Abstract
The special AT-rich DNA binding protein (SATB2) is a nuclear matrix-associated protein and an important transcription factor for biological development, gene regulation and chromatin remodeling. Aberrant regulation of SATB2 has been found to highly correlate with various types of cancers including lung, colon, prostate, breast, gastric and liver. Recent studies have revealed that a subset of small non-coding RNAs, termed microRNAs (miRNAs), are important regulators of SATB2 function. As post-transcriptional regulators, miRNAs have been found to have fundament importance maintaining normal cellular development. Evidence suggests that multiple miRNAs, including miR-31, miR-34, miR-182, miR-211, miR-599, are capable of regulating SATB2 in cancers of the lung, liver, colon and breast. This review examines the molecular functions of SATB2 and miRNAs in the text of cancer development and potential strategies for cancer therapy with a focus on systemic miRNA delivery.
Special AT-rich DNA binding protein (SATB2) plays important roles in biological development including cell differentiation as well as disease development. However, altered SATB2 expression has been linked to many forms of cancer. This study will highlight the interrelationship between microRNAs and SATB2 in cancer development.
Introduction
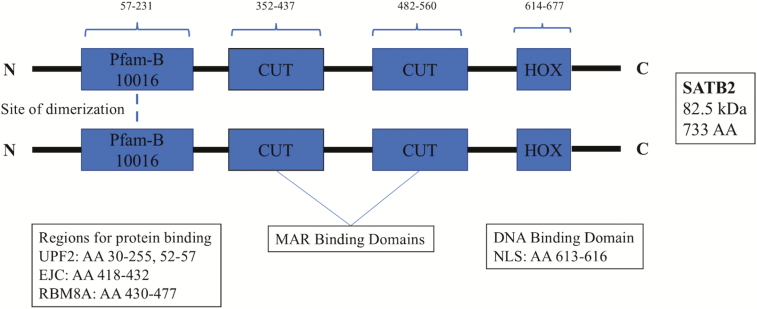
Gene expression is a tightly regulated, complex process, which ensures the efficient and specific functionality of the different cells that make up an organism. One of the means by which gene expression is regulated during biological processes such as embryonic development or long-term disease pathogenesis is by DNA–protein interactions (1). The special AT-rich DNA binding protein (SATB2) is an 82.5 kDa protein composed of 733 amino acids (Figure 1) (2,3). This protein is transcribed from a 191 kb gene consisting of 11 exons and is located in a gene-poor region along the long arm of chromosome 2 (2q32-2q33) (2,4). Transcription occurs telomerically and can produce one variant via alternative splicing (2,4). Originally identified as KIAA1034, this protein was renamed after it was found to be strikingly similar to the matrix/scaffold attachment region protein SATB1 (1,5). Both SATB1 and SATB2 are members of the CUT superclass of homeobox genes that are known to act as transcription factors and general transcriptional regulators during development (6,7). The CUT superclass can be further divided into three classes: CUX, ONECUT and SATB. The SATB class includes SATB1 and SATB2, which are highly divergent from the other members of the CUT superclass (6,8). The two SATB proteins are structurally similar, with each containing two CUT domains upstream of a homeodomain (2,6,8). Although all three of these major motifs function as DNA binding sites, studies on homeobox proteins have demonstrated that the CUT domains and homeodomains function synergistically to increase DNA binding affinity (7). In addition, there is a large structural region that is ~81% conserved between the two proteins that corresponds to a Pfam-B_10016 domain (2). Interestingly, this domain has been detected in all orthologous SATB proteins and is necessary for the formation of functional dimers (2,9). The SATB proteins are also unique because in addition to serving as transcription factors, they also are major regulators of gene expression by interacting with chromatin to alter higher order structures at matrix/scaffold attachment regions within the nucleus (5). These SATB-induced changes in chromatin structure have effects on global gene expression and help naive cells to differentiate and mature during development (1,5,10).
Figure 1.
Linearized SATB2 protein structure. The linearized protein structure demonstrates site of dimerization and binding domains.
Function of SATB2
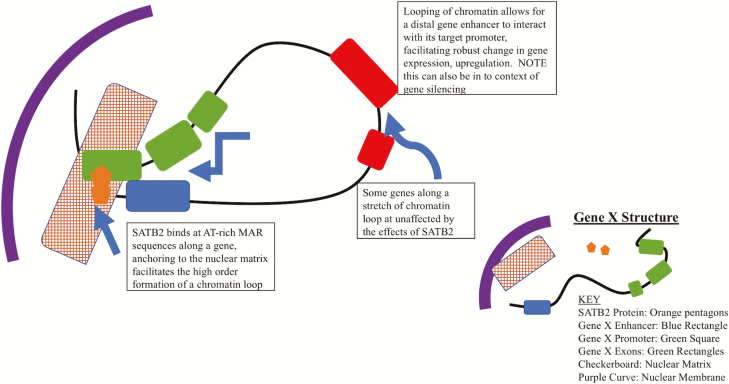
Originally identified as the cleft palate gene, patients presenting cleft palate have been found to be SATB2 haploinsufficient due to a transcription-interrupting sequence translocation (2,4,11). Further research demonstrated that SATB2 is necessary for embryonic development. Studies have shown that there is temporal- and spatial-dependent expression of SATB2 in developing mouse embryos that lead to proper cranial–facial formation throughout the gestational time frame (3,12). From a molecular standpoint, SATB2 binds to nuclear matrix attachment regions and is involved in transcriptional regulation as well as chromatin remodeling (5,10,13). The nuclear matrix is one of the key structures found in the cell nucleus and functions to (i) support the integrity of the entire nuclear structure (similar to a cytoskeleton) and (ii) provide anchoring regions for DNA to facilitate transcription (14). The organization and structure of the nuclear matrix are highly ordered to expedite and maintain efficient gene expression and may be responsible for the differential gene expression and chromatin structure observed in cancer and normal cells (15). Adding to the complexity of gene expression, the nuclear matrix and the anchoring of DNA to matrix attachment regions provide spatiality and topography to DNA regions (16). Positioning studies have discovered that in the nuclear space, regions of DNA containing AT-rich sequences are almost always found to be located on the periphery of the nuclear chromatin, whereas GC-rich regions, which are hallmarks of promoter structure, are found internally in nuclear chromatin (14,17). Expression of genes at the nuclear matrix is carried out by the formation of complex 3D structures of chromatin, dubbed chromatin loops. Chromatin loops are a level of genomic organization that allows for the spatial connection of DNA regions that are far from one another (18). Along with looping, the attachment of matrix attachment regions to the matrix alters the state of supercoiling along DNA, making regions more accessible to DNA binding proteins and subsequent physical bending and twisting to interact with other cis-regions of DNA (16,19). The formation of higher order chromatin loops at the nuclear matrix can provide 3D interaction between a gene promoter and a distal enhancer to carry out robust gene expression (Figure 2) (16) or between a gene region and an insulator to block gene expression (20). In the developing central nervous system, along with other developing tissues, SATB2 has been found to be associated with a specific chromatin-remodeling complex. Studies have demonstrated that SATB2 forms a complex and binds sequences, which are AT rich, anchoring them to the nuclear matrix to facilitate higher order chromatin remodeling and fine-tuned gene expression (5). This is found in developing cells and most likely is linked to the differential gene expression that is necessary for naive cellular differentiation.
