Abstract
Transactivation response element RNA-binding protein (TRBP; TARBP2) is known to play important roles in human immunodeficiency virus (HIV) replication and microRNA biogenesis. However, recent studies implicate TRBP in a variety of biological processes as a mediator of cross-talk between signal transduction pathways. Here, we provide the first evidence that TRBP is required for efficient neurosphere formation and for the expression of neural stem cell markers and Notch target genes in primary neural progenitor cells in vitro. Consistent with this, introduction of TRBP into the mouse embryonic brain in utero increased the fraction of cells expressing Sox2 in the ventricular zone. We also show that TRBP physically interacts with the Notch transcriptional coactivation complex through C promoter-binding factor 1 (CBF1; RBPJ) and strengthens the association between the Notch intracellular domain (NICD) and CBF1, resulting in increased NICD recruitment to the promoter region of a Notch target gene. Our data indicate that TRBP is a novel transcriptional coactivator of the Notch signaling pathway, playing an important role in neural stem cell regulation during mammalian brain development.
KEY WORDS: TRBP, Notch, Embryonic neural stem cells, Transcriptional activation, Central nervous system development
Summary: Transactivation response element RNA-binding protein (TRBP) acts as a transcriptional coactivator of the Notch pathway and regulates neural stem cells during brain development in mice.
INTRODUCTION
Mammalian brain development requires the tightly regulated spatiotemporal control of neural stem cell proliferation and differentiation for proper neural cell composition, laminar organization and size of a mature brain (Rakic, 1995; Breunig et al., 2011). This regulation is achieved by numerous cellular signaling pathways triggered by various cues, such as secretory factors and cell surface ligands (Yoon and Gaiano, 2005; Kohwi and Doe, 2013).
Transactivation response element RNA-binding protein (TRBP; also known as TARBP2) was initially identified as a human immunodeficiency virus type 1 (HIV-1) TAR RNA-binding partner (Gatignol et al., 1991; Dorin et al., 2003), and received much attention because of its activity in enhancing the catalytic activity of Dicer, a key regulator in microRNA biogenesis (Chendrimada et al., 2005; Haase et al., 2005). However, more recently, TRBP has been proposed to have microRNA-independent functions in various cellular events, including cell growth, oncogenesis and antiviral response by mediating cross-talk between signal transduction pathways (Daniels and Gatignol, 2012). Interestingly, a search for TRBP using Eurexpress, a transcriptome atlas database for the mouse embryo (www.eurexpress.org), showed in situ hybridization results of expression in the ventricular zone, but its role there has not yet been determined.
Notch controls crucial functions in many tissues in adults as well as at various developmental stages (Yoon and Gaiano, 2005). In the developing mammalian central nervous system, Notch controls maintenance and cell fate choices of neural stem cells (Yoon et al., 2004). In this study, we establish an unexpected functional link between TRBP and the Notch signaling pathway in the regulation of neural stem cell characteristics during mammalian brain development.
