Abstract
Caudal fin regeneration is characterized by a proliferation boost in the mesenchymal blastema that is controlled precisely in time and space. This allows a gradual and robust restoration of original fin size. However, how this is established and regulated is not well understood. Here, we report that Yap, the Hippo pathway effector, is a chief player in this process: functionally manipulating Yap during regeneration dramatically affects cell proliferation and expression of key signaling pathways, impacting regenerative growth. The intracellular location of Yap is tightly associated with different cell densities along the blastema proximal-distal axis, which correlate with alterations in cell morphology, cytoskeleton and cell-cell contacts in a gradient-like manner. Importantly, Yap inactivation occurs in high cell density areas, conditional to F-actin distribution and polymerization. We propose that Yap is essential for fin regeneration and that its function is dependent on mechanical tension, conferred by a balancing act of cell density and cytoskeleton activity.
KEY WORDS: Zebrafish, Fin regeneration, Hippo/Yap, F-actin, Cell density
Highlighted article: Yap, a Hippo pathway effector, is required to regulate cell proliferation and is regulated by changes in cell density and cell morphology to control fin regeneration.
INTRODUCTION
The ability of adult vertebrates to regenerate lost or injured organs is restricted to few examples in nature. The zebrafish (Danio rerio) is one of these impressive cases, being able to regrow a fully functional and anatomically similar organ through epimorphic regeneration (Morgan, 1901; Poss et al., 2000). In particular, caudal fin regeneration proceeds in three main phases: wound healing, blastema formation and outgrowth. Upon amputation, wound healing starts by immediate migration of the epidermis adjacent to the stump, which will give rise to a specialized wound epidermis (Poleo et al., 2001; Lee et al., 2009). The blastema will then begin to form, entailing the migration of differentiated intra-ray mesenchymal cells towards the stump that de-differentiate and proliferate in a lineage-restricted fashion (Knopf et al., 2011; Sousa et al., 2011; Stewart and Stankunas, 2012). Once the blastema is completely formed, it is divided into regions: a distal region associated with little proliferation and stem cell-like properties and a proximal region where most of the proliferation and differentiation events occur. Then in the outgrowth phase, these regions become more distinct, with a non-proliferative distal-most blastema tip, a medial region where most of the cell proliferation occurs and a proximal area where differentiation takes place (Nechiporuk and Keating, 2002; Wehner et al., 2014).
Remarkably, upon amputation the caudal fin regenerates the precise amount of tissue that was lost, at the correct location. This indicates that a positional memory instructs the blastema cells according to their proximo-distal fin localization (Lee et al., 2005). Coupled to this property, the regenerative process occurs independently of the number of amputations applied and animal age (Azevedo et al., 2011). Such properties indicate a tight growth control program, involving precise coordination between proliferation and positional information along the regenerating caudal fin. Although it is as yet unclear how these two central processes are molecularly controlled, they are likely to involve the integration of various signals. To date, fibroblast growth factor (FGF) is the only morphogen that has been shown to promote a proliferation rate increase in a proximal-distal gradient-like manner (Lee et al., 2005). Recently, inhibition of the phosphatase Calcineurin and of potassium channel activity were shown to be necessary for proportionate growth of the fin during development and regeneration (Perathoner et al., 2014; Kujawski et al., 2014). Clearly, further clarification of the cellular mechanisms that restrain uncontrolled proliferation is required in order to understand what regulates the final size of the renewed organ.
One signaling pathway that has arisen as a candidate for growth control during regeneration is the conserved Hippo pathway, which is essential for proper regulation of developmental organ growth in Drosophila and vertebrates (Pan, 2010). This kinase cascade can be activated by multiple inputs and ultimately converges in phosphorylation and inactivation of its effectors, the transcriptional activator Yap and its paralog Taz, by excluding one or both from the nucleus (Huang et al., 2005; Dong et al., 2007). In the nucleus, Yap and Taz bind to different partners, such as members of the TEAD family of transcription factors, together stimulating the transcription of multiple target genes (Mahoney et al., 2005; Zhao et al., 2008).
Different studies have implicated the Hippo pathway in repair mechanisms. Several reports have shown a role for this pathway as a mediator of intestinal and heart repair, as well as limb bud regeneration (Cai et al., 2010; Staley and Irvine, 2010; Hayashi et al., 2014). Moreover, extracellular matrix (ECM) stiffness, cell morphology and the actin cytoskeleton have been shown to be involved in Yap/Taz activation in vitro (Dupont et al., 2011). Activation of Yap and Taz can also be cell density dependent (Zhao et al., 2007), possibly through the action of adherens junctions (Schlegelmilch et al., 2011). These findings demonstrate that cells are able to interpret physical signals from their surroundings through Yap and Taz, but transduction of these mechanical cues into actual signaling is still little understood in vivo. Importantly, in zebrafish organ regeneration, including that of the caudal fin, where accurate growth must be controlled, this pathway has never been addressed. Furthermore, the study of mechanotransduction, cytoskeleton and adhesion has remained limited in this in vivo context (Santos-Ruiz et al., 2005).
Here, we explore the hypothesis that changes in tissue tension and cell density, inherent to wounding and regeneration of the caudal fin, trigger a series of events that contribute to regeneration involving the Hippo pathway. Our results indicate that Yap is indispensable for proliferation in the blastema and necessary for regeneration to proceed. Interestingly, we found that Yap inactivation correlates with high cell density areas and localization of α-Catenin and F-actin. This suggests that tension changes, which result from heterogeneous cell densities and are sensed by the junctions and cytoskeleton, influence growth within the blastema. We propose that blastema tissue is constrained by mechanical forces that are mechanotransduced by Yap, contributing to final size recovery during epimorphic regeneration.
RESULTS
Yap intracellular localization varies according to the stage and region of the blastema
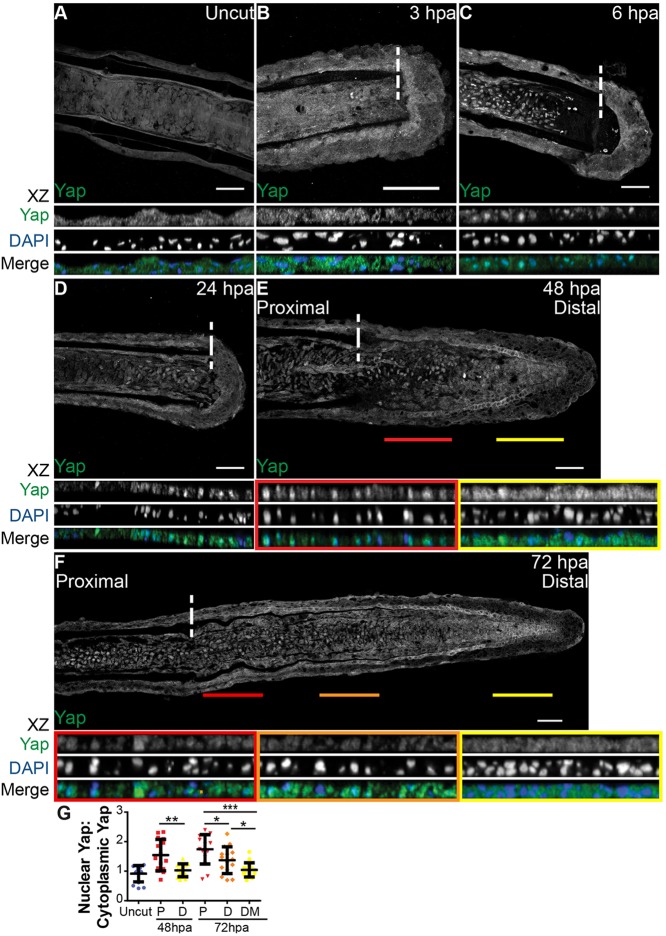
A hallmark of Yap and Taz activation is their translocation from the cytoplasm to the nucleus (Yagi et al., 1999; Zhao et al., 2007; Oh and Irvine, 2008). To understand whether Yap is activated during the caudal fin regenerative process, we assessed its protein expression in longitudinal sections of fin rays by immunofluorescence. This characterization showed striking intracellular dynamics at several regeneration stages. At 6 h post amputation (hpa), mesenchymal cells in the segment next to the amputation plane displayed Yap in the nucleus, in contrast to uncut and 3 hpa fins, in which Yap was uniformly present in the cytoplasm (Fig. 1A-C,G). This suggests that Yap becomes activated early in the regenerative process, during wound healing. The nuclear localization of Yap was maintained at 24 hpa, as mesenchymal cells form the blastema (Fig. 1D). By 48 hpa, when the blastema was complete, we detected different regions of intracellular Yap: in proximal regions Yap was mostly nuclear, whereas distally it was mainly cytoplasmic (Fig. 1E,G). This correlates with known proliferative regions in the blastema (Nechiporuk and Keating, 2002): the proximal region contains most of the proliferative events in contrast with the distal region. Finally, during blastema outgrowth at 72 hpa, these regions were more defined, with Yap more cytoplasmic in distal-most regions and progressively becoming nuclear towards proximal regions (Fig. 1F,G). This led us to believe that Yap could be playing an active role in controlling proliferation during caudal fin regeneration. Of note, the average intensity ratios between nuclear and cytoplasmic Yap in 48 hpa and 72 hpa distal-most regions were similar to uncut fin values, suggesting that Yap is inactive in these blastema areas (Fig. 1G).
Fig. 1.