Figure 2.
SATB2-induced chromatin looping provides topological organization and enhanced gene expression. The figure shows how SATB2 binds to the MAR sequence along a gene and facilitates chromatin looping, which brings distal genomic loci into close spatial proximity.
Role of SATB2 in cancer
The complexity of cancer biology and tumor development makes the identification of specific therapeutic targets a difficult task. Tumors are known to be a heterogeneous population of cells, characterized by their unchecked growth, evasion of anti-growth signals and apoptosis, as well as the ability to migrate and invade other regions of the body through the lymphatic and circulatory systems (21,22). Within this heterogeneous tumor are cancer stem cells (CSCs), which are thought to be the archetypal cell driving the continued progression of malignant cancers. Similar to embryonic stem cells, CSCs have the ability to differentiate and have a high capacity for self-renewal (23). This makes this cell type perhaps deadlier than ‘mature cancer cells’ within a tumor. The standard CSC model is based on four major premises: (i) tumor heterogeneity is hierarchical and is often similar to the tissue origin of cancer in question; (ii) the hierarchy of tumor biology is fueled by CSCs, rather than mature cancer cells; (iii) CSCs have strong ties to their identity and drive tumor development, as studies have shown that mature cancer cells have a limited capacity to induce tumor growth in xenograft models (23,24). This also demonstrates limited plasticity within the tumor; and (iv) CSCs are very resistant to traditional cancer therapies, causing secondary and recurring cancers to occur following treatment (23). Studies in tumor biology have identified the reactivation and strong expression of many embryonic genes in a variety of cancers, which further supports the CSC theory (23). Genome-wide DNA methylation profiling of colorectal stem and cancer cells also demonstrated promoter-specific hypermethylation and subsequent gene silencing (25). Because of the detection of embryonic gene reactivation in cancer, this has been a topic of great interest in studies involving the mechanisms of tumorigenesis.
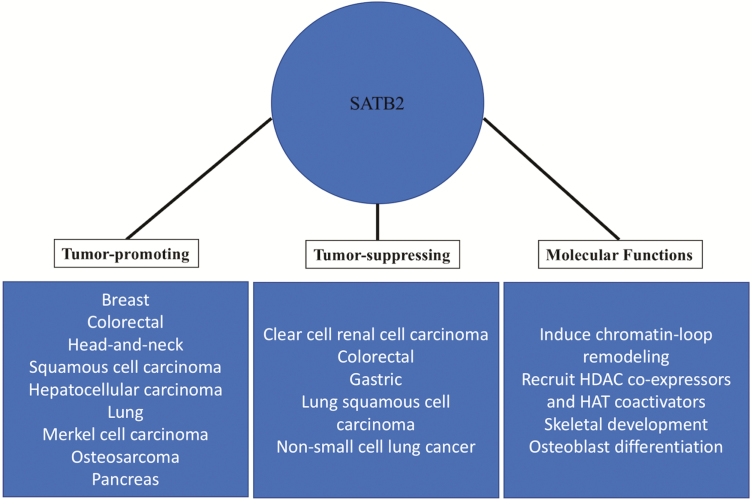
SATB2 is known to function as a both tumor suppressor and promoter (Figure 3). As a strong predictor of colorectal cancer (CRC), SATB2 serves as a diagnostic marker for this type of cancer (26–28). CRC is one of the most predominant and lethal forms of cancer in the world, with ~1.4 million new cases being diagnosed and 700 000 associated deaths per year (29). Survival rates for CRC depend on the form and stage at detection, as late stage, and more aggressive cancers are difficult to treat; however, increased screening for prevention has improved early-stage intervention. There is a need for the development and validation of biomarkers for early detection, so that therapeutic interventions and prognoses for CRC can be positively affected. Utilizing immunostaining and other molecular-based detection methods, it has been demonstrated that SATB2 and another protein, cytokeratin 20, are present in almost 95% of all CRC (29). Importantly, SATB2 protein expression is highly enriched in CRC CSCs and mature cells, based on published research and supported by data from The Cancer Genome Atlas (https://cancergenome.nih.gov/) (29,30). In adult tissues, one of the only regions where SATB2 is expressed is the upper gut where there is a frequent need for renewal of the intestinal lining (30–32). A study involving SATB2’s role in the development of CRC based on CSC involvement demonstrated that SATB2 plays a major role in maintaining the pluripotency of CSCs as they drive cancer progression. Promoter analysis of five major stem cell factors, OCT4, c-MYC, NANOG, SOX2 and KLF4, revealed SATB2 binding sites, suggesting that SATB2 binding may facilitate the re-expression of these genes, which maintain stemness, during CRC development and progression (33). As stated earlier, the expression of SATB2 is necessary for proper development, and its expression in adult tissues is essentially non-existent, with exceptions being in the cerebral cortex region of the brain, as noted by data compiled in the human protein atlas (https://www.proteinatlas.org/ENSG00000119042-SATB2/tissue) (3). SATB2 is also involved in the wingless/integrated (WNT) signaling pathway, serving as an upstream activator. This critical signaling pathway is prominent in development and is dysregulated in cancer (33,34). In CRC models, it has been found that aberrant expression of SATB2 can activate the WNT pathway. WNT signaling brought about by SATB2 expression directly mediated the activation of the β-catenin/TCF-LEF pathway and upregulated malignant cellular transformation, as well as metastatic properties. Some of these properties include the upregulation of epithelial–mesenchymal transition-associated genes, SNAIL, SLUG, ZEB1, as well as a shift from E-cadherin expression to N-cadherin (33,35). These hallmarks of epithelial-mesenchymal transition are thought to be a precursor to metastasis and identify a direct pathway in which SATB2 aberrantly upregulates stem cell-associated genes.
Figure 3.
Dual role of SATB2 in carcinogenesis and its molecular functions. The figure summarizes the molecular functions of SATB2 as well as its oncogenic and tumor-suppressive roles under different cancer contexts.