RESULTS AND DISCUSSION
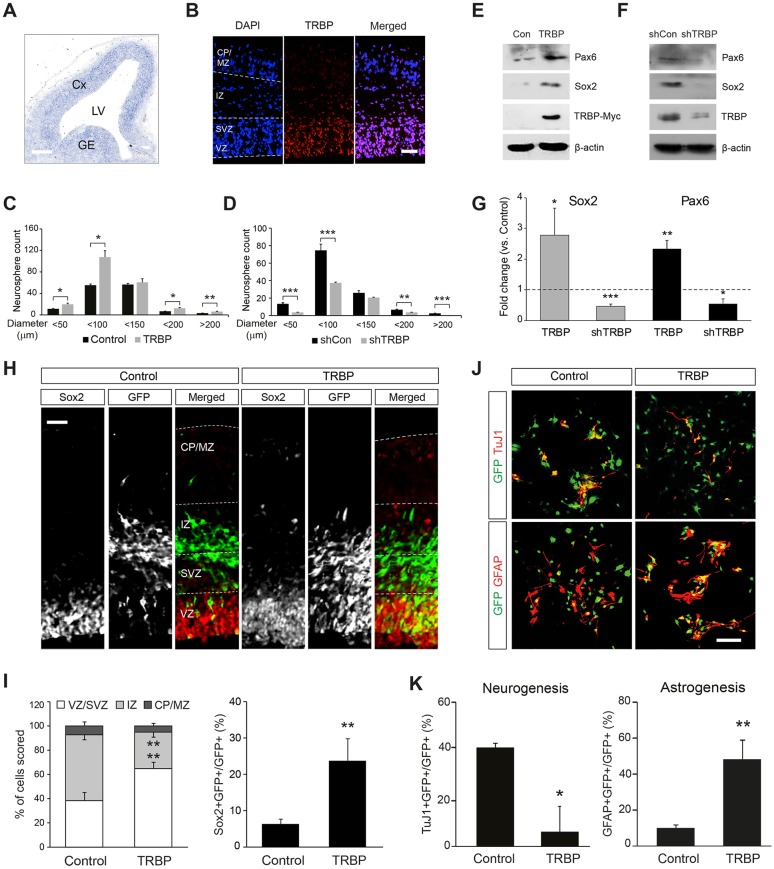
We first confirmed TRBP expression in the mouse embryonic brain by in situ hybridization and immunofluorescence staining. Both techniques revealed TRBP expression in the germinal zones lining the lateral ventricles in the embryonic day (E) 13.5 forebrain, implying a potential role in neural progenitor populations (Fig. 1A,B). To test the functions of TRBP in embryonic neural progenitor cells in vitro, we began with neurospheres (Singec et al., 2006) isolated after transducing E14.5 neural progenitors with a TRBP-expressing retroviral vector MSIG (Han et al., 2015). Expression of TRBP gave rise to an increase in the number and size of neurospheres compared with control virus (Fig. 1C), suggesting that TRBP enhances the self-renewal capacity of neural progenitor cells in vitro. Increased expression of the neural stem cell markers Sox2 and Pax6 (Ellis et al., 2004; Pevny and Nicolis, 2010) supported this idea (Fig. 1E,G). Consistent with these results, TRBP knockdown by short hairpin RNA (shRNA) reduced the size and number of neurospheres and the expression of Sox2 and Pax6 (Fig. 1D,F,G). Providing important confirmation of our results in an in vivo context, we found that cells expressing the TRBP vector were more likely to express Sox2 and be located in the ventricular/subventricular zone (VZ/SVZ) after in utero electroporation (Fig. 1H,I). Interestingly, we also noted that TRBP increased astrocyte production at the expense of neurogenesis under differentiating conditions in vitro (Fig. 1J,K); this is reminiscent of Notch effects in the embryonic central nervous system (Yoon and Gaiano, 2005; Jang et al., 2014).
Fig. 1.
TRBP enhances embryonic neural stem cell properties. (A,B) Trbp gene expression pattern in the E13.5 mouse forebrain was determined by (A) in situ hybridization and (B) immunofluorescence after antigen retrieval using proteinase K digestion. (C,D) Neurosphere assay using mouse E14.5 primary neural progenitors transduced with a retroviral vector expressing (C) TRBP or (D) shRNA specific to TRBP (shTRBP). (E-G) Western blot analysis for Pax6 and Sox2 proteins of neural progenitor cell lysates infected with (E) TRBP-expressing or (F) shTRBP-expressing retrovirus. Quantification is shown in G. (H,I) Double immunolabeling of E15.5 brain sections electroporated in utero with TRBP-expressing plasmid at E13.5 using anti-GFP (reporter, green) and Sox2 (red) antibodies (H). Quantification is shown in I. (J,K) Representative immunostaining using anti-GFP (green) together with anti-βIII-tubulin (clone TuJ1; neuron, red) or GFAP (astrocyte, red) antibodies after in vitro differentiation of E14.5 neural progenitor cells. Quantification is shown in K. Scale bars: 200 µm in A; 50 µm in B,H,J. CP, cortical plate; Cx, cortex; GE, ganglionic eminence; IZ, intermediate zone; LV, lateral ventricles; MZ, marginal zone; SVZ, subventricular zone; VZ, ventricular zone. All error bars represent s.d. *P<0.05, **P<0.01, ***P<0.001, Student's t-test.