Yap is present and highly dynamic during fin regeneration. (A-F) Representative immunostaining with anti-Yap in caudal fin longitudinal sections at several regenerative stages. (A) Uncut control; (B) 3 hpa; (C) 6 hpa; (D) 24 hpa; (E) 48 hpa; (F) 72 hpa. xz projections of mesenchymal regions highlight Yap intracellular localization. Distal (yellow) and proximal (red) lines at 48 hpa (E), and distal-most (yellow), distal (orange) and proximal (red) lines at 72 hpa (F), correspond to the mesenchymal areas in the medial blastema where xz projections were made (shown below). Dashed lines indicate amputation plane. n=10-15 sections; 5 fish/condition. Scale bars: 50 µm. (G) Quantification of Yap intracellular localization by expressing a ratio between average intensities of nuclear Yap:cytoplasmic Yap of xz projections from blastemas at different time points. Higher ratios correspond to higher intensities of nuclear Yap. P corresponds to xz of proximal (red); D to distal (orange); DM to distal-most regions (yellow). *P<0.05, **P<0.01, ***P<0.001; two-tailed, non-parametric Mann–Whitney test. n=15 sections, 5 fish/condition. Mean±s.d. are shown.
To determine whether other Hippo pathway members were present during zebrafish caudal fin regeneration, we analyzed their expression in different regenerative stages through in situ hybridization and quantitative PCR (qPCR) (supplementary material Fig. S1A,B). The core components of the pathway (stk3, sav1, lats2, yap1, wwtr1, nf2b, frmd6) and possible Yap DNA-binding partners (tead1a and tead4) were expressed in the blastema. However, Hippo pathway core components were not upregulated upon amputation, indicating that transcription does not play a key role in regulation of the pathway. Subsequently, we addressed the expression of phosphorylated active forms of Mst1/2 (Mst2 corresponds to zebrafish Stk3) and Lats1/2 as well as total Nf2 in 72 hpa blastemas by immunohistochemistry. We detected increased phosphorylated Mst1/2 and Lats1/2 in distal areas (supplementary material Fig. S1C,D,F,G) and presence of Nf2 throughout the blastema (supplementary material Fig. S1E). This suggests that Hippo pathway components capable of inactivating Yap are active specifically in distal regions where Yap is mainly cytoplasmic. These results show that this pathway is conserved in zebrafish and activated in the blastema during caudal fin regeneration.
Yap controls proliferation levels during regeneration
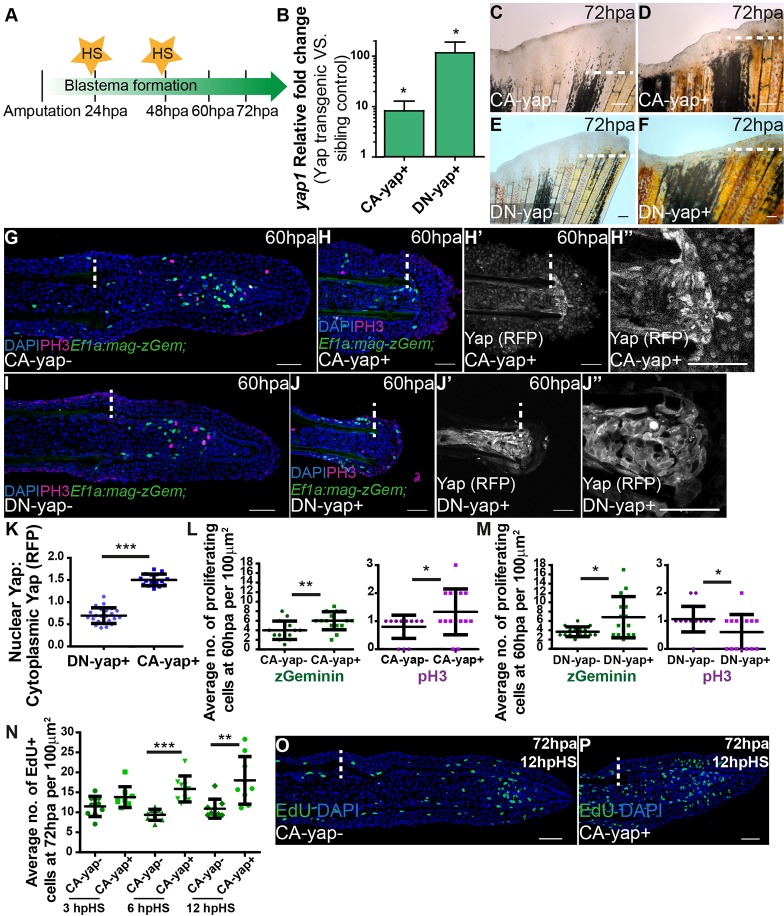
To determine whether Yap controls proliferation during regeneration, we used heat-shock transgenic techniques to manipulate Yap functionally. This allowed us to constitutively activate Yap (hsp70:RFP-CAyap, referred to as CA-yap; supplementary material Fig. S2A) and to dominantly inactivate Yap (hsp70:RFP-DNyap, referred to as DN-yap; supplementary material Fig. S2A). DN-yap transgenic fish were validated by addressing the previously demonstrated requirement of Yap for cardiac precursor cell migration, using morpholinos (supplementary material Fig. S2B-M) (Fukui et al., 2014; Miesfeld and Link, 2014). Moreover, when we performed immunostainings against RFP and Yap in both transgenic lines, we observed colocalization (supplementary material Fig. S3A-F).
Upon single heat-shock, yap1 expression was increased sixfold in CA-yap and 115-fold in DN-yap transgenic fish (Fig. 2B). In both cases, by applying a daily heat-shock during the blastema-forming phases (24 and 48 hpa; Fig. 2A), regeneration was impaired at 72 hpa (Fig. 2C-F). To understand whether the phenotypes could be due to proliferation defects, we applied the same protocol to transgenics CA-yap/DN-yap combined with Ef1a:mag-zGeminin, allowing for live detection of the cell cycle phases S/G2/M (Sugiyama et al., 2009). Immunohistochemistry analysis of Geminin together with the mitosis marker phospho-Histone 3 (pH3) revealed more proliferation in double transgenics Ef1a:mag-zGeminin; CA-yap+, resulting in deformed but smaller blastemas by 60 hpa compared with siblings (Fig. 2G,H,L). In Ef1a:mag-zGeminin; DN-yap+ double transgenics, proliferation was reduced causing undersized blastemas. This is probably due to cell cycle delay at G2/M phases (Xia et al., 2002), as cells were able to enter the cell cycle and express Geminin, but not proceed to mitosis, shown by the reduction of pH3 (Fig. 2I,J,M). To understand further the effects of Yap manipulation in proliferation, we performed single heat-shocks in 72 hpa CA-yap transgenics injected with 5-ethynyl-2′-deoxyuridine (EdU), the S-phase cell cycle marker (Choi et al., 2013). Importantly, we detected a progressive increase in EdU-positive cells in CA-yap+ blastemas by 6 h post heat-shock (hpHS) and 12 hpHS, but not in siblings or 3 hpHS (Fig. 2N-P; supplementary material Fig. S4A-D). To address the small blastema phenotypes in both Yap transgenics, we performed terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), which detects cell death (Cole and Ross, 2001). Interestingly, there was also a gradual increase of cell death in CA-yap+, starting at 18 hpHS, after the main proliferative phase post heat-shock (6 and 12 hpHS). This did not occur in siblings or DN-yap+ transgenics, indicating that in the latter case, reduction in blastema size is likely to be due to less proliferation (supplementary material Fig. S4E-N). Interestingly, ectopic cell death is observed almost exclusively in the CA-yap+ epidermis, indicating a possible impairment of the wound epidermis (perhaps affecting its signaling function), resulting in small blastema phenotypes in CA-Yap+ transgenics.
Fig. 2.
Yap influences proliferation in the blastema. (A) Experimental outline of heat-shock protocol used to access Yap functionality during regeneration. After amputations, fish were allowed to regenerate for 24 h, the time at which the first heat-shock was performed. A second heat-shock at 48 hpa was applied and phenotypes were assessed at 60 hpa or 72 hpa. The same protocol was applied to sibling controls. (B) qPCR determination of yap1 expression levels in blastemas of Yap transgenics versus respective siblings upon single heat-shock at 72 hpa. RNA extraction was performed at 2 hpHS. *P<0.01; two-tailed, non-parametric paired Wilcoxon test, logarithmic scale, base 10. (C-F) Representative brightfield images of Yap transgenics and siblings at 72 hpa after the protocol shown in A was performed. (C) CA-yap control; (D) CA-yap positive; (E) DN-yap control; (F) DN-yap positive. n=5 fish/condition. Scale bars: 200 µm. (G-J) Representative immunofluorescence with anti-pH3 in 60 hpa longitudinal sections of double transgenics Ef1α:mag-zGeminin; CA-yap/DN-yap and siblings after the protocol shown in A was performed. (G) CA-yap control; (H) CA-yap positive; (I) DN-yap control; (J) DN-yap positive. (H′,H″,J′,J″) Corresponding transgenic CA-Yap and DN-Yap RFP expression. Siblings do not show RFP expression. Scale bars: 50 µm. (K) Quantification of Yap-RFP intracellular localization in CA-yap and DN-yap transgenics by expressing a ratio between average intensities of nuclear Yap:cytoplasmic Yap of xz projections of respective mesenchymal cells. ***P<0.001; two-tailed, non-parametric Mann–Whitney test. n=16-23 sections, 3 fish/condition. (L,M) Quantification of average proliferation labeled with Geminin and pH3 occurring per 100 µm2 in Ef1α:mag-zGeminin; CA-yap/DN-yap and siblings, at 60 hpa. *P<0.05, **P<0.01; two-tailed, non-parametric Mann–Whitney test. n=15 sections, 3 fish/condition. (N) Quantification of average EdU-positive cells occurring per 100 µm2 in CA-yap and siblings, at 72 hpa in different times post heat-shock. **P<0.01, ***P<0.001; two-tailed, non-parametric Mann–Whitney test. n=9 sections, 3 fish/condition. (O,P) Representative immunofluorescence with EdU in 72 hpa longitudinal sections of CA-yap and siblings after 12 hpHS. (O) CA-yap control; (P) CA-yap positive. Scale bars: 50 µm. Dashed lines indicate amputation plane. Mean±s.d. are shown.