MicroRNAs
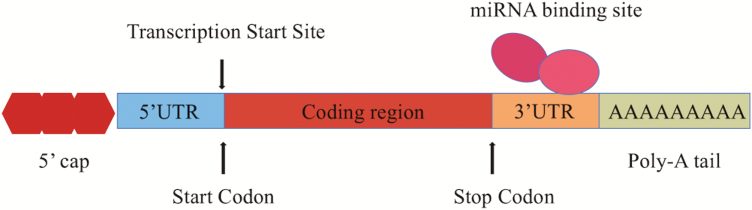
Epigenetic control of gene expression pathways by microRNAs (miRNAs) has been of increasing interest because these trans-acting factors show promise for therapeutic delivery and effect (36). MiRNAs were first discovered in Caenorhabditis elegans in 1993 and are predicted to account for 1–5% of the human genome (37–39). These molecules are highly conserved, long single-stranded non-coding RNAs that are 22–26 nucleotides in length and are involved in post-transcriptional gene silencing (40–42). MiRNAs are able to induce mRNA degradation and inhibit subsequent protein translation by binding to the 3′-untranslated region (UTR) of target messenger RNA (mRNA), as shown in Figure 4. Due to an imperfect short binding recognition sequence, each miRNA has the potential to bind hundreds of target mRNAs. In plants, miRNAs have few targets because of their extended complementarity (43,44). This is in direct contrast with animals, whose miRNAs can have multiple targets and together regulate global changes in gene expression. The lack of specific complementarity between mRNAs and miRNAs in animals resides in a short seven-nucleotide sequence of complementarity (43). Specifically, the seven nucleotides on the target mRNA can base pair to positions 2–8 of the miRNA at the 5′-end, also known as the ‘seed’ (39). It is important to note that animal miRNAs have significant diversification because the ‘seed’ region can readily change; thus, miRNA targets can be easily amassed and lost (45). In addition, evidence suggests that miRNA expression can be influenced by changes in transcriptional control, epigenetics, as well as through errors in miRNA biogenesis (36,46,47). The consequences of dysregulated miRNAs also depend on the type and function of the target gene affected. Target genes are categorized into three main groups based on level of activity upon miRNA binding: switch, tuning and neutral (40,43). Switch genes are those that are turned off by miRNAs, tuning genes are unaffected by miRNA regulation, and neutral genes have no particular impact on the cell. In other words, miRNAs may target a multitude of genes, but the effects may be inconsequential if the genes regulated are considered tuning or neutral. Therefore, it is important to sift through the target gene pool to uncover switch genes to understand the exact roles of individual miRNAs.
Figure 4.
Illustration of miRNA binding site on SATB2 mRNA. The illustration depicts SATB2 mRNA and miRNA interaction. SATB2 has multiple binding sites for miRNAs, which are exclusively found in the 3′-UTR.
Dysregulation of key miRNAs can influence important oncogenic or tumor suppressor gene expression patterns and promote malignant cellular transformation (48–50). Abnormal expression levels of miRNAs have been reported in multiple types of cancers, including colon, lung, gastric and breast (51). Specifically, changes in miRNA copy number and or gene location have been shown to promote sustained cell proliferation, evasion of apoptosis, angiogenesis and activation of invasion and metastasis (36). In the context of SATB2, it is important to take into consideration both cellular context and degree of miRNA regulation. For example, although miR-31 has been shown to regulate SATB2 in multiple types of cancer tissues including lung, breast and colorectal, miR-211 has only been shown to be deregulated in liver cancer (52–55). In addition to tissue-dependent miRNA regulation, it is also important to determine the degree of which each miRNA regulates SATB2 expression. In other words, although SATB2 can be regulated by a diverse pool of miRNAs, future studies are needed to determine which and why some miRNAs may have more extensive effects than others.
MicroRNA and SATB2 function
With exception to the cerebral cortex, colon, rectum, appendix, hippocampus and caudate, SATB2 is usually not expressed in adult tissues. Thus, most studies on the biological functions of SATB2 with consideration to miRNA regulation are limited to its role in osteogenic differentiation. Studies performed using murine bone marrow stromal cells showed that overexpression of SATB2 can lead to differential expression of a myriad of miRNAs, 10 downregulated and 18 upregulated (56). GOs and KEGG pathway analyses showed that these miRNAs were involved in various biological processes including osteogenic pathways (WNT signaling and MAPK pathways), mesenchymal cell differentiation and skeletal development (56). Specifically, findings suggest that miR-27a and SATB2 are inversely correlated and that they are able to regulate the same set of genes involved in osteogenesis including BMP2, BMPR1A and Smad9. Furthermore, during MC3T3-E1 osteoblast differentiation, ~60 miRs were found upregulated. Among these deregulated miRs was the miR-23a~27a~24-2 cluster, which have been shown to be associated with both normal hematopoietic differentiation and leukemia (57–60). Researchers have found that Runx2, a prominent transcriptional regulator for bone formation, negatively regulated the expression of these miRNAs, and interestingly, each of these miRs targeted SATB2 during osteogenesis by binding to its 3′-UTR. A logical reasoning behind the upregulation of these miRs is to prevent sustained bone formation and differentiation (57,61,62). Other miRNAs found to inhibit SATB2 during osteogenic differentiation include miR-34 (63) and miR-205 (64). SATB2 is also known to play an important role in osteogenic differentiation of bone mesenchymal stem cells (BMSCs). BMSCs have the ability to differentiate into various types of cells including adipocytes, chondrocytes and osteoblasts, making them suitable candidates for stem cell therapy (64–66). Recent studies reveal that osteogenic differentiation of BMSCs can be regulated by miRNAs via SATB2 repression (55,63,64,67). Specifically, bioinformatics analysis and luciferase reporter assays performed by Hu et al. demonstrated that miR-205, a known tumor suppressor, is able to bind to SATB2 3′-UTR and repress its expression (64,68). These studies suggest that not only can miRNAs target multiple genes, but also a single gene can correspondingly be targeted by multiple miRNAs. The following section summarizes all currently available studies on specific miRNAs and their regulation of SATB2 in different types of cancers (Table 1).
Table 1.