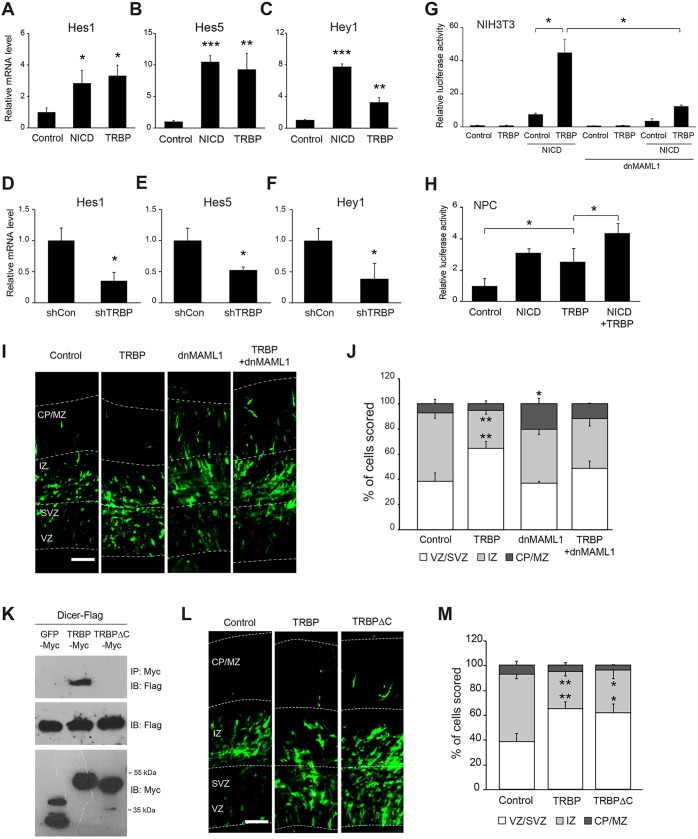
These results led us to examine whether TRBP regulates the Notch signaling pathway. Indeed, in primary neural progenitor cells TRBP increased mRNA expression of the Notch targets Hes1 and Hes5 to levels comparable to those driven by the Notch intracellular domain (NICD; a constitutively active form of Notch) (Yang et al., 2011), and Hey1 mRNA expression to a level ∼50% of NICD-stimulated levels (Fig. 2A-C). TRBP shRNA-infected cells had a greater than 50% reduction in the levels of Hes1, Hes5 and Hey1 mRNAs (Fig. 2D-F). However, unlike the qRT-PCR results using primary neural progenitor cells, TRBP alone did not activate the Notch reporter driven by a C promoter-binding factor 1 (CBF1)-responsive element (four CBF1 binding sites and the basal SV40 promoter) in NIH3T3 cells (Fig. 2G), having undetectable levels of Notch activity. Instead, TRBP dramatically increased promoter activity only when NICD was present, suggesting that upregulation of Notch target gene transcription by TRBP occurs at the promoter level and is NICD dependent. Moreover, co-expression of a dominant-negative form of mastermind-like 1 (dnMAML1), a key component of the Notch coactivation complex (Wu et al., 2000; Weng et al., 2003), disrupted the effects of TRBP (Fig. 2G,I,J), indicating that TRBP action requires core Notch coactivational machinery. qRT-PCR revealed that gene expression levels of Notch coactivational complex members were unchanged by the gain- or loss-of-function of TRBP in primary neural progenitor cells (Fig. S1A,B), suggesting that the enhanced Notch target promoter activation by TRBP was not due to the increased gene expression of a coactivational complex member. In primary neural progenitor cells, forced expression of TRBP alone increased Notch-dependent promoter activity, presumably because of endogenous NICD present in these cells, and co-transfection with NICD-expressing vector enhanced activity even further (Fig. 2H), consistent with the results shown in Fig. 2A-C. Facilitated in vivo activation of the Hes5 promoter was also observed when a Discosoma Red (DsRed) reporter driven by the Hes5 promoter was introduced together with the TRBP plasmid into the brain (Fig. S2).