Additionally, we observed differences in Yap-RFP intracellular dynamics: in CA-yap, Yap-RFP is predominantly nuclear (Fig. 2H′,H″), whereas in DN-yap, Yap-RFP is more cytoplasmic (Fig. 2J′,J″). These observations were supported by quantifying the average intensity ratios between nuclear and cytoplasmic Yap-RFP, in both functional transgenics (Fig. 2K). Ratios in CA-yap+ transgenics were comparable to ratios found in areas of wild-type blastemas in which Yap is mainly active, whereas DN-yap+ ratios showed similar values to those obtained in inactive Yap zones (compare Fig. 2K with Fig. 1G), confirming the intracellular localization of CA-Yap and DN-Yap proteins, as they functionally localize to the expected intracellular compartment. Together, these results suggest that proliferation needs to be tightly regulated for regeneration to occur.
Yap regulates the expression of known targets and regeneration factors
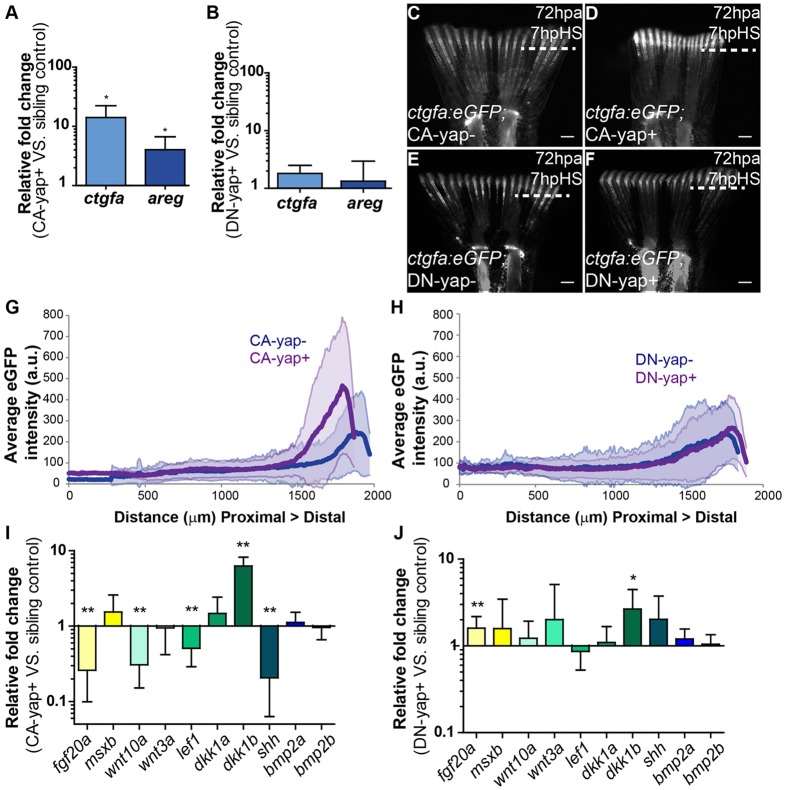
To ascertain whether manipulating Yap affects its activation state by induction of transcription of downstream genes, we searched for bona fide Yap transcriptional targets in the blastema by performing qPCR upon heat-shock induction in Yap functional transgenics. We found that connective tissue growth factor A (ctgfa) and amphiregulin (areg), two of Yap's well-characterized targets (Zhao et al., 2008; Zhang et al., 2009), were upregulated in CA-yap+ transgenics, but not in DN-yap+, compared with control siblings (Fig. 3A,B). Because ctgf is duplicated in zebrafish (Fernando et al., 2010), we confirmed that the observed effect is ctgfa specific (supplementary material Fig. S3G). Upregulation of ctgfa in response to Yap activation was also shown through in situ hybridization in CA-yap transgenics versus siblings, upon single heat-shock (supplementary material Fig. S3H,I). In addition, we monitored ctgfa activation dynamics in vivo with a reporter line (ctgfa:eGFP; supplementary material Fig. S2A) coupled to Yap transgenics. We observed that by 7 hpHS, the expression of ctgfa was upregulated in 72 hpa blastemas of double transgenics ctgfa:eGFP; CA-yap+ compared with siblings (Fig. 3C,D,G). Conversely, in ctgfa:eGFP; DN-yap+ transgenics, ctgfa expression remained equal to controls, in agreement with qPCR results (Fig. 3E,F,H). Furthermore, evidence of Yap activation in double transgenics ctgfa:eGFP; CA-yap+ was obtained by performing immunofluorescence against Yap and GFP in the same fins used for in vivo experiments (supplementary material Fig. S3J-M). We observed that at 7 hpHS Yap was more nuclear in those fins, confirming the heat-shock efficiency. These results lead to the conclusion that the regeneration phenotypes observed in the functional transgenics are specific to Yap manipulation and that ctgfa is likely to be a direct target of Yap in zebrafish, as it is in higher vertebrates. Of note, ctgf can also be a transcriptional target of other pathways, namely WNT and TGFβ (Luo et al., 2004; Fujii et al., 2012). This possibly explains the observation that GFP expression of the ctgfa:eGFP reporter occurs even in mesenchymal regions where Yap is not active, indicating the existence of other means of ctgfa regulation not involving Yap (supplementary material Fig. S3J-K).
Fig. 3.
Transcriptional gene regulation induced by Yap. (A,B) qPCR determination of ctgfa and areg expression levels (relative to sibling controls) in blastemas of CA-yap positive (A) and DN-yap positive (B) transgenics upon single heat-shock at 72 hpa. *P<0.01; two-tailed, non-parametric paired Wilcoxon test, logarithmic scale, base 10. (C-F) Representative ctgfa expression in double transgenics ctgfa:eGFP; CA-yap/DN-yap and siblings upon heat-shock at 72 hpa, at 7 hpHS. (C) CA-yap control; (D) CA-yap positive; (E) DN-yap control; (F) DN-yap positive. n=5 fish/condition. Scale bars: 500 µm. Dashed lines indicate amputation plane. (G,H) Quantification of average eGFP intensity (in arbitrary units, a.u.) of individual rays including blastemas along the PD axis of double transgenics ctgfa:eGFP; CA-yap/DN-yap and siblings. (G) CA-yap positive and siblings; (H) DN-yap+ and siblings. n=80-90 rays, 5 fish/condition; shadows indicate the s.e.m. for each curve. (I,J) qPCR determination of fgf20a, msxb, wnt10a, wnt3a, lef1, dkk1a, dkk1b, shh, bmp2a and bmp2b expression levels (relative to sibling controls) in CA-yap positive (I) and DN-yap positive (J) transgenics upon single heat-shock at 72 hpa. *P<0.05, **P<0.01; two-tailed, non-parametric paired Wilcoxon test, logarithmic scale, base 10. All RNA extractions were performed at 2 hpHS. Mean±s.d. are shown.
To explore these observations further, we analyzed the expression of several genes involved in regeneration by performing qPCR upon heat-shock in Yap transgenics. We found that in CA-yap+ transgenics, a number of these factors were significantly downregulated, namely fgf20a, wnt10a, lef1 and shh, whereas dkk1b was highly upregulated (Fig. 3I). By contrast, DN-yap+ transgenics presented the opposite tendency of expression, with fgf20a upregulated (Fig. 3J). Of interest, msxb, bmp2a and bmp2b were unaltered upon Yap manipulations (Fig. 3I,J). This indicates that Yap has the ability to regulate a number of key regeneration factors. The regeneration impairment occurring in CA-yap and DN-yap transgenics is likely to be caused by a combination of changes in expression of those factors and cell proliferation.
Yap does not affect ctgfa expression in uncut fins
Fins grow throughout the life of adult fish, depending on a homeostasis process involving some of the genes necessary for epimorphic regeneration (Wills et al., 2008). To test whether in uninjured fins activation of Yap could lead to proliferation phenotypes related to those observed during regeneration, we applied the same heat-shock protocol as before (Fig. 2A) to both Yap transgenics and siblings in uncut situations. Remarkably, in uncut fins CA-yap induction led to no morphological changes (supplementary material Fig. S5A,B). Also, upregulation of ctgfa was not detected by 7 hpHS in uncut double transgenics ctgfa:eGFP; CA-yap (supplementary material Fig. S5C,D), indicating that CA-yap expression is not sufficient to stimulate the transcription of its target gene. This indicates the existence of a robust inhibitory mechanism for homeostatic proliferation in adult fins. The same results were observed in DN-yap uncut transgenics (supplementary material Fig. S5E-H). Interestingly, applying the same single heat-shock protocol to ctgfa:eGFP; CA-yap double transgenics in which only half of the caudal fin was amputated, ctgfa expression was upregulated by 7 hpHS, in contrast to the uncut half fin (supplementary material Fig. S5I,J).