Summary of existing literature on the role of SATB2 and miRNAs in carcinogenesis
| MicroRNA | Cancer type | Direction of regulation | SATB2 direction of regulation | Cell type | Reference |
|---|---|---|---|---|---|
| miR-31 | Lung | Down | Up | BEAS-2B | (52) |
| miR-31 | Endometrial | Down | Up | CAFs | (72) |
| miR-31 | Breast | Down | Up | Triple-negative breast cancer tissues and BCa MDA-MB-231, HBL- 100, MCF-7 | (54) |
| miR-31 | Colorectal | Up | Down | SW480, DLD-1 | (55) |
| miR-449a | Colorectal | Down | Up | CT26 | (76) |
| miR-182 | Colorectal | Up | Down | 293T, DLD-1, HCT116, SW480, SW260, Lovo | (67) |
| miR-34 | Colorectal | Down | Down | Human CRC tissues, HT-29, Colo-320, SW480, W620, HCT-15 | (85) |
| miR-875 | Lung | Up | Down | H157, A549 | (88) |
| miR-599 | Lung | Up | Down | Human lung tissues, A549, A427 | (97) |
| miR-211 | Liver | Down | Up | HepG2, SMMC7721 | (53) |
MicroRNA regulation of SATB2 in carcinogenesis
MicroRNA-31 is one of the most abundant miRNAs and is commonly studied in disease development (69). Studies suggest that miR-31 is a prominent regulator of SATB2 (70). In stomach, ovarian, prostate and breast cancers, miR-31 is found downregulated, whereas colorectal, liver and head-and-neck tumors demonstrate upregulated miR-31 expression (71). Arsenic, a potent environmental carcinogen, has also been found to repress miR-31 expression and subsequently promote the translation of SATB2 protein in human bronchial epithelial (BEAS-2B) cells (52). This study suggests that both upregulation of SATB2 and suppression of miR-31 are correlated with the malignant transformation of BEAS-2B cells. Furthermore, exogenous expression of miR-31 led to reduced SATB2 expression as well as reduced cell migration, invasion and anchorage-independent growth. Additionally, cancer-associated fibroblasts (CAFs) have been found to excrete excess extracellular matrix and alter the tumor microenvironment, which contributes to tumor rigidity (71). One study compared the differences in miRNA expression between normal endometrial fibroblasts and CAFs. Results suggest that miR-31 was most significantly downregulated, whereas SATB2 was upregulated in CAFs (72). Further results indicate that miR-31 and SATB2 mainly contributed to cell migration, suggesting a role in metastatic capacity of tumor cells.
Triple-negative breast cancer is a subtype of breast cancer (BCa), characterized by a lack of estrogen receptor, progesterone receptor and human epithelial growth factor receptor-2 (HER2) expression on the surface of tumor cells (54). To investigate the prognostic value of miR-31 in BCa, a study led by Luo et al. examined its expression level in triple-negative breast cancer tissues and BCa MDA-MB-231, HBL-100 and MCF-7 cell lines. Results reveal significant downregulation of miR-31 in both biopsies and cell lines. In addition, overexpression of miR-31 demonstrated reduced cell migration and invasion, but no change in cell proliferation (54). These data suggest that rather than affecting cellular growth, miR-31 principally acts as an anti-metastatic miRNA in BCa (54). Furthermore, a bioinformatic analysis and luciferase reporter assay revealed SATB2 as a direct target of and inversely correlated with miR-31. SATB2 is potentially oncogenic, and its upregulation in BCa cell lines reinforces this idea, demonstrating the gene’s ability to promote cell proliferation, migration and invasion. Taken together, these results suggest that miR-31 functions to reduce cell migration and invasion in BCa cells by suppressing SATB2 expression, whereas cell proliferation induced by SATB2 may be independent of miR-31 regulation.
MiRNAs in general act more as a double-edge sword, as depending on tissue type or environmental situation, these small non-coding RNAs can be beneficial or detrimental to gene expression and regulation (73). In contrast to the above studies, miR-31 upregulation and SATB2 downregulation are correlated with increased CRC aggressiveness (55). MiR-31 overexpression and SATB2 downregulation in SW480 and DLD-1 cells demonstrated increased cellular migration, invasive capacity and anchorage-independent growth. In addition, Balb/C-nu/nu athymic nude mice injected with miR-31 overexpressing SW480 cells experienced a significantly higher rate of tumor formation and metastasis when compared with SW480 control.
CRC is the third most common type of cancer (74–76), and several studies have linked poor disease prognosis and metastasis with SATB2 expression (55,67,76,77). It is important to note that although colon and rectal cancers have entirely separate etiologies, most studies tend to group them together. In an effort to discover an upstream regulator of SATB2 in CRC, Sun et al. examined several known cancer-related miRNAs in CT26 cells (76). Data suggest that compared with prominent tumor suppressor miRNAs, miR-143/145, miR-449a was most significantly reduced in CT26 cells (78,79). Previous studies have indicated that miR-449a can downregulate several types of cancers including lung and colon cancers, by suppressing cell survival, migration, invasion and proliferation (80–84). Experiments using multiple CRC cell lines have demonstrated decreased miR-499a and increased SATB2 mRNA expression, suggesting an inverse relationship. To examine the effect of miR-499a upregulation and subsequent SATB2 downregulation, the researchers constructed HCT116s overexpressing miR-499a. The results of this study suggest that overexpression of miR-499a is associated with diminished cell proliferation, stimulation of apoptosis and reduced tumor volume (76). On the other hand, using human embryonic kidney 293T cells as well as human CRC cell lines, DLD-1, HCT116, SW480, SW260 and Lovo, one study demonstrated that increase in miR-182 and decrease in SATB2 resulted in enhanced cell proliferation, migration and invasion (67). The opposing direction of SATB2 regulation in CRC may be partly due to the difference in cell lines used or the difference in the degree of cell transformation.
Another miRNA involved in regulating SATB2 in CRC is miR-34c-5p, a subtype of miR-34c (85). Although the study suggests that SATB2 is significantly reduced in CRC tissues, immunohistochemistry assays showed differing SATB2 expression, varying from negative to strongly positive, although most resulted in negative to weakly positive expression levels. Moreover, despite demonstrating the inverse relationship between miR-34 and SATB2, both were found downregulated in CRC tissues, which suggests that miR-SATB2 interaction may be more intricate that seemed.
MiRNAs have also been shown to play an important role in the development of non-small cell lung cancer (NSCLC) through the regulation of key target genes (86–95). Tumor biopsies from NSCLC patients have been found to highly express miR-875-5p, which acts as a tumor suppressor in prostate cancer (96). In addition, the study reported increased H157 and A549 cell migration and invasion, as well as reduced SATB2 expression at the protein level (88). In a similar study, Tian et al. found expression of miR-599 was significantly upregulated in NSCLC patients (97). In vitro experimentation using the NSCLC cell lines A549 and A427 further confirmed the correlation between overexpression of miR-599 and increased cell proliferation, invasion and migration. In addition, results from luciferase reporter assays suggest that miR-599 is able to bind to the 3′-UTR of SATB2 and thereby reduce SATB2 mRNA translation (97). Data from RT-PCR indicate that miR-599 overexpression can also significantly reduce SATB2 mRNA levels. This evidence suggests that miR-599 can bind to SATB2 mRNA, induce its mRNA degradation and, in turn, promote proliferation and invasion of NSCLC cells.