Fig. 2.
TRBP increases Notch-dependent transcription. (A-F) mRNA expression levels of each indicated Notch target gene as measured by qRT-PCR, 2 days after E14.5 primary neural progenitor cells were transduced with retroviral vectors expressing (A-C) TRBP or (D-F) shTRBP. (G,H) Luciferase assay was used to detect the activation of a Notch reporter consisting of firefly luciferase fused to a CBF1-responsive element. (G) NIH3T3 or (H) E14.5 primary neural progenitor cells were transfected with a Notch reporter and the indicated expression vectors. Cells were harvested at 2 days post-transfection for luciferase assays. (I,L) Anti-GFP immunolabeling of E15.5 brain sections electroporated in utero with the indicated expression vectors at E13.5. (J,M) Quantification of I and L, respectively. (K) Co-IP assays using HEK 293T cells showing that a TRBP mutant (TRBPΔC) is deficient in binding to Dicer. Scale bars: 50 µm. Error bars represent s.d. *P<0.05, **P<0.01, ***P<0.001, Student's t-test.
Since the best-known function of TRBP is to enhance Dicer activity, we tested whether Dicer is involved in TRBP enhancement of Notch pathway activation by generating a TRBP mutant lacking Dicer binding ability (TRPBΔC; Fig. 2K) based on a previous report (Daniels et al., 2009). This TRBP mutant still efficiently increased Hes5 mRNA production and Sox2 protein levels (Fig. S3A,B) in neural progenitor cells in vitro, and induced cell localization to the VZ/SVZ in vivo (Fig. 2L,M) at wild-type levels, strongly indicating that TRBP enhancement of Notch pathway activation is Dicer independent. Previous studies also support our observations: the expression of Hes genes was not altered in Dicer-ablated embryonic brains (Peng et al., 2012), and neural progenitors lacking Dicer did not precociously exit the VZ and still retained progenitor marker expression in vivo (Knuckles et al., 2012).
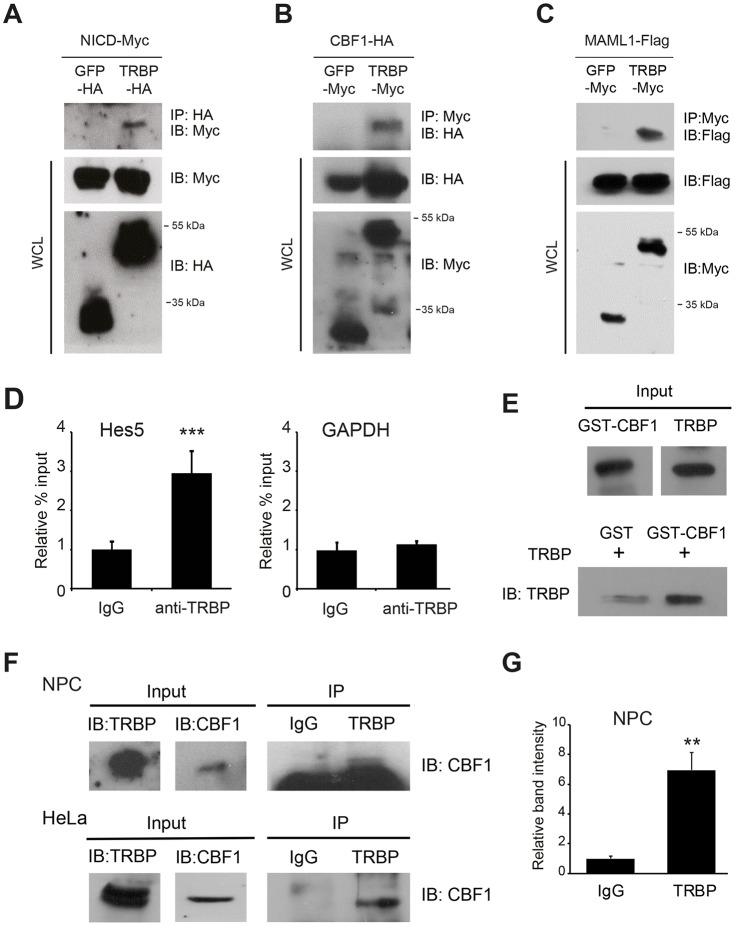
Since TRBP is a nuclear protein and simultaneously increases the transcription of several Notch target genes, one possible mechanism explaining these observations might involve direct modulation of the transcriptional machinery. To test this hypothesis, we used co-immunoprecipitation (co-IP) assays with the HEK 293T cell line to explore the potential interactions between TRBP and the Notch transcriptional activation complex. Myc-tagged NICD co-immunoprecipitated with TRBP-HA protein, whereas no band was detected from GFP-HA-transfected cells (Fig. 3A). Likewise, clear association of TRBP with CBF1 (RBPJ) or MAML1 was also observed (Fig. 3B,C). TRBP co-immunoprecipitated well with CBF1 in RNase-treated samples, suggesting that TRBP interacts with Notch components in an RNA-independent manner (Fig. S4A,B).
