inherited peripheral neuropathies: outcomes from a cohort of 50 families. Clin Genet. 2018;93:301–309. doi: 10.1111/cge.13101. [DOI] [PubMed] [Google Scholar]
- 29.Kohler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology Project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohler S, Vasilevsky NA, Engelstad M, et al. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017;45:D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Snijders A, Segraves R, et al. High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization. Am J Hum Genet. 2005;76:312–326. doi: 10.1086/427762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kromeyer-Hauschild K, Wabitsch M, Kunze D, et al. Perzentile für den Body-mass-Index für das Kindes- und Jugendalter unter Heranziehung verschiedener deutscher Stichproben. Monatsschrift Kinderheilkunde. 2001;149:807–818. [Google Scholar]
- 33.Hempel M, Cremer K, Ockeloen CW, et al. De novo mutations in CHAMP1 cause intellectual disability with severe speech impairment. Am J Hum Genet. 2015;97:493–500. doi: 10.1016/j.ajhg.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JH, Shinde DN, Reijnders MRF, et al. De novo mutations in SON sisrupt RNA splicing of genes essential for brain development and metabolism, causing an intellectual-disability syndrome. Am J Hum Genet. 2016;99:711–719. doi: 10.1016/j.ajhg.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E1.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E2.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIMorg: Online Mendelian Inheritance in Man (OMIM(R)), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E4.Voigt M, Fusch C, Olbertz D. Analyse des Neugeborenenkollektivs der Bundesrepublik Deutschland 12 Mitteilung: Vorstellung engmaschiger Perzentilwerte (-kurven) für die Körpermaße Neugeborener. Geburtsh Frauenheilk. 2006;66:956–970. [Google Scholar]
- E5.Zhang X, Snijders A, Segraves R, et al. High-resolution mapping of genotype-phenotype relationships in cri du chat syndrome using array comparative genomic hybridization. Am J Hum Genet. 2005;76:312–326. doi: 10.1086/427762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMETHODS
Method
Study design
The 50 index patients were admitted consecutively to the Department of Pediatrics, University Medical Center Hamburg-Eppendorf for complex neuropediatric diagnostic work-up (German surgical procedure code OPS 1–942). All patients underwent the following standardized program of investigations:
The diagnostic procedures, including trio WES, were billed as “complex neuropediatric diagnostic work-up with extended genetic work-up” (OPS 1–942.2) or “complex neuropediatric diagnostic work-up with neurometabolic laboratory testing and/or infectiological/autoimmune inflammatory laboratory testing with extended genetic work-up” (OPS 1–942.3).
Documentation of genotype—WES technique
The exome sequencing was performed at the Institute of Human Genetics, Helmholtz Center Munich. All known coding DNA fragments from a patient and their biological parents were enriched with the SureSelect Human All Exon 50Mb V5 Kit (Agilent, Santa Clara, CA, USA) and sequenced using the HiSeq2500 system (Illumina, San Diego, CA, USA). The reads were assigned to the reference genome “human genome assembly hg19” (UCSC Genome Browser) with the aid of the Burrows-Wheeler Aligner (BWA, v.0.5.87.5). The detection of genetic variants (deviations from the norm) was achieved by means of SAMtools (v0.1.18), PINDEL (v 0.2.4t), and ExomeDepth (v1.0.0). In this way 95 to 99% of the exome sequences were covered at least 20-fold.
Documentation of genotype—classification of variants and genes
The genetic data generated by exome sequencing were analyzed at the Institute of Human Genetics, University Medical Center Hamburg-Eppendorf.
The disease relevance of a given identified variant was evaluated with the aid of a number of variables: MAF, assessment by bioinformatic prediction programs, comparison with databases (e.g., Database of Exome Aggregation Consortium, ExAC [e1]; Online Mendelian Inheritance of Man, OMIM [e2]), and current knowledge of the coded protein and its function.
A variant in a gene was classified as causing disease (i.e., as a mutation) if:
We defined known disease genes as those whose association with disease had been demonstrated by published clinical–genetic and/or functional data.