One final example of miR-associated SATB2 activity in cancer is the association of miR-211 with SATB2 in liver cancer. In hepatocellular carcinoma, miR-211 was found significantly reduced, whereas SATB2 expression elevated (53). Overexpression of SATB2 and repression of miR-211 were correlated with increased cell proliferation and migration in HepG2 and SMMC7721 cells. To examine the relationship between miR-211 and SATB2, miR-211 mimics were ectopically expressed, leading to less cellular growth and invasion through inhibition of SATB2 expression. These data were further supported by luciferase reporter assays, which illustrate direct binding between miR-211 and the 3′-UTR of SATB2.
To date, none of the 18 differentially regulated miRNAs found in SATB2-overexpressed BMSCs were found to play a role in carcinogenesis. This seems to indicate that the miRNAs responsible for regulating SATB2 during normal osteogenic differentiation are most likely different from those found to induce cancer development. However, this assumption will require further experimental validation especially because miR-34 has been shown to regulate both osteogenic differentiation and CRC by targeting SATB2 (63).
Exogenous miRNA delivery
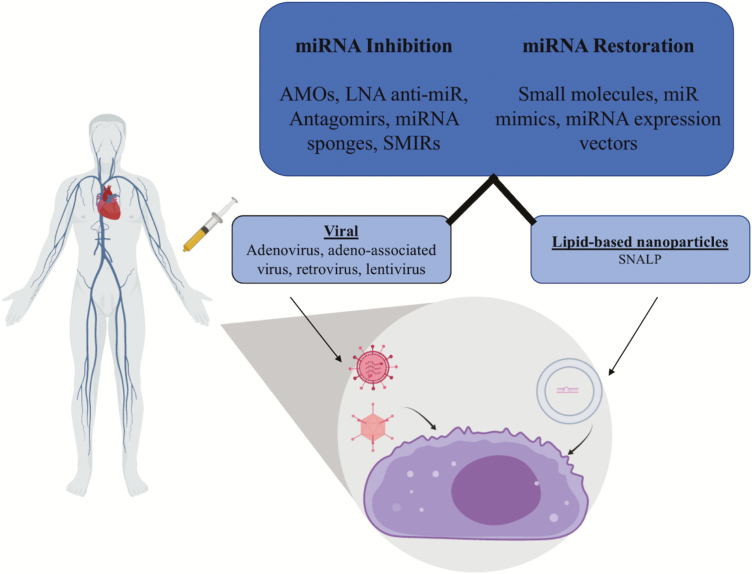
Research into the identification of the mechanisms and targets of miRNAs in gene regulation have led to the concept that exogenous microRNA delivery could be a potential therapeutic intervention strategy to combat the development and progression of cancer. The development of nano-delivery systems can improve the stability and transport of future drugs through systemic circulation to desired target sites (Figure 5) (98). Furthermore, sequence specificity of miRNAs to aberrantly expressed mRNAs provide a more direct and efficient targeting mechanism for therapeutic effect (99). The development and testing of viral delivery or lipid-based nanoparticles have been shown to have limited to no toxicity and thus could provide a strong delivery system for nano-based therapeutics (88,98,100). Conventional liposomal nanocapsules have been shown to have efficient chemistry in regards to the positive development of the therapeutic index of nucleic acid delivery. However, in vivo roadblocks, including low encapsulation efficiency, instability in biological fluids and toxicity issues have demonstrated the necessity for a more efficient and effective delivery system (101). A biological advantage to using engineered lipid-based particles is the ability to manipulate their chemistry to react in the cellular environment to allow for maximum effect, utilizing cellular pH or xenobiotic response molecules (88). A study utilizing a novel delivery system, stable nucleic acid lipid particles, to deliver miR-34a mimetics demonstrated an effective means of nucleic acid delivery to tumor tissue in vivo, evident by inhibition of tumor growth in a mouse model (102). This was defined in molecular terms by disruption of undesirable ERK and Akt signaling, which, when left unchecked, provide molecular stimulation for unchecked growth and survival of cancer cells (103). In addition, the miR-34 family can be induced by the TP53 protein, which is a classic tumor suppressor gene (104). Bypassing mutations or epigenetics alterations to TP53 with direct administration of miR-34 mimetics is a potential intervention strategy to a set of cancers in which cell cycle checkpoints are dysregulated. Importantly, it was also described that the delivery of miR-34 mimetics by stable nucleic acid lipid particles could reduce the capacity for aberrant WNT signaling (105), which was described previously to be regulated by SATB2 expression. Further understanding of SATB2-associated pathways, specific anti-sense oligonucleotide binding efficiency, as well as vector packaging and delivery will aid in the development of more efficient therapeutic strategies.
Figure 5.
Illustration of miRNA delivery. miRNA delivery can be used to either inhibit target mRNAs or restore miRNA supply. The figure illustrates two prominent ways, viral and lipid-based nanoparticles, of systemic miRNA delivery in the human body.
Engineered viral delivery is another method by which miRNAs could be delivered as a therapeutic strategy. Recombinant viral vectors expressing three major types of miR interactors have been tested for efficacy: miR mimetics, antagomirs and miR sponges. These small molecules contain many miR binding sites, which allow for molecular trapping, sequestration and functional inhibition (106). Modification of viruses such as lentivirus and retrovirus may allow for the long-term expression of microRNA in cells, provoking therapeutic effect. Studies involving microRNA delivery via many different virus types have demonstrated effective expression of these miRNAs, which can influence the expression and modulation of cellular state. One study utilized an engineered retrovirus to deliver miR-138 into mouse embryonic fibroblast cells. Effective viral delivery allowed for the transcription of this microRNA, which enhanced the pluripotency of stem cells, demonstrating a positive effect with limited toxicity in vitro (107). This was accomplished by the activity of exogenous miR-138, which targets the 3′-UTR of p53 and inhibits the proliferation of stem cells (107). A second study in animal model used lentivirus to transduce miR-15a/16, which improved gene expression and improved the prognosis of chronic lymphocytic leukemia in this model (108). These methods of miR delivery appear highly effective because of the transcriptional machinery that viruses use when involved in infection. However, one major concern is the insertion of virally expressed sequences into the patient’s genome, which could cause widespread genomic instability and potential activation and enhancement of oncogenic genes, which could lead to a worse prognosis or the development of secondary cancers or immune deficiency (109). Thus, the development of safe particles that are highly specific toward a cell type or disease is imperative for providing a safe genetic therapeutic effect.