Fig. 3.
TRBP interacts with the Notch transcriptional coactivation complex. (A-C) Representative western blots showing co-IP of TRBP with each component of the Notch coactivational complex using the HEK 293T cell line: (A) NICD, (B) CBF1 and (C) MAML1. (D) ChIP assay using primary neural progenitor chromatin extracts and anti-TRBP antibody, and subsequent real-time PCR analysis for the genomic Hes5 and Gapdh promoter regions. (E) GST pull-down assay using E. coli-expressed recombinant GST-CBF1 and TRBP proteins. (F,G) Co-IP assays were performed with anti-TRBP and anti-CBF1 antibodies to show endogenous interactions between TRBP and CBF1 in primary mouse progenitors and HeLa cells (F). Quantification of neural progenitor results is shown in G. NPC, neural progenitor cell; WCL, whole-cell lysate. Error bars represent s.d. **P<0.01, ***P<0.001, Student's t-test.
Consistent with the co-IP results, a chromatin immunoprecipitation (ChIP) assay using chromatin extracts derived from E14.5 primary neural progenitor cells and anti-TRBP antibody revealed that endogenous TRBP protein is recruited to the genomic Hes5 promoter region (Fig. 3D). Furthermore, the glutathione S-transferase (GST)-tagged CBF1 protein was able to pull down TRBP in a cell-free in vitro binding assay (Fig. 3E), suggesting that CBF1 is a direct binding target of TRBP. The interaction between endogenous TRBP and CBF1 was also confirmed in primary mouse neural progenitors and HeLa cells by co-IP assays with anti-TRBP and anti-CBF1 antibodies (Fig. 3F,G).
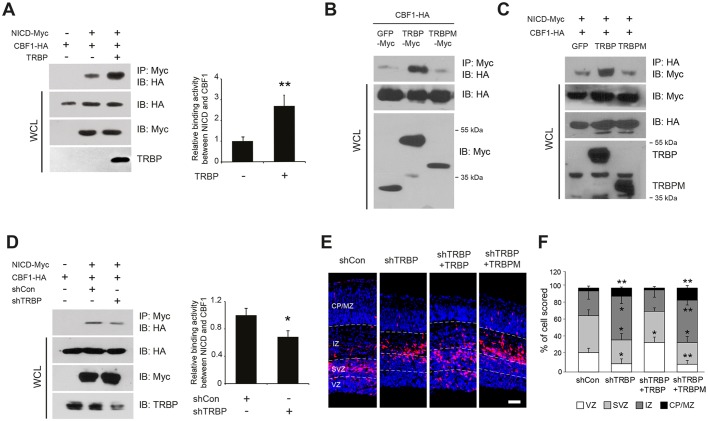
Finally, we examined the functional consequences of TRBP association with the Notch transactivational complex using the HEK 293T cell line. Overexpression of TRBP caused an increase in association between CBF1 and NICD by up to 2.5-fold (Fig. 4A), and a mutant TRBP protein (TRBPM) deficient in dsRNA-binding domains and binding activity to CBF1 lost this ability (Fig. 4B,C). Furthermore, this interaction was significantly weakened by TRBP knockdown (Fig. 4D). Considering the sizable amount of residual TRBP protein in TRBP shRNA-expressing cells, this degree of reduction strongly indicates that TRBP plays an important role in stabilizing the Notch coactivational complex at physiological levels.