The uninjured adult caudal fin tissue is known for its ability to silence transgenes (Thummel et al., 2006). To confirm that the previous results were not consequences of such an effect, we applied single heat-shocks in Yap transgenics with 72 hpa half fin amputations and performed yap1 in situ hybridization. In both CA-yap+ and DN-yap+ there was a clear yap1 upregulation upon transgene activation compared with yap1 expression observed in siblings (supplementary material Fig. S5K-N), both in amputated and uncut fins (supplementary material Fig. S5K′-N′,K″-N″). This indicates that Yap transgenes can be transcribed in uninjured, homeostatic situations, but fin cells appear to be in a non-responsive state. During epimorphic regeneration, however, cells appear to be prone to respond to Yap activity, causing specific phenotypes.
Cell density along the blastema associates with the localization of active Yap
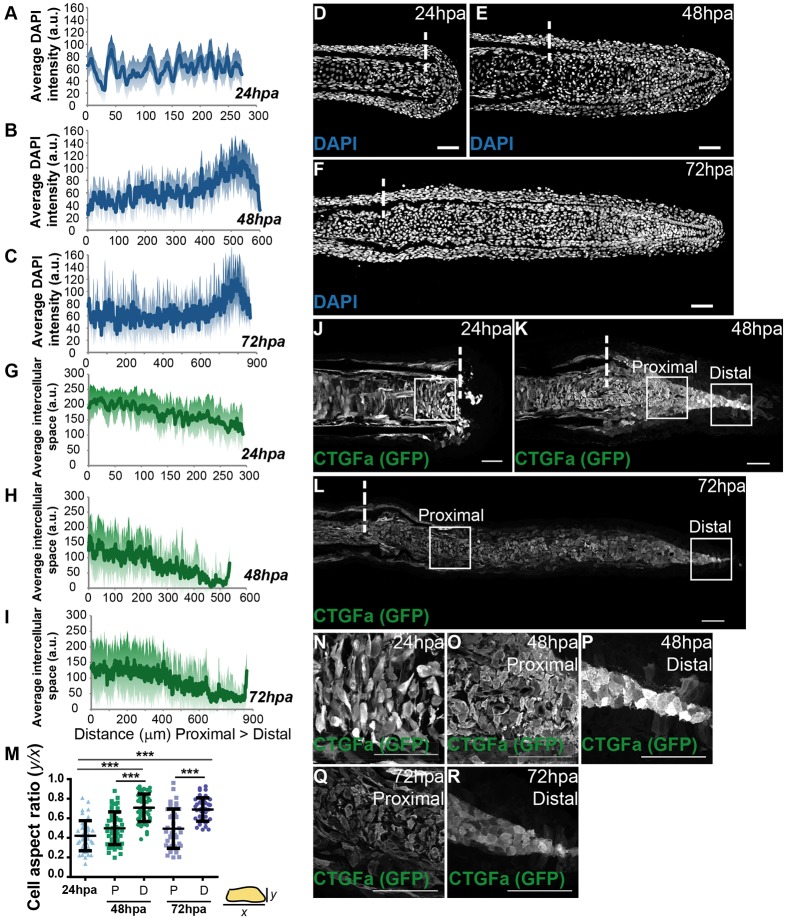
One possible explanation for the regulation of Yap intracellular dynamics in the blastema is that changes in cell matrix rigidity and/or cell density triggered by the amputation can modify tissue tension, thus affecting cell morphology as well as cell area (Zhao et al., 2007; Dupont et al., 2011; Wada et al., 2011; Aragona et al., 2013). These changes could be sensed at the cell membrane, through its junctions and cytoskeleton, controlling Yap activation and proliferation levels (Schlegelmilch et al., 2011; Fernández et al., 2011). To test this hypothesis, we established a cell density map along the blastema proximal-distal (PD) axis by measuring the average intensity of mesenchymal nuclei in different regeneration stages. We found that at 24 hpa cell density was homogeneous along the PD axis (Fig. 4A,D). Strikingly, at 48 and 72 hpa, when the blastema is complete, cell density was differential: higher in distal regions compared with proximal regions of the same samples (Fig. 4B,C,E,F). The high cell density regions correlated with distal areas where Yap is mainly cytoplasmic, thus mostly inactive, suggesting that cell density could control Yap inactivation through in vivo contact inhibition of proliferation (compare Fig. 4E,F with Fig. 1E,F). To determine whether these cell density changes translated into effects on cell morphology, we performed immunofluorescence against GFP in ctgfa:eGFP transgenics, as this marker labels all mesenchymal cells, at the same stages as before. We observed that at 24 hpa mesenchymal cells were variable in shape, presenting many protrusions, in agreement with their migratory phenotype during blastema formation (Fig. 4J,N). When quantifying the cell aspect ratio, which provides a measure of cell roundness (representing the relationship between their x and y axes), we found that 24 hpa mesenchymal cells were more elongated, exhibiting an average aspect ratio of 0.42 (Fig. 4M). Moreover, by quantifying the space between mesenchymal cells along the PD axis, we observed that 24 hpa cells were uniformly spread throughout the blastema (Fig. 4G). At 48 and 72 hpa, the distal mesenchymal cells became more compact (Fig. 4H,I,K,L,O-R), reflecting the density increase without a major change in cell size along the PD axis. Regarding morphology, these distal cells showed significantly less variability in shape, being rounder and lacking protrusions (average aspect ratio of 0.71 and 0.67, for 48 and 72 hpa, respectively), compared with cells of the corresponding proximal region (0.50 and 0.48, for 48 and 72 hpa, respectively) or 24 hpa blastemas (Fig. 4M). This correlation between cell density and cell morphology along the blastema suggests that cells adapt their shape to tension changes and that these mechanical cues might affect Yap activation.
Fig. 4.
Mesenchymal cell density and morphology are altered according to the regenerative stage. (A-C) Quantification of average DAPI intensity (in arbitrary units, a.u.) in mesenchymal cells along the PD axis of blastemas at 24 hpa (A), 48 hpa (B) and 72 hpa (C). n=7 sections, 3 fish/condition. (D-F) Representative DAPI-stained longitudinal sections of blastemas at 24 hpa (D), 48 hpa (E) and 72 hpa (F). (G-I) Quantification of average space (a.u.) between mesenchymal cells along the PD axis at 24 hpa (G), 48 hpa (H) and 72 hpa (I). n=9 sections; 3 fish/condition. (J-L) Representative anti-GFP-stained longitudinal sections of ctgfa:eGFP transgenics at 24 hpa (J), 48 hpa (K) and 72 hpa (L). (M) Quantification of average cell aspect ratio of mesenchymal cells at 24, 48 and 72 hpa, in which y is the minor axis of the cell and x the major cell axis. A perfect circular shape corresponds to a ratio between y and x of 1. P corresponds to proximal; D to distal regions. ***P<0.0001; two-tailed, non-parametric Mann–Whitney test. n=45 cells/condition; 5 cells randomly selected/image; 9 sections; 3 fish/condition. (N-R) High magnification images of the boxed areas in J-L highlight cell morphology of blastema cells at 24 hpa (N); 48 hpa proximally (O) and distally (P); and 72 hpa proximally (Q) and distally (R). Dashed lines indicate amputation plane. Scale bars: 50 µm. Medial blastema areas were considered for all measurements: shadows indicate the s.e.m. for each curve.