One potential method of administration of SATB2-interacting miRNA is through the use of engineered standard retrovirus. The retrovirus genus consists of a number of species, including lentivirus. Interestingly, a standard retrovirus (not including lentivirus) specifically infects actively dividing cells (110). This mechanism could be utilized to target cancer cells, and perhaps in particular CSCs of which a hallmark is unchecked cell division (21). It also limits the potential off-target effects brought about by circulation of a virus in the body. The use of retroviruses to deliver both miRNA and miRNA inhibitors has been very successful in the laboratory setting (107). Engineering safe retroviruses that can act to amplify and deliver miRNAs that target SATB2 could help to improve prognosis by downregulation of some of the aberrantly activated embryonic and growth signals that are upregulated by SATB2. Limiting SATB2-induced stemness in cancer cells could allow for combination therapy (including surgery on some solid tumors) that could, in turn, limit the biological load of active cancer cells in the body. Retroviral delivery poses some concerns and thus must be studied and engineered to ensure safe manipulation of the genome. The most pressing concern is the ability of retrovirus to integrate directly into the host genome. This insertion into the patient genome could provide some detrimental level of mutagenesis and, for example, lead to prolonged oncogene activation (109,111). This, of course, would be contradictory to the therapeutic purpose of the retroviral therapy and is the reason why most current research into viral engineering for therapy has focused on the use of lentiviral vectors (111).
Conclusion
It is clear that SATB2 is aberrantly transcribed in many cancer types, though its downstream effects on gene expression and the inhibition or enhancement of different cancers are extremely complex. In addition, the complexity of these target effects, as well as potential off-target effects of miRNA expression, is a major concern when connecting the mechanistic approach to SATB targeting to that of a cancer therapy. It is evident that much more work needs to be done to validate SATB2 as a target and marker of cancer development, and to gain more insight into its targets and specific effects on regulation. In addition, this review also presents detailed evidence for the duality of SATB2’s involvement in cancer, mainly in the context of miRNA pathways. A summary of currently available studies shows that SATB2 is capable of acting as both tumor suppressor and promoter in the same or different cancer types. For example, SATB2 has been found to both enhance and suppress CRC (67,76). However, it is important to note that SATB2 was regulated by two distinct miRNAs: miR-449a and miR-182, in the two studies. This suggests that even in the same type of cancer, SATB2 may be regulated by a multitude of miRNAs and pathways, which further highlights the complexity of SATB2’s involvement in cancer. Future study is needed to identify the many roles that SATB2 plays in regards to tissue-specific cancers, and how targeting this gene or associated pathways will affect systemic expression of SATB2 in the hopes of developing a safe and effective biomarker or therapeutic intervention strategy.
Funding
This research was funded by the following NIH grants: ES000260, ES022935, ES023174, and ES02613.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- Bca
breast cancer
- BMSCs
bone mesenchymal stem cells
- CAF
cancer-associated fibroblasts
- CRC
colorectal cancer
- CSC
cancer stem cell
- mRNA
messenger RNA
- NSCLC
non-small cell lung cancer
- SATB2
special AT-rich DNA binding protein 2
- UTR
untranslated region
References
- 1. Szemes M., et al. (2006) Isolation and characterization of SATB2, a novel AT-rich DNA binding protein expressed in development- and cell-specific manner in the rat brain. Neurochem. Res., 31, 237–246. [DOI] [PubMed] [Google Scholar]
- 2. FitzPatrick D.R., et al. (2003) Identification of SATB2 as the cleft palate gene on 2q32-q33. Hum. Mol. Genet., 12, 2491–2501. [DOI] [PubMed] [Google Scholar]
- 3. Zhao X., et al. (2014) The role of SATB2 in skeletogenesis and human disease. Cytokine Growth Factor Rev., 25, 35–44. [DOI] [PubMed] [Google Scholar]
- 4. Bengani H., et al. ; UK10K Consortium (2017) Clinical and molecular consequences of disease-associated de novo mutations in SATB2. Genet. Med., 19, 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Britanova O., et al. (2005) Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci., 21, 658–668. [DOI] [PubMed] [Google Scholar]
- 6. Bürglin T.R., et al. (2002) Loss and gain of domains during evolution of cut superclass homeobox genes. Int. J. Dev. Biol., 46, 115–123. [PubMed] [Google Scholar]
- 7. Harada R., et al. (1994) Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J. Biol. Chem., 269, 2062–2067. [PubMed] [Google Scholar]
- 8. Bürglin T.R. (2011) Homeodomain subtypes and functional diversity. Subcell. Biochem., 52, 95–122. [DOI] [PubMed] [Google Scholar]
- 9. Galande S., et al. (2001) SATB1 cleavage by caspase 6 disrupts PDZ domain-mediated dimerization, causing detachment from chromatin early in T-cell apoptosis. Mol. Cell. Biol., 21, 5591–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gyorgy A.B., et al. (2008) SATB2 interacts with chromatin-remodeling molecules in differentiating cortical neurons. Eur. J. Neurosci., 27, 865–873. [DOI] [PubMed] [Google Scholar]
- 11. Brewer C.M., et al. (1999) A locus for isolated cleft palate, located on human chromosome 2q32. Am. J. Hum. Genet., 65, 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dobreva G., et al. (2006) SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell, 125, 971–986. [DOI] [PubMed] [Google Scholar]
- 13. Dickinson L.A., et al. (1992) A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell, 70, 631–645. [DOI] [PubMed] [Google Scholar]
- 14. Rynearson A.L., et al. (2011) Nuclear structure, organization, and oncogenesis. J. Gastrointest. Cancer, 42, 112–117. [DOI] [PubMed] [Google Scholar]
- 15. Lever E., et al. (2010) The role of nuclear organization in cancer. J. Pathol., 220, 114–125. [DOI] [PubMed] [Google Scholar]
- 16. von Kries J.P., et al. (1991) A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell, 64, 123–135. [DOI] [PubMed] [Google Scholar]
- 17. Georgiev G.P., et al. (1991) A. E. Braunstein Plenary Lecture. Nuclear skeleton, DNA domains and control of replication and transcription. Eur. J. Biochem., 200, 613–624. [DOI] [PubMed] [Google Scholar]
- 18. Kadauke S., et al. (2009) Chromatin loops in gene regulation. Biochim. Biophys. Acta, 1789, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luchnik A.N., et al. (1985) DNAaseI-hypersensitive minichromosomes of SV40 possess an elastic torsional strain in DNA. Nucleic Acids Res., 13, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan H., et al. (2018) The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res., 28, 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 22. Krizhanovsky V., et al. (2009) Stem cells: the promises and perils of p53. Nature, 460, 1085–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Batlle E., et al. (2017) Cancer stem cells revisited. Nat. Med., 23, 1124–1134. [DOI] [PubMed] [Google Scholar]