Fig. 4.
TRBP reinforces association between NICD and CBF1. (A,D) The binding affinity between NICD and CBF1 upon (A) overexpression or (D) knockdown of TRBP was examined by co-IP assays using the HEK 293T cell line. Co-immunoprecipitated CBF1-HA protein levels were normalized to CBF1-HA protein levels in the respective whole-cell lysates. (B,C) Co-IP assays show that a TRBP mutant (TRBPM) lacking dsRNA-binding domains and deficient in binding to CBF1 (B) does not enhance the NICD-CBF1 interaction (C). TRBPM was identified through domain-mapping experiments. (E,F) Immunolabeling of E15.5 brain sections electroporated in utero with the indicated expression vectors at E13.5 using anti-GFP and Alexa Fluor 555-conjugated secondary antibodies (E). Quantification is shown in F. Scale bar: 50 µm. All error bars represent s.d. *P<0.05, **P<0.01, Student's t-test.
The importance of TRBP interaction with CBF1 was further confirmed in vivo. Cell exit from the VZ/SVZ was prominent 2 days post-electroporation of an shRNA vector against TRBP at E13.5, and this change was reversed by wild-type TRBP and not by TRBPM (Fig. 4E,F), indicating that TRBP-induced augmentation of neural stem cell properties is strictly CBF1 binding dependent.
Because both TRBP and CBF1 are nuclear and are ubiquitously expressed in many types of tissues (Lu and Lux, 1996; Daniels and Gatignol, 2012), it is plausible to hypothesize that TRBP may be a constitutive component of the Notch coactivation complex. Our data showing interaction between TRBP and CBF1 in non-neural human HeLa cells (Fig. 3F) provides support for this idea. Examination of the role of TRBP in cellular events such as oncogenesis or hematopoiesis, in which Notch signaling plays a pivotal role, would yield insight into the potentially ubiquitous importance of this Notch coactivation complex member.
MATERIALS AND METHODS
Animals
All animal studies were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee at Sungkyunkwan University (SKKUIACUC-2016-04-0005-2). Timed-pregnant CD1 mice were obtained from Koatech (Korea) and used for neural progenitor cell preparation and in utero electroporation.
In situ hybridization
In situ hybridization was conducted according to standard protocols (Siegenthaler et al., 2009). Digoxigenin-labeled sense and antisense riboprobes were generated from pGEM-T Easy vectors (Promega) containing cDNA sequences corresponding to residues 1-623 of mouse TRBP.
Co-immunoprecipitation (co-IP)
For immunoprecipitations of tagged proteins, transfected HEK 293T cells obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) were lysed in immunoprecipitation lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA) with protease inhibitor (Sigma). Total proteins were incubated with primary antibody against a tag [mouse anti-Myc (Santa Cruz Biotechnology, sc-40X; 1:1000), mouse anti-HA (Abcam, ab16918; 1:500) or mouse anti-FLAG (Sigma, F1804; 1:500)] and protein G-conjugated beads (Invitrogen) at 4°C overnight. Beads were washed three times with lysis buffer and 2× sample buffer was added. Samples were then boiled for 10 min, and beads were removed by centrifuging at 17,000 g for 30 min. Proteins were resolved by SDS-PAGE and subjected to immunoblotting using an anti-tag antibody. ImageJ software (http://rsb.info.nih.gov/ij/) was used to quantify band intensity of the immunoblot. Immunoprecipitations of endogenous TRBP with anti-TRBP antibody (a kind gift from Dr V. N. Kim, Seoul National University, Seoul, Korea) using mouse embryonic neural progenitors or HeLa cells, and subsequent immunoblotting with anti-CBF1 antibody (Cosmo Bio, ZRBP2; 1:100) were performed in a similar manner.