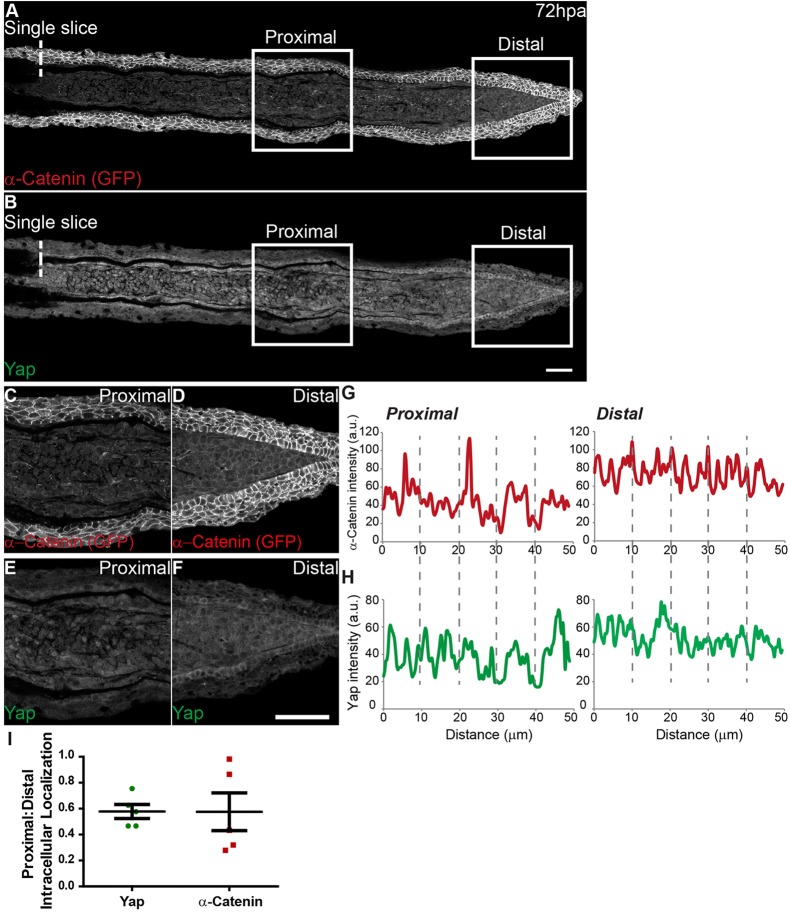
α-Catenin correlates with Yap intracellular localization
To find adhesion- and cytoskeleton-associated proteins that might act as cell density sensors in the blastema and mediators of Yap activation, we performed systematic immunohistochemistry in 48 and 72 hpa blastemas searching for expression differences along the PD axis (supplementary material Table S1). This led to the identification of several adhesion proteins present in specific cell types (supplementary material Fig. S6). Interestingly, α-Catenin was the only junctional protein localized in blastemas in a PD-dependent manner. By performing immunostainings against Yap and GFP in α-Catenin-Citrine protein trap transgenics (Žigman et al., 2010) at 72 hpa, we observed that expression of endogenous α-Catenin correlated with Yap intracellular dynamics (Fig. 5A,B). In particular, the junctional localization of α-Catenin in the distal, dense and round mesenchymal cells corresponds to areas where Yap is more cytoplasmic (Fig. 5D,F,G,H, Distal); conversely, in proximal areas where Yap is more nuclear and cells are sparse and protrusive, α-Catenin was not present (Fig. 5C,E,G,H, Proximal). Moreover, by quantifying intensity ratios between PD regions, we observed that 50% of Yap translocates from nucleus to cytoplasm. Interestingly, 50% of α-Catenin changes intracellular location when comparing proximal to distal blastema regions (Fig. 5I). This suggests that α-Catenin could constitute an in vivo mechanosensor of cell density during regeneration, owing to its apparent response to an increase in cell-cell contacts in the distal blastema, an area that corresponds to Yap inactivation.
Fig. 5.
α-Catenin accumulates in distal blastema regions where Yap is mainly cytoplasmic. (A-F) Representative immunofluorescence with anti-Yap and anti-GFP antibodies in 72 hpa longitudinal sections of α-Catenin transgenics. Owing to stronger expression intensity, transgenics were used instead of the α-Catenin antibody. (A) α-Catenin; (B) corresponding Yap expression. (C-F) High magnification images of the boxed areas in A,B showing α-Catenin (C, proximal; D, distal) and Yap (E, proximal; F, distal) expression. (G,H) Corresponding proximal and distal intensity profiles (in arbitrary units, a.u.) of the medial mesenchymal cells shown in C-F showing average intensity of α-Catenin (G) and Yap (H). (I) Quantification of changes in intracellular localization of Yap and α-Catenin across the PD axis by expressing a ratio between average intensities of proximal:distal Yap or α-Catenin of xz projections of respective mesenchymal cells. Mean±s.d. are shown. Dashed lines indicate amputation plane. n=5 sections, 3 fish. Scale bars: 50 µm.
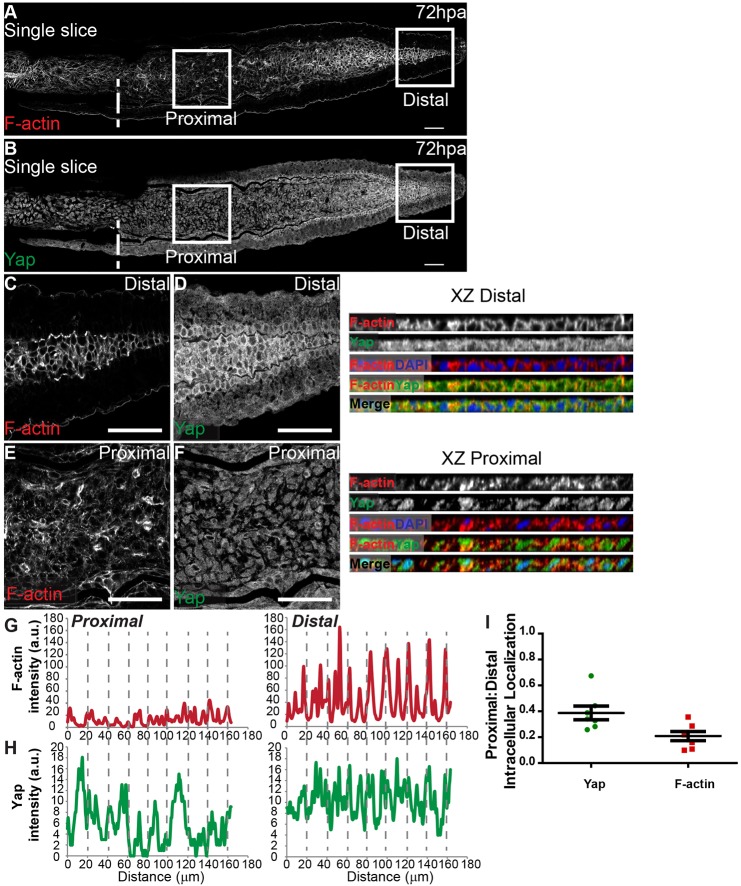
F-actin controls Yap activation
The cytoskeleton plays an active role in mediating mechanical forces to which cells are exposed, hence we investigated F-actin localization in the blastema. Phalloidin staining showed that F-actin also underwent changes along the PD axis in 72 hpa blastemas (Fig. 6A,B): it localizes to the cortex in distal cells, where cell density is higher and Yap more cytoplasmic (Fig. 6C,D,G,H, XZ Distal), whereas proximally, F-actin is present throughout the cell cytoplasm in areas of lower density and nuclear Yap (Fig. 6E,F,G,H, XZ Proximal). By quantifying F-actin and Yap PD intensity ratios, we observed that changes in F-actin intracellular location along the PD axis were even greater than those observed for Yap (Fig. 6I). F-actin also colocalized with junctional α-Catenin in distal blastemas (supplementary material Fig. S7A-F).
Fig. 6.
Differential PD expression of F-actin associates with Yap intracellular location. (A-F) Representative immunofluorescence with anti-Yap antibody and phalloidin (F-actin) in 72 hpa longitudinal sections. (A) F-actin; (B) corresponding Yap expression. (C-F) High magnification images of the boxed areas in A,B showing F-actin (C,E) and Yap (D,F) localization in proximal (E,F) and distal (C,D) regions. Single color and merged xz projections of distal (C,D) and proximal (E,F) blastemas highlight intracellular localization. (G,H) Corresponding proximal and distal intensity profiles (in arbitrary units, a.u.) of xz projections represented showing average intensity of F-actin (G) and Yap (H). (I) Quantification changes in intracellular localization of Yap and F-actin across the PD axis by expressing a ratio between average intensities of proximal:distal Yap or F-actin of xz projections of respective mesenchymal cells. Mean±s.d. are shown. Dashed lines indicate amputation plane. n=7 sections, 5 fish. Scale bars: 50 µm.
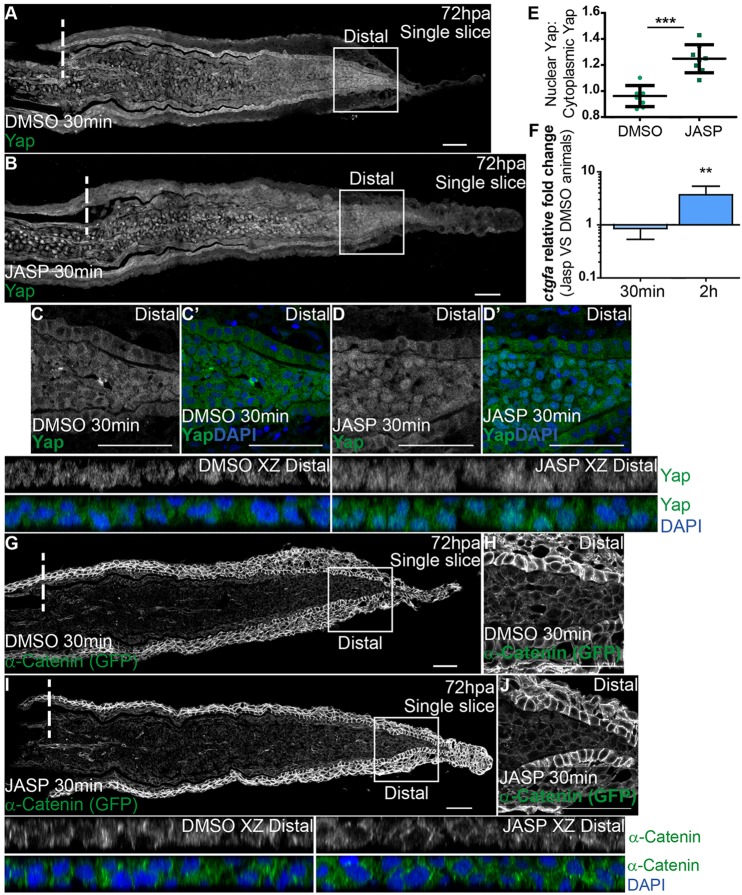
To determine whether F-actin is involved in regulating Yap activity in vivo during regeneration, we performed intraperitoneal injections of jasplakinolide (Jasp), an inducer of F-actin polymerization and stabilization (Bubb et al., 1994; Reddy et al., 2013), in 72 hpa α-Catenin transgenics. To confirm that Jasp injection was leading to F-actin interference, we performed immunostainings against phosphorylated Ezrin/Radixin/Moesin. This antibody was used because it detects F-actin similarly to phalloidin, with the benefit of not occupying the same binding site of Jasp (Bubb et al., 1994). In contrast to DMSO-injected controls, Jasp-injected fins lost the characteristic cortical F-actin localization in blastema distal tips, indicating that Jasp effectively acts on F-actin (supplementary material Fig. S7G-J). Next, we accessed Yap intracellular localization by immunostaining and observed that it was affected as early as 30 min after Jasp injection. Nuclear translocation of Yap was induced throughout the blastema, in contrast to DMSO-injected controls (Fig. 7A,B); this was particularly evident in distal regions of Jasp-treated animals (Fig. 7C-E, compare DMSO XZ Distal with JASP XZ Distal). To confirm that this Yap nuclear translocation had an impact on its activation state, we performed qPCR for its target gene, ctgfa, in Jasp- versus DMSO-injected animals. This analysis showed that at 30 min after Jasp injection, ctgfa transcription levels were not readily affected in the blastema; however, by 2 h post Jasp injection ctgfa was significantly upregulated compared with DMSO animals (Fig. 7F). This suggests that disrupting F-actin dynamics overrides the mechanical cues provided by high cell density in the blastema distal tip, exerting influence on Yap activation. We also found that distal accumulation of α-Catenin is not altered upon Jasp treatment (Fig. 7G-J), indicating that α-Catenin is not directly affected by F-actin manipulation in the blastema. Altogether, these observations show that F-actin is an upstream regulator of Yap in vivo, controlling its activation during regeneration.