A variant in a gene was classified as probably causing disease if:
A gene was classified as a candidate gene if:
Phenotyping results
Sociodemographic data
For 49 patients (98%) data were available on the consanguinity of the parents. The parents’ ethnicity was documented for 45 patients (90%). The parents came from 12 different countries, including Germany (23 [46%]), Turkey (5 [10%]), Iran, Afghanistan, and Kosovo (each 3 [6%]). Nine patients (18%) had consanguineous parents. The consanguineous couples were from Germany (1 [2%]), Turkey, Iran, Afghanistan (each 2 [4%]), Egypt, and Pakistan (each 1 [2%]).
Perinatal abnormalities
Abnormal features of gestation or delivery had been reported for 18 patients (36%): an abnormal amount of amniotic fluid in nine cases (18%), vaginal bleeding in three (6%), intrauterine growth retardation (IUGR) in seven (14%), and premature delivery in five (10%). Other complications (e.g., placental insufficiency, gestational diabetes) were documented in individual cases. Nine patients showed abnormal measurements at birth: One patient was small for gestational age (SGA) (birth weight, length, and head circumference ?-2 standard deviations [SD] according to Voigt et al. [e4]), and isolated microcephaly was reported for two patients. In five patients (10%) birth weight and/or body length were ?-2 SD, and in one patient birth weight and head circumference were >+ 2 SD.
Imaging, functional analysis, and metabolic analysis
The EEG results were abnormal in 20 (43%) of 47 patients, for example “pathological waking EEG with generalized susceptibility to seizures and activation by hyperventilation and photostimulation” or “pathological waking EEG with inconstant bilateral temporal foci and left temporal susceptibility to seizures.” The findings on cMRI were abnormal in 29 (58%) of 50 patients, for instance “midline deformity with hypoplasia of the corpus callosum and absent septum pellucidum. Prominent cerebellar tonsils, but currently no Chiari I. Residual hemorrhages” or “suspicion of small right frontal subdural hematoma with no appreciable space-occupying effect. Increasing diffuse leptomeningeal and subarachnoid contrast-enhancing substrate over time, primarily compatible with a progressive angiomatous process with leptomeningeal accentuation.” Ophthalmological examination revealed abnormalities in 23 (62%) of 37 children, e.g., “left: optic nerve hypoplasia,” “right/left: mild tortuosity of veins,” or “right/left: hyperopia, astigmatism (etable 2). Blood gas analysis (BGA), performed in 45 index patients, revealed elevated lactate concentrations in 13 cases (29%) and striking base excess in nine patients (20%) (decreased levels in two children, elevated levels in seven). Of the 49 children who underwent cerebrospinal fluid analysis, one (2%) showed an elevated glucose concentration, 12 (25%) had abnormal lactate levels (low in two children, high in 10), and three children (6%) displayed elevated concentrations of protein. The majority of patients underwent extended metabolic screening, including analysis of amino acids in plasma and cerebrospinal fluid together with determination of acylcarnitines in dried blood spots and measurement of organic acids in urine. In no patient was there a constellation of biochemical findings pointing to a specific disease.
Genetic analysis had been carried out previously in 33 children (66%). Four had undergone chromosome analysis; five, array-CGH analysis; 10, chromosome analysis plus array-CGH analysis; five, individual or panel genetic analysis; and nine, individual or panel genetic analysis plus chromosome analysis or array-CGH analysis. None of these earlier analyses had revealed any abnormalities (e.g., chromosome analysis; normal male karyotype 46,XY; array-CGH analysis: no abnormal findings; ARX genetic analysis: no abnormal findings).
At least detailed questioning regarding the medical history of the index patient and family members
Extended clinical assessment
Wide-ranging examinations of blood, urine, and cerebrospinal fluid parameters
Cranial magnetic resonance imaging (cMRI) at 1.5 T or 3 T
Electroencephalography (EEG)
Trio WES
It affected a known disease gene
The very same variant had previously been described as causing disease, or the variant was comparable in type with known disease-causing variants in the same gene
There was overlap in phenotype between our patient and the published patients with causative mutations in the same gene
It affected a candidate gene (see below)
More than one prediction program (Polyphen 2, SIFT and CADD) classified it as pathogenic
In the case of a de novo mutation, it was not listed in the ExAC database
Based on the trio WES results it was the only one of the patient’s genes in which a rare variant was found, or based on the trio WES results and bioinformatic prediction algorithms it was classified as probably causing disease
Previously published data (e.g., functional in-vivo or in-vitro studies) pointed to disease association in humans or there was, via Genematcher (3), contact to other study groups who had also detected variants in the same gene in patients with overlapping symptoms