GST pull-down assays
The Trbp and Cbf1 genes were cloned into the bacterial expression vector pGEX-4T1 (GE Healthcare Life Science), which has a GST tag at the N-terminus and a thrombin cleavage site, and GST fusion proteins were expressed in E. coli BL21 cells by the IPTG induction method and purified as described (Lee et al., 2006). Recombinant TRBP protein without the GST tag was prepared by incubating GST-TRBP protein with 1 U/µl thrombin (Sigma) at room temperature overnight, followed by inactivation with 1 mM PMSF. GST alone or GST-CBF1 protein coupled to glutathione-agarose beads (Sigma) was incubated with thrombin-cleaved TRBP protein at 4°C overnight. The mixtures were washed three times with PBS and eluted with glutathione elution buffer [50 mM Tris-HCl pH 8.0, 10 mM reduced glutathione (Sigma)]. The samples were then subjected to SDS-PAGE followed by western blot analysis.
Statistical analysis
Statistical tests were performed using SPSS version 23 (IBM). All data represent three or more independent experiments.
Acknowledgements
We thank Dr V. N. Kim for the kind gift of anti-TRBP antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
S.-H.B., D.H., J.K., M.K., J.Y.C. and H.X.N. carried out experiments and analyzed data. K.Y. designed the study and wrote the manuscript with contribution from S.J.P.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT and Future Planning (2015R1A2A2A01005687).
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.139493.supplemental
References
- Breunig J. J., Haydar T. F. and Rakic P. (2011). Neural stem cells: historical perspective and future prospects. Neuron 70, 614-625. 10.1016/j.neuron.2011.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada T. P., Gregory R. I., Kumaraswamy E., Norman J., Cooch N., Nishikura K. and Shiekhattar R. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740-744. 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. M. and Gatignol A. (2012). The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiol. Mol. Biol. Rev. 76, 652-666. 10.1128/MMBR.00012-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. M., Melendez-Peña C. E., Scarborough R. J., Daher A., Christensen H. S., El Far M., Purcell D. F. J., Lainé S. and Gatignol A. (2009). Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 10, 38 10.1186/1471-2199-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorin D., Bonnet M. C., Bannwarth S., Gatignol A., Meurs E. F. and Vaquero C. (2003). The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J. Biol. Chem. 278, 4440-4448. 10.1074/jbc.M208954200 [DOI] [PubMed] [Google Scholar]
- Ellis P., Fagan B. M., Magness S. T., Hutton S., Taranova O., Hayashi S., McMahon A., Rao M. and Pevny L. (2004). SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 26, 148-165. 10.1159/000082134 [DOI] [PubMed] [Google Scholar]
- Gatignol A., Buckler-White A., Berkhout B. and Jeang K. T. (1991). Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 251, 1597-1600. 10.1126/science.2011739 [DOI] [PubMed] [Google Scholar]
- Haase A. D., Jaskiewicz L., Zhang H., Lainé S., Sack R., Gatignol A. and Filipowicz W. (2005). TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6, 961-967. 10.1038/sj.embor.7400509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Byun S.-H., Park S., Kim J., Kim I., Ha S., Kwon M. and Yoon K. (2015). YAP/TAZ enhance mammalian embryonic neural stem cell characteristics in a Tead-dependent manner. Biochem. Biophys. Res. Commun. 458, 110-116. 10.1016/j.bbrc.2015.01.077 [DOI] [PubMed] [Google Scholar]