Fig. 7.
F-actin controls Yap intracellular dynamics. (A-D′) Representative immunofluorescence with anti-Yap antibody in 72 hpa longitudinal sections of α-Catenin transgenics injected with jasplakinolide (JASP; B) and DMSO controls (A). (C-D′) High magnification images of the boxed areas in A,B showing Yap and DAPI in DMSO (C,C′) and JASP (D,D′) conditions. Corresponding xz projections of the distal blastemas shown in C,D highlight intracellular localization (DMSO XZ Distal, JASP XZ Distal). (E) Quantification of Yap intracellular localization by expressing a ratio between average intensities of nuclear Yap:cytoplasmic Yap of xz projections from distal blastemas in DMSO or JASP conditions, at 30 min post injection. Higher ratios correspond to higher intensities of nuclear Yap. ***P<0.001, two-tailed, non-parametric Mann–Whitney test. n=8 sections, 4 fish/condition. (F) qPCR determination of ctgfa levels in JASP versus DMSO animals, at 30 min and 2 h post injection, time points when RNA was extracted from blastemas. **P<0.01; two-tailed, non-parametric paired Wilcoxon test, logarithmic scale, base 10. (G-J) α-Catenin (anti-GFP) expression in animals injected with DMSO (G,H) or JASP (I,J). (H,J) High magnification images of the boxed areas in G,I showing α-Catenin in distal blastemas of animals injected with DMSO (H) or JASP (J). Corresponding xz projections of the distal blastemas shown in H,J highlight intracellular localization of α-Catenin and DAPI. Intraperitoneal injections were performed in 72 hpa animals, 30 min pre-fixation of blastemas. n=12 sections, 4 fish/condition. Mean±s.d. are shown. Dashed lines indicate amputation plane. Scale bars: 50 µm.
DISCUSSION
Our study identifies an in vivo mechanism that regulates Yap activity within the zebrafish caudal fin blastema based on cell density differences along the regenerating tissue. The mesenchymal cells seem to respond to a mechanotransduction process that involves changes in cell morphology, junction assembly and cytoskeleton remodeling, which together lead to a graded control of tissue growth via Yap, the Hippo pathway effector.
Once formed, the blastema exhibits spatial compartmentalization (Nechiporuk and Keating, 2002). We show that Yap intracellular localization associates with these areas and could account for the different levels of proliferation described. Remarkably, the same Yap dynamics also correlates with the degree of cell density along the blastema PD axis. In distal high cell density domains, Yap is mainly cytoplasmic (inactive), whereas in proximal lower cell density domains, Yap is largely nuclear (active). The functional relevance of Yap intracellular localization was confirmed by genetic manipulation: expression of a constitutively active form of Yap leads to more proliferation, whereas expression of a Yap dominant-negative construct results in the opposing phenotype. The effects of Yap manipulation are not only restricted to proliferation, but also influence the expression of several signaling factors. Importantly, the size of the blastema is reduced in both cases, indicating that Yap is on top of a complex network of tissue growth regulation during regeneration, not limited to proliferation control.
Our observations of different levels of cell density (i.e. confluence) in the blastema suggest a mechanism of contact inhibition of proliferation, which to our knowledge has not been described before in regeneration or other in vivo tissue repair contexts. Recently, in vitro studies have proposed that Yap activation is hierarchically controlled by different upstream cues depending on culture confluence (Aragona et al., 2013). Interestingly, in our in vivo system, the distal mesenchymal cells that exhibit high density and cytoplasmic Yap show multiple features that might contribute to Yap inactivation. These cells present rounder cell morphology, show α-Catenin buildup, have increased activated Mst1/2 and Lats1/2, and display changes in F-actin localization. This suggests that a combination of several mechanical and signaling mechanisms exist to inactivate Yap robustly in the distal blastema. The initial step in the cascade of events leading to Yap inactivation in the distal tip needs further investigation, but it is conceivable that it depends primarily on the increase of density and cell contacts. The cause for cells accumulating distally is as yet unclear, possibly being a structural consequence of the surrounding epithelial layers that impose physical constraints to the mesenchyme.
The membrane recruitment of α-Catenin as a consequence of high cell density, resembles the process described in the mouse epidermis (Schlegelmilch et al., 2011); importantly, it may lead to the cortical repositioning of F-actin that in turn drives Yap inactivation. Interestingly, if we consider that fewer substrate contacts with the ECM are a consequence of high cell density, in which blastema cells become rounder, this should also cause F-actin relocation to the cell cortex and exclusion of Yap from the nucleus. We can only speculate about the possible mechanical contribution of ECM stiffness towards Yap inactivation, but it is tempting to associate blastema high cell density zones with possible soft substrates in which there is low mechanical tension conferred by the ECM. This would enable the system to have multiple ways of directing F-actin to the cell cortex and consequently inactivating Yap distally (Gumbiner and Kim, 2014).
Importantly, we show for the first time that F-actin plays a major role in mediating the influence of the blastema environment on Yap subcellular distribution in vivo.
F-actin has been shown to act as a mechanotransducer in other systems (Romet-Lemonne and Jégou, 2013; Heisenberg and Bellaïche, 2013); therefore, it may be directly involved in sensing the mesenchymal cell density state and translating that into Yap activation. Additionally, α-Catenin may act upstream of F-actin, possibly as a primary mechanosensor that responds to density changes, as its recruitment in distal blastemas is unaffected by F-actin manipulation; nevertheless, further studies are necessary to understand this mechanism.
High cell density in the blastema appears to play an instructive role in the distal inactivation of Yap; however, it is unlikely that low cell density is the initial Yap activation trigger, but is rather a required permissive step. The first signs of Yap activation were observed at 6 hpa when cell migration towards the stump had not started; hence, blastema density is unlikely to be considerably lower than in the uncut mesenchymal tissue where Yap is inactive (Fig. 1A-C). Yap activation could be linked to the presence of F-actin in the mesenchyme or with secreted growth factors, such as WNT, which is activated soon after amputation and is known for interplay with the Hippo pathway (Imajo et al., 2012; Rosenbluh et al., 2012; Stoick-Cooper et al., 2007).
Our finding that Yap signaling appears to be inhibited in uninjured, fully differentiated caudal fins supports the idea that Yap has a specific function during regeneration, when there is a need for precise control of tissue growth. This apparent prerequisite of a less differentiated environment is consistent with observations in other systems, in which experimental manipulation of Yap leads to phenotypes in tissues that are not fully differentiated. This is the case during embryonic development, in adult contexts involving stem cell niches and in cancer models, all situations in which cell plasticity is enhanced (Hiemer and Varelas, 2013). This is suggestive of multiple levels of Yap regulation in differentiated tissues.
The coordination of a number of signaling pathways and morphogens during fin regeneration is essential to guarantee robustness in restoration of correct final size. Importantly, our work proposes that mechanical forces, conferred by heterogeneous cell densities within the blastema, are also crucial for regeneration. By a process of mechanotransduction, mediated by α-Catenin and F-actin, Yap regulation, and therefore blastema growth, are tightly balanced. This work thus reinforces a central physiological role for this Hippo pathway member in vivo.
MATERIALS AND METHODS
Ethics statement
All experiments were approved by the Animal User and Ethical Committees at Instituto Medicina Molecular and Instituto Gulbenkian Ciência, according to European Union directives and Portuguese law (Directive 2010/63/EU, Decreto-Lei 113/2013).