- 24. Schatton T., et al. (2009) Identification and targeting of cancer stem cells. Bioessays, 31, 1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanley M.P., et al. (2017) Genome-wide DNA methylation profiling reveals cancer-associated changes within early colonic neoplasia. Oncogene, 36, 5035–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dragomir A., et al. (2014) The role of SATB2 as a diagnostic marker for tumors of colorectal origin: results of a pathology-based clinical prospective study. Am. J. Clin. Pathol., 141, 630–638. [DOI] [PubMed] [Google Scholar]
- 27. Yu W., et al. (2017) SATB2/β-catenin/TCF-LEF pathway induces cellular transformation by generating cancer stem cells in colorectal cancer. Sci. Rep., 7, 10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y.J., et al. (2018) SATB2 is a promising biomarker for identifying a colorectal origin for liver metastatic adenocarcinomas. EBioMedicine, 28, 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Magnusson K., et al. (2011) SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am. J. Surg. Pathol., 35, 937–948. [DOI] [PubMed] [Google Scholar]
- 30. Berg K.B., et al. (2017) SATB2 as an immunohistochemical marker for colorectal adenocarcinoma: a concise review of benefits and pitfalls. Arch. Pathol. Lab. Med., 141, 1428–1433. [DOI] [PubMed] [Google Scholar]
- 31. Lindskog C., et al. (2014) Analysis of candidate genes for lineage-specific expression changes in humans and primates. J. Proteome Res., 13, 3596–3606. [DOI] [PubMed] [Google Scholar]
- 32. Wu F., et al. (2016) SATB2 expression increased anchorage-independent growth and cell migration in human bronchial epithelial cells. Toxicol. Appl. Pharmacol., 293, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu W., et al. (2017) Cellular transformation of human mammary epithelial cells by SATB2. Stem Cell Res., 19, 139–147. [DOI] [PubMed] [Google Scholar]
- 34. Calvo R., et al. (2000) Embryonic genes in cancer. Ann. Oncol., 11 (suppl. 3), 207–218. [DOI] [PubMed] [Google Scholar]
- 35. Brabletz T., et al. (2018) EMT in cancer. Nat. Rev. Cancer, 18, 128–134. [DOI] [PubMed] [Google Scholar]
- 36. Peng Y., et al. (2016) The role of microRNAs in human cancer. Signal Transduct. Target. Ther., 1, 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macfarlane L.A., et al. (2010) MicroRNA: biogenesis, function and role in cancer. Curr. Genomics, 11, 537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rajewsky N. (2006) L(ou)sy miRNA targets? Nat. Struct. Mol. Biol., 13, 754–755. [DOI] [PubMed] [Google Scholar]
- 39. Rajewsky N. (2006) MicroRNA target predictions in animals. Nat. Genet., 38 (suppl.), S8–S13. [DOI] [PubMed] [Google Scholar]
- 40. Bartel D.P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 41. Kim V.N. (2005) MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol., 6, 376–385. [DOI] [PubMed] [Google Scholar]
- 42. Kim V.N. (2005) Small RNAs: classification, biogenesis, and function. Mol. Cells, 19, 1–15. [PubMed] [Google Scholar]
- 43. Flynt A.S., et al. (2008) Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet., 9, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rhoades M.W., et al. (2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- 45. Berezikov E. (2011) Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet., 12, 846–860. [DOI] [PubMed] [Google Scholar]
- 46. Han L., et al. (2007) DNA methylation regulates microRNA expression. Cancer Biol. Ther., 6, 1284–1288. [DOI] [PubMed] [Google Scholar]
- 47. Saito Y., et al. (2006) Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle, 5, 2220–2222. [DOI] [PubMed] [Google Scholar]
- 48. Ben-Hamo R., et al. (2015) MicroRNA regulation of molecular pathways as a generic mechanism and as a core disease phenotype. Oncotarget, 6, 1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Monk M., et al. (2001) Human embryonic genes re-expressed in cancer cells. Oncogene, 20, 8085–8091. [DOI] [PubMed] [Google Scholar]
- 50. Sotiropoulou G., et al. (2009) Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA, 15, 1443–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ji W., et al. (2010) Targeting microRNAs in cancer gene therapy. Genes, 8, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen Q.Y., et al. (2018) Role of miR-31 and SATB2 in arsenic-induced malignant BEAS-2B cell transformation. Mol. Carcinog., 57, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang G., et al. (2015) miR-211 suppresses hepatocellular carcinoma by downregulating SATB2. Oncotarget, 6, 9457–9466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luo L.J., et al. (2016) MiR-31 inhibits migration and invasion by targeting SATB2 in triple negative breast cancer. Gene, 594, 47–58. [DOI] [PubMed] [Google Scholar]
- 55. Yang M.H., et al. (2013) Elevated microRNA-31 expression regulates colorectal cancer progression by repressing its target gene SATB2. PLoS One, 8, e85353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gong Y., et al. (2014) MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol. Cell. Biochem., 387, 227–239. [DOI] [PubMed] [Google Scholar]
- 57. Hassan M.Q., et al. (2010) A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl Acad. Sci. USA, 107, 19879–19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang S., et al. (2008) Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int. J. Cancer, 123, 972–978. [DOI] [PubMed] [Google Scholar]
- 59. Liu T., et al. (2009) MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett., 273, 233–242. [DOI] [PubMed] [Google Scholar]
- 60. Mertens-Talcott S.U., et al. (2007) The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res., 67, 11001–11011. [DOI] [PubMed] [Google Scholar]
- 61. Gaur T., et al. (2010) Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol., 340, 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li Z., et al. (2009) Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem., 284, 15676–15684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wei J., et al. (2012) miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J. Cell Biol., 197, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hu N., et al. (2015) Regulative effect of Mir-205 on osteogenic differentiation of bone mesenchymal stem cells (BMSCs): possible role of SATB2/Runx2 and ERK/MAPK pathway. Int. J. Mol. Sci., 16, 10491–10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bianco P., et al. (2001) Bone marrow stromal stem cells: nature, biology, and potential applications. Stem Cells, 19, 180–192. [DOI] [PubMed] [Google Scholar]
- 66. Ding X., et al. (2013) The sonic hedgehog pathway mediates brain plasticity and subsequent functional recovery after bone marrow stromal cell treatment of stroke in mice. J. Cereb. Blood Flow Metab., 33, 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang M.H., et al. (2014) MicroRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J. Transl. Med., 12, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Elgamal O.A., et al. (2013) Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS One, 8, e76402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Stepicheva N.A., et al. (2016) Function and regulation of microRNA-31 in development and disease. Mol. Reprod. Dev., 83, 654–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brocato J., et al. (2015) SATB1 and 2 in colorectal cancer. Carcinogenesis, 36, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stuelten C.H., et al. (2010) miR-31 in cancer: location matters. Cell Cycle, 9, 4608–4609. [DOI] [PubMed] [Google Scholar]