- Jang J., Byun S.-H., Han D., Lee J., Kim J., Lee N., Kim I., Park S., Ha S., Kwon M. et al. (2014). Notch intracellular domain deficiency in nuclear localization activity retains the ability to enhance neural stem cell character and block neurogenesis in mammalian brain development. Stem Cells Dev. 23, 2841-2850. 10.1089/scd.2014.0031 [DOI] [PubMed] [Google Scholar]
- Knuckles P., Vogt M. A., Lugert S., Milo M., Chong M. M. W., Hautbergue G. M., Wilson S. A., Littman D. R. and Taylor V. (2012). Drosha regulates neurogenesis by controlling Neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 15, 962-969. 10.1038/nn.3139 [DOI] [PubMed] [Google Scholar]
- Kohwi M. and Doe C. Q. (2013). Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823-838. 10.1038/nrn3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Hur I., Park S.-Y., Kim Y.-K., Suh M. R. and Kim V. N. (2006). The role of PACT in the RNA silencing pathway. EMBO J. 25, 522-532. 10.1038/sj.emboj.7600942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F. M. and Lux S. E. (1996). Constitutively active human Notch1 binds to the transcription factor CBF1 and stimulates transcription through a promoter containing a CBF1-responsive element. Proc. Natl. Acad. Sci. USA 93, 5663-5667. 10.1073/pnas.93.11.5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C., Li N., Ng Y.-K., Zhang J., Meier F., Theis F. J., Merkenschlager M., Chen W., Wurst W. and Prakash N. (2012). A unilateral negative feedback loop between miR-200 microRNAs and Sox2/E2F3 controls neural progenitor cell-cycle exit and differentiation. J. Neurosci. 32, 13292-13308. 10.1523/JNEUROSCI.2124-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L. H. and Nicolis S. K. (2010). Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 42, 421-424. 10.1016/j.biocel.2009.08.018 [DOI] [PubMed] [Google Scholar]
- Rakic P. (1995). A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 18, 383-388. 10.1016/0166-2236(95)93934-P [DOI] [PubMed] [Google Scholar]
- Siegenthaler J. A., Ashique A. M., Zarbalis K., Patterson K. P., Hecht J. H., Kane M. A., Folias A. E., Choe Y., May S. R., Kume T. et al. (2009). Retinoic acid from the meninges regulates cortical neuron generation. Cell 139, 597-609. 10.1016/j.cell.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singec I., Knoth R., Meyer R. P., Maciaczyk J., Volk B., Nikkhah G., Frotscher M. and Snyder E. Y. (2006). Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat. Methods 3, 801-806. 10.1038/nmeth926 [DOI] [PubMed] [Google Scholar]
- Weng A. P., Nam Y., Wolfe M. S., Pear W. S., Griffin J. D., Blacklow S. C. and Aster J. C. (2003). Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of Notch signaling. Mol. Cell. Biol. 23, 655-664. 10.1128/MCB.23.2.655-664.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Aster J. C., Blacklow S. C., Lake R., Artavanis-Tsakonas S. and Griffin J. D. (2000). MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484-489. 10.1038/82644 [DOI] [PubMed] [Google Scholar]
- Yang M.-S., Hong J.-S., Kim S.-T., Lee K.-Y., Park K. W., Kwon S.-T., Kweon D.-H., Koh Y. H., Gaiano N. and Yoon K. (2011). Among gamma-secretase substrates Notch1 alone is sufficient to block neurogenesis but does not confer self-renewal properties to neural stem cells. Biochem. Biophys. Res. Commun. 404, 133-138. 10.1016/j.bbrc.2010.11.080 [DOI] [PubMed] [Google Scholar]
- Yoon K. and Gaiano N. (2005). Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 8, 709-715. 10.1038/nn1475 [DOI] [PubMed] [Google Scholar]
- Yoon K., Nery S., Rutlin M. L., Radtke F., Fishell G. and Gaiano N. (2004). Fibroblast growth factor receptor signaling promotes radial glial identity and interacts with Notch1 signaling in telencephalic progenitors. J. Neurosci. 24, 9497-9506. 10.1523/JNEUROSCI.0993-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]