Zebrafish lines and fin amputation
All zebrafish (Danio rerio) lines used were maintained in a re-circulating system with a 14 h/day, 10 h/night cycle at 28°C. Experiments were performed in 3- to 9-month-old wild-type AB strain adults. All fin amputations were performed in fish anesthetized in 160 mg/ml MS-222 (Sigma) using a scalpel as described (Poss et al., 2000). Regeneration proceeded until defined time points at 33°C, except for heat-shock experiments. For those, transgenics and siblings were maintained at 28°C and heat-shocked once daily at 38°C for 1 h, by water bath incubation. Subsequently, fish were transferred to 28°C until desired time points. All live imaging was performed in anesthetized fish, with images acquired using a Zeiss V12-Lumar with a Zeiss digital camera. For transgenic generation, see supplementary Materials and Methods.
Chemical treatments
For jasplakinolide (Jasp) (Santa Cruz Biotechnology) treatments, 72 hpa ctnna-Citrine fish were injected intraperitoneally with 10 µl/g of 1 mM Jasp working solution (final concentration of 7.1 µg Jasp/g) in dimethyl sulfoxide (DMSO; Sigma). Fish were injected 30 min or 2 h prior to fixation using 30 G U-100 insulin syringes (BD Micro-Fine). Control fish were injected in parallel with an equivalent volume of DMSO. EdU injections were performed as described (Blum and Begemann, 2011) in 72 hpa CA-yap transgenics and siblings. Fish were injected 1 h prior to fixation as described above. Fins were fixed in 4% paraformaldehyde (Sigma) in PBS overnight and processed for cryosectioning, or were pooled for RNA extraction directly in Trizol reagent.
Total RNA isolation and quantitative real-time PCR (qPCR)
For all gene expression analyses, regenerated tissue from five caudal fins, including one ray segment of the stump, were harvested per experiment and pooled for each sample. Tissue from heat-shock transgenics and siblings was retrieved 2 h after completion of the heat-shock. Tissue from Jasp- and DMSO-injected animals was retrieved at 30 min or 2 h post injection. All samples were analyzed in biological and technical triplicate for each gene. RNA was extracted using Trizol reagent (Invitrogen) and treated with DnaseI (Roche) according to the manufacturer's protocol. cDNA was synthesized from 1 μg total RNA using the Transcriptor High Fidelity cDNA Synthesis Kit (Roche), following the oligo dT protocol. qPCR was performed using a Roche LightCycler 480 and FastStart Essential DNA Green Master Mix. Cyclic conditions were: 15 min at 95°C followed by 55 amplification cycles, each consisting of 30 s at 95°C, 15 s at 68°C. Gene expression values were normalized using the elongation factor 1α (ef1α, NM_131263; eef1a1l1 – Zebrafish Information Network) housekeeping gene and fold change was calculated using the ΔΔCt method. Results were plotted using GraphPad Prism software and two-tailed, non-parametric paired Wilcoxon tests were performed between the several conditions. Primer sequences are listed in supplementary material Table S2.
Antisense morpholino oligonucleotides (MOs) microinjections
A fluorescent standard Control-MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and a splicing block zYapE2I2 Vivo-MO (5′-GCAACATTAACAACTCACTTTAGGA-3′) (adapted from Hu et al., 2013) were purchased from Gene Tools, LLC and 6 ng of each morpholino were injected into 1-cell-stage wild-type embryos. Embryos were left to develop at 28°C until the desired stage and fixed in 4% paraformaldehyde at 4°C overnight.
Embryo heat-shock
Embryos collected from incrossing DN-yap siblings and incrossing DN-yap positive transgenics were placed into 50 ml falcons and heat activated in a bath at 37°C for 20 min. Following the heat-shock, embryos were transferred into petri dishes and left to develop at 28°C until the desired stage and fixed in 4% paraformaldehyde at 4°C overnight.
Image analysis
For all image analyses, maximum intensity z-stack projections were made using ImageJ software, except when noted. For concatenation of images along the PD axis of the same longitudinal section, the ImageJ plugin 3D Stitching was used. xz projections were performed with Dynamic Reslice in ImageJ, using synchronized channels. All error bars in graphs correspond to the standard deviation of the mean. For proliferation, Yap intracellular localization quantification, cell density, cell morphology and intercellular mesenchymal space measurements, see supplementary Materials and Methods.
In situ hybridization
Whole-mount in situ hybridizations in caudal fins and embryos were performed as described (Sousa et al., 2011; Thisse and Thisse, 2008). Genes were PCR cloned by TA overhangs in PGEM-T-easy (Promega) using primers (supplementary material Table S2) and 5 days post fertilization zebrafish total cDNA. cmlc2 probe was a gift (Yelon et al., 1999). Digoxygenin-labeled antisense RNA probes for all studied genes were synthesized as described (Henrique et al., 1995) (supplementary material Table S3). Images of in situ hybridizations were obtained with a Leica Z6APO stereomicroscope, using a Leica DFC490 digital camera or a Zeiss V12-Lumar with a Zeiss digital camera.
Immunofluorescence
This protocol was adapted from Mateus et al. (2012) with the following modifications: after overnight fixation with 4% paraformaldehyde, fins were saturated in 30% sucrose (Sigma) in PBS overnight, then embedded in 7.5% gelatin (Sigma)/15% sucrose in PBS and subsequently frozen in liquid nitrogen. Longitudinal sections were cut at 12 µm using a Microm cryostat and maintained at −20°C afterwards. Sections on slides were thawed for 15 min at room temperature, washed twice in PBS at 37°C for 10 min, washed once in 0.1 M glycine (Sigma) in PBS for 10 min, followed by acetone permeabilization and onwards as described. For EdU detection, directions from the kit Click-iT (C10637, Invitrogen) were followed. For TUNEL detection, directions from In situ Cell Death Detection Kit, Fluorescein (Roche) were followed. Cryosections were counterstained with DAPI (0.001 mg/ml in PBS, Sigma). For phalloidin staining (1:200, conjugated with Alexa Fluor 568, Invitrogen), upon fixation there was no methanol transfer and fins proceeded directly to PBS-30% sucrose. Sections were mounted with DAKO Fluorescent Mounting Media and imaged using a Zeiss LSM710 confocal microscope with a C-Apochromat 40× water objective. The antibodies used are listed in supplementary material Table S4.
Acknowledgements
We are grateful to Kenneth Poss for support and sharing Yap transgenics. We thank Lara M. Carvalho and Aida Barros for fish care; Lara C. Carvalho, Sara Sousa and Maria Gagliardi for reading the manuscript; and Telmo Pereira for data analysis.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
R.M. performed all experiments with the help of R.L., G.B., F.V. and A.F.; R.M., R.L., G.B. and A.J. conceived and designed experiments and performed data analysis; R.M. and G.B. established the ctgfa:eGFP line and Y.F. the hsp70:RFP-CAyap and hsp70:DNyap lines; R.M. and A.J. prepared the manuscript.
Funding
This work was supported by funding from Fundação para a Ciência e Tecnologia [SFRH/BD/62126/2009,PTDC/BEX-BID/1176/2012]; and Agence Nationale de la Recherche [ANR-11-BSV5-0021].