- 72. Aprelikova O., et al. (2010) The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts. Cell Cycle, 9, 4387–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kwan J.Y., et al. (2016) The complexity of microRNAs in human cancer. J. Radiat. Res., 57 (suppl. 1), i106–i111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fearon E.R. (2011) Molecular genetics of colorectal cancer. Annu. Rev. Pathol., 6, 479–507. [DOI] [PubMed] [Google Scholar]
- 75. Jemal A., et al. (2011) Global cancer statistics. CA. Cancer J. Clin., 61, 69–90. [DOI] [PubMed] [Google Scholar]
- 76. Sun X., et al. (2017) miR-449a inhibits colorectal cancer progression by targeting SATB2. Oncotarget, 8, 100975–100988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang S., et al. (2009) Down-regulated expression of SATB2 is associated with metastasis and poor prognosis in colorectal cancer. J. Pathol., 219, 114–122. [DOI] [PubMed] [Google Scholar]
- 78. Bauer K.M., et al. (2012) Effects of the miR-143/-145 microRNA cluster on the colon cancer proteome and transcriptome. J. Proteome Res., 11, 4744–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Borralho P.M., et al. (2011) miR-143 overexpression impairs growth of human colon carcinoma xenografts in mice with induction of apoptosis and inhibition of proliferation. PLoS One, 6, e23787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ding M., et al. (2015) MicroRNA-449a suppresses non-small cell lung cancer. Cell Biochem. Biophys., 71, 1255–1259. [DOI] [PubMed] [Google Scholar]
- 81. Luo W., et al. (2013) MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS One, 8, e64759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ren X.S., et al. (2014) Tumor-suppressive microRNA-449a induces growth arrest and senescence by targeting E2F3 in human lung cancer cells. Cancer Lett., 344, 195–203. [DOI] [PubMed] [Google Scholar]
- 83. Ye W., et al. (2014) MiR-449a functions as a tumor suppressor in endometrial cancer by targeting CDC25A. Oncol. Rep., 32, 1193–1199. [DOI] [PubMed] [Google Scholar]
- 84. Zhou Y., et al. (2014) MicroRNA-449a reduces cell survival and enhances cisplatin-induced cytotoxicity via downregulation of NOTCH1 in ovarian cancer cells. Tumour Biol., 35, 12369–12378. [DOI] [PubMed] [Google Scholar]
- 85. Gu J., et al. (2018) SATB2 targeted by methylated miR-34c-5p suppresses proliferation and metastasis attenuating the epithelial-mesenchymal transition in colorectal cancer. Cell Prolif., 51, e12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. He B., et al. (2016) Bioinformatics and microarray analysis of miRNAs in aged female mice model implied new molecular mechanisms for impaired fracture healing. Int J Mol Sci 17, 1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huang T., et al. (2016) MicroRNA-186 suppresses cell proliferation and metastasis through targeting MAP3K2 in non-small cell lung cancer. Int. J. Oncol., 49, 1437–1444. [DOI] [PubMed] [Google Scholar]
- 88. Lin X., et al. (2017) Nanoparticle delivery of miR-34a eradicates long-term-cultured breast cancer stem cells via targeting C22ORF28 directly. Theranostics, 7, 4805–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mizuguchi Y., et al. (2016) Dysregulated miRNA in progression of hepatocellular carcinoma: a systematic review. Hepatol. Res., 46, 391–406. [DOI] [PubMed] [Google Scholar]
- 90. Pan J.Y., et al. (2017) miR-134: a human cancer suppressor? Mol. Ther. Nucleic Acids, 6, 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sun C.C., et al. (2016) Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids, 5, e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sun C.C., et al. (2016) MicroRNA-346 facilitates cell growth and metastasis, and suppresses cell apoptosis in human non-small cell lung cancer by regulation of XPC/ERK/Snail/E-cadherin pathway. Aging (Albany, NY), 8, 2509–2524. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93. Sun C.C., et al. (2016) Hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. Oncotarget, 7, 21510–21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sun C.C., et al. (2016) The novel miR-9600 suppresses tumor progression and promotes paclitaxel sensitivity in non-small-cell lung cancer through altering STAT3 expression. Mol. Ther. Nucleic Acids, 5, e387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang H.B., et al. (2016) miR-143 suppresses the proliferation of NSCLC cells by inhibiting the epidermal growth factor receptor. Exp. Ther. Med., 12, 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. El Bezawy R., et al. (2017) miR-875-5p counteracts epithelial-to-mesenchymal transition and enhances radiation response in prostate cancer through repression of the EGFR-ZEB1 axis. Cancer Lett., 395, 53–62. [DOI] [PubMed] [Google Scholar]
- 97. Tian W., et al. (2017) The miR-599 promotes non-small cell lung cancer cell invasion via SATB2. Biochem. Biophys. Res. Commun., 485, 35–40. [DOI] [PubMed] [Google Scholar]
- 98. Dawidczyk C.M., et al. (2014) Nanomedicines for cancer therapy: state-of-the-art and limitations to pre-clinical studies that hinder future developments. Front. Chem., 2, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fernandez-Piñeiro I., et al. (2017) Nanocarriers for microRNA delivery in cancer medicine. Biotechnol. Adv., 35, 350–360. [DOI] [PubMed] [Google Scholar]
- 100. Chen Y., et al. (2015) In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev., 81, 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Scognamiglio I., et al. (2014) Transferrin-conjugated SNALPs encapsulating 2′-O-methylated miR-34a for the treatment of multiple myeloma. Biomed. Res. Int., 2014, 217365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Di Martino M.T., et al. (2014) In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS One, 9, e90005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lal A., et al. (2011) Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet., 7, e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Garzon R., et al. (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discov., 9, 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hashimi S.T., et al. (2009) MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood, 114, 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ebert M.S., et al. (2010) MicroRNA sponges: progress and possibilities. RNA, 16, 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ye D., et al. (2012) MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells, 30, 1645–1654. [DOI] [PubMed] [Google Scholar]
- 108. Kasar S., et al. (2012) Systemic in vivo lentiviral delivery of miR-15a/16 reduces malignancy in the NZB de novo mouse model of chronic lymphocytic leukemia. Genes Immun., 13, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Giri B.R., et al. (2018) Roles of microRNAs in T cell immunity: implications for strategy development against infectious diseases. Med. Res. Rev. [DOI] [PubMed] [Google Scholar]
- 110. Yang N. (2015) An overview of viral and nonviral delivery systems for microRNA. Int. J. Pharm. Investig., 5, 179–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Herrera-Carrillo E., et al. (2017) Improving miRNA delivery by optimizing miRNA expression cassettes in diverse virus vectors. Hum. Gene Ther. Methods, 28, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]