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.119701/-/DC1
References
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S. and Piccolo S. (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047-1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Azevedo A. S., Grotek B., Jacinto A., Weidinger G. and Saúde L. (2011). The regenerative capacity of the zebrafish caudal fin is not affected by repeated amputations. PLoS ONE 6, e22820 10.1371/journal.pone.0022820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum N. and Begemann G. (2011). Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development 116, 107-116. 10.1242/dev.065391 [DOI] [PubMed] [Google Scholar]
- Bubb M. R., Senderowicz A. M., Sausville E. A., Duncan K. L. and Korn E. D. (1994). Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 269, 14869-14871. [PubMed] [Google Scholar]
- Cai J., Zhang N., Zheng Y., de Wilde R. F., Maitra A. and Pan D. (2010). The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383-2388. 10.1101/gad.1978810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W.-Y., Gemberling M., Wang J., Holdway J. E., Shen M.-C., Karlstrom R. O. and Poss K. D. (2013). In vivo monitoring of cardiomyocyte proliferation to identify chemical modifiers of heart regeneration. Development 140, 660-666. 10.1242/dev.088526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole L. K. and Ross L. S. (2001). Apoptosis in the developing zebrafish embryo. Dev. Biol. 240, 123-142. 10.1006/dbio.2001.0432 [DOI] [PubMed] [Google Scholar]
- Dickmeis T., Plessy C., Rastegar S., Aanstad P., Herwig R., Chalmel F., Fischer N. and Strähle U. (2004). Expression profiling and comparative genomics identify a conserved regulatory region controlling midline expression in the zebrafish embryo. Genome Res. 14, 228-238. 10.1101/gr.1819204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A. and Pan D. (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120-1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. et al. (2011). Role of YAP/TAZ in mechanotransduction. Nature 474, 179-183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Fernández B. G., Gaspar P., Brás-Pereira C., Jezowska B., Rebelo S. R. and Janody F. (2011). Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337-2346. 10.1242/dev.063545 [DOI] [PubMed] [Google Scholar]
- Fernando C. A., Conrad P. A., Bartels C. F., Marques T., To M., Balow S. A., Nakamura Y. and Warman M. L. (2010). Temporal and spatial expression of CCN genes in zebrafish. Dev. Dyn. 239, 1755-1767. 10.1002/dvdy.22279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Toyoda T., Nakanishi H., Yatabe Y., Sato A., Matsudaira Y., Ito H., Murakami H., Kondo Y. and Kondo E. et al. (2012). TGF-β synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. J. Exp. Med. 209, 479-494. 10.1084/jem.20111653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H., Terai K., Nakajima H., Chiba A., Fukuhara S. and Mochizuki N. (2014). S1P-Yap1 signaling regulates endoderm formation required for cardiac precursor cell migration in zebrafish. Dev. Cell 31, 128-136. 10.1016/j.devcel.2014.08.014 [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. and Kim N.-G. (2014). The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 127, 709-717. 10.1242/jcs.140103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Tamura K. and Yokoyama H. (2014). Yap1, transcription regulator in the Hippo signaling pathway, is required for Xenopus limb bud regeneration. Dev. Biol. 388, 57-67. 10.1016/j.ydbio.2014.01.018 [DOI] [PubMed] [Google Scholar]
- Heisenberg C.-P. and Bellaïche Y. (2013). Forces in tissue morphogenesis and patterning. Cell 153, 948-962. 10.1016/j.cell.2013.05.008 [DOI] [PubMed] [Google Scholar]
- Henrique D., Adam J., Myat A., Chitnis A., Lewis J. and Ish-Horowicz D. (1995). Expression of a Delta homologue in prospective neurons in the chick. Nature 375, 787-790. 10.1038/375787a0 [DOI] [PubMed] [Google Scholar]
- Hiemer S. E. and Varelas X. (2013). Stem cell regulation by the Hippo pathway. Biochim. Biophys. Acta 1830, 2323-2334. 10.1016/j.bbagen.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Hu J., Sun S., Jiang Q., Sun S., Wang W., Gui Y. and Song H. (2013). Yes-Associated Protein (Yap) is required for early embryonic development in zebrafish (Danio rerio). Int. J. Biol. Sci. 9, 267-278. 10.7150/ijbs.4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K. and Pan D. (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421-434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Imajo M., Miyatake K., Iimura A., Miyamoto A. and Nishida E. (2012). A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 31, 1109-1122. 10.1038/emboj.2011.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaźwińska A., Badakov R. and Keating M. T. (2007). Activin-betaA signaling is required for zebrafish fin regeneration. Curr. Biol. 17, 1390-1395. 10.1016/j.cub.2007.07.019 [DOI] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S. et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713-724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Kujawski S., Lin W., Kitte F., Börmel M., Fuchs S., Arulmozhivarman G., Vogt S., Theil D., Zhang Y. and Antos C. L. (2014). Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev. Cell 28, 573-587. 10.1016/j.devcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Lee Y., Grill S., Sanchez A., Murphy-Ryan M. and Poss K. D. (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173-5183. 10.1242/dev.02101 [DOI] [PubMed] [Google Scholar]
- Lee Y., Hami D., De Val S., Kagermeier-Schenk B., Wills A. A., Black B. L., Weidinger G. and Poss K. D. (2009). Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 331, 270-280. 10.1016/j.ydbio.2009.05.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Q., Kang Q., Si W., Jiang W., Park J. K., Peng Y., Li X., Luu H. H., Luo J., Montag A. G. et al. (2004). Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J. Biol. Chem. 279, 55958-55968. 10.1074/jbc.M407810200 [DOI] [PubMed] [Google Scholar]
- Mahoney W. M., Hong J.-H., Yaffe M. B. and Farrance I. K. G. (2005). The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem. J. 388, 217-225. 10.1042/BJ20041434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus R., Pereira T., Sousa S., de Lima J. E., Pascoal S., Saúde L. and Jacinto A. (2012). In vivo cell and tissue dynamics underlying zebrafish fin fold regeneration. PLoS ONE 7, e51766 10.1371/journal.pone.0051766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesfeld J. B. and Link B. A. (2014). Establishment of transgenic lines to monitor and manipulate Yap/Taz-Tead activity in zebrafish reveals both evolutionarily conserved and divergent functions of the Hippo pathway. Mech. Dev. 133, 177-188. 10.1016/j.mod.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H. (1901). Regeneration. New York: Macmillan Company; London, Macmillan Company. [Google Scholar]
- Nechiporuk A. and Keating M. T. (2002). A proliferation gradient between proximal and msxb-expressing distal blastema directs zebrafish fin regeneration. Development 129, 2607-2617. [DOI] [PubMed] [Google Scholar]
- Oh H. and Irvine K. D. (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081-1088. 10.1242/dev.015255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19, 491-505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perathoner S., Daane J. M., Henrion U., Seebohm G., Higdon C. W., Johnson S. L., Nüsslein-Volhard C. and Harris M. P. (2014). Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 10, e1004080 10.1371/journal.pgen.1004080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poleo G., Brown C. W., Laforest L. and Akimenko M.A. (2001). Cell proliferation and movement during early fin regeneration in zebrafish. Dev. Dyn. 221, 380-390. 10.1002/dvdy.1152 [DOI] [PubMed] [Google Scholar]
- Poss K. D., Shen J. and Keating M. T. (2000). Induction of lef 1 during zebrafish fin regeneration. Dev. Dyn. 219, 282-286. [DOI] [PubMed] [Google Scholar]
- Reddy P., Deguchi M., Cheng Y. and Hsueh A. J. W. (2013). Actin cytoskeleton regulates hippo signaling. PLoS ONE 8, e73763 10.1371/journal.pone.0073763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romet-Lemonne G. and Jégou A. (2013). Mechanotransduction down to individual actin filaments. Eur. J. Cell Biol. 92, 333-338. 10.1016/j.ejcb.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Rosenbluh J., Nijhawan D., Cox A. G., Li X., Neal J. T., Schafer E. J., Zack T. I., Wang X., Tsherniak A., Schinzel A. C. C. et al. (2012). β-catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell 151, 1457-1473. 10.1016/j.cell.2012.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Ruiz L., Santamaría J. A. and Becerra J. (2005). Cytoskeletal dynamics of the teleostean fin ray during fin epimorphic regeneration. Differentiation 73, 175-187. 10.1111/j.1432-0436.2005.00016.x [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K., Mohseni M., Kirak O., Pruszak J., Rodriguez J. R., Zhou D., Kreger B. T., Vasioukhin V., Avruch J., Brummelkamp T. R. et al. (2011). Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell 144, 782-795. 10.1016/j.cell.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa S., Afonso N., Bensimon-Brito A., Fonseca M., Simões M., Leon J., Roehl H., Cancela M. L. and Jacinto A. (2011). Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138, 3897-3905. 10.1242/dev.064717 [DOI] [PubMed] [Google Scholar]
- Staley B. K. and Irvine K. D. (2010). Warts and yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 20, 1580-1587. 10.1016/j.cub.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S. and Stankunas K. (2012). Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev. Biol. 365, 339-349. 10.1016/j.ydbio.2012.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Gomez A. W., Armstrong B. E., Henner A. and Stankunas K. (2014). Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 6, 482-498. 10.1016/j.celrep.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoick-Cooper C. L., Weidinger G., Riehle K. J., Hubbert C., Major M. B., Fausto N. and Moon R. T. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479-489. 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- Sugiyama M., Sakaue-Sawano A., Iimura T., Fukami K., Kitaguchi T., Kawakami K., Okamoto H., Higashijima S.-I. and Miyawaki A. (2009). Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl. Acad. Sci. USA 106, 20812-20817. 10.1073/pnas.0906464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C. and Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69. 10.1038/nprot.2007.514 [DOI] [PubMed] [Google Scholar]
- Thummel R., Burket C. T. and Hyde D. R. (2006). Two different transgenes to study gene silencing and re-expression during zebrafish caudal fin and retinal regeneration. ScientificWorldJournal 6 Suppl. 1, 65-81. 10.1100/tsw.2006.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K.-I., Itoga K., Okano T., Yonemura S. and Sasaki H. (2011). Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907-3914. 10.1242/dev.070987 [DOI] [PubMed] [Google Scholar]
- Wehner D., Cizelsky W., Vasudevaro M. D., Ozhan G., Haase C., Kagermeier-Schenk B., Röder A., Dorsky R. I., Moro E., Argenton F. et al. (2014). Wnt/β-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 6, 467-481. 10.1016/j.celrep.2013.12.036 [DOI] [PubMed] [Google Scholar]
- Wills A. A., Kidd A. R., Lepilina A. and Poss K. D. (2008). Fgfs control homeostatic regeneration in adult zebrafish fins. Development 135, 3063-3070. 10.1242/dev.024588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Qi H., Li Y., Pei J., Barton J., Blackstad M., Xu T. and Tao W. (2002). LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene 21, 1233-1241. 10.1038/sj.onc.1205174 [DOI] [PubMed] [Google Scholar]
- Yagi R., Chen L.-F., Shigesada K., Murakami Y. and Ito Y. (1999). A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551-2562. 10.1093/emboj/18.9.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D., Horne S. A. and Stainier D. Y. R. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23-37. 10.1006/dbio.1999.9406 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ji J.-Y., Yu M., Overholtzer M., Smolen G. A., Wang R., Brugge J. S., Dyson N. J. and Haber D. A. (2009). YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 11, 1444-1450. 10.1038/ncb1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L. et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747-2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C.-Y., Chinnaiyan A. M. et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962-1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žigman M., Trinh L. A., Fraser S. E. and Moens C. B. (2010). Zebrafish neural tube morphogenesis requires Scribble-dependent oriented cell divisions. Curr. Biol. 21, 79-86. 10.1016/j.cub.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]