ABSTRACT
Nervous and neuroendocrine systems mediate environmental conditions to control a variety of life history traits. Our goal was to provide mechanistic insights as to how neurosecretory signals mediate division of labor in the honey bee (Apis mellifera). Worker division of labor is based on a process of behavioral maturation by individual bees, which involves performing in-hive tasks early in adulthood, then transitioning to foraging for food outside the hive. Social and nutritional cues converge on endocrine factors to regulate behavioral maturation, but whether neurosecretory systems are central to this process is not known. To explore this, we performed transcriptomic profiling of a neurosecretory region of the brain, the pars intercerebralis (PI). We first compared PI transcriptional profiles for bees performing in-hive tasks and bees engaged in foraging. Using these results as a baseline, we then performed manipulative experiments to test whether the PI is responsive to dietary changes and/or changes in juvenile hormone (JH) levels. Results reveal a robust molecular signature of behavioral maturation in the PI, with a subset of gene expression changes consistent with changes elicited by JH treatment. In contrast, dietary changes did not induce transcriptomic changes in the PI consistent with behavioral maturation or JH treatment. Based on these results, we propose a new verbal model of the regulation of division of labor in honey bees in which the relationship between diet and nutritional physiology is attenuated, and in its place is a relationship between social signals and nutritional physiology that is mediated by JH.
KEY WORDS: Honey bee, Neurosecretory, Pars intercerebralis, Transcriptomics
Summary: Transcriptomic profiling of the pars intercerebralis in the honey bee brain provides mechanistic insights into how neurosecretory signals mediate social behavior.
INTRODUCTION
A major goal in biology is to understand the mechanisms that underlie complex traits, including animal social behavior (Robinson et al., 2008). Social behaviors such as courtship, kin recognition and social foraging are tightly linked to reproduction and survival and as such, are regulated by the interplay between an organism's internal physiological state and its environment (West-Eberhard, 2003). Understanding how internal and environmental signals interact with specific neural substrates to regulate social behaviors remains an important question. A large body of literature indicates that specialized sensory and neural mechanisms have evolved to process social information (Insel and Fernald, 2004).
Neurosecretory cells in the brain play a central role in integrating social and physiological information in both vertebrates and invertebrates. In mammals, for example, neurosecretory cells in the hypothalamus receive pheromonal information through neuronal inputs from the vomeronasal system. The hypothalamus subsequently regulates the production of hormones from the pituitary gland, eliciting changes in reproductive and aggressive behaviors (Dulac and Torello, 2003). Similarly in insects, neurosecretory cells in the brain innervate endocrine glands known as the corpora cardiaca and the corpora allata (Carrow et al., 1984; Eichmüller et al., 1991; Ludwig et al., 2002). The majority of these neurosecretory cells are located along the dorsal midline of the insect brain, in a region known as the pars intercerebralis (PI). The PI consists of an unpaired cluster of neurons, which is bordered laterally by the mushroom body calyces and ventrally by the central body (Strausfeld, 1976; Ludwig et al., 2002; de Velasco et al., 2007). The neurosecretory cells in the PI are specifically described as monopolar neurons whose dendrites project broadly into the medial protocerebrum and whose axons join the first nerve towards the corpora cardiaca (Hartenstein, 2006).
Similar to the hypothalamus in mammals, neurosecretory cells in the PI regulate the production of neuropeptides and hormones involved in growth and reproduction (Hodková, 1976; Shiga and Numata, 2000; Rulifson et al., 2002; Flatt et al., 2005), as well as several behaviors including locomotion, sleep and aggressive behaviors (Belgacem and Martin, 2002; Foltenyi et al., 2007; Crocker and Sehgal, 2010; Davis et al., 2014). However, the role of PI neurons in regulating division of labor in social insects has not been investigated.
We explored whether the PI mediates worker division of labor in honey bees, a process based on the behavioral maturation of individual adult workers. Bees perform within-hive tasks such as brood care (‘nursing’) for the first 2–3 weeks of life, and then switch to foraging tasks outside (Winston, 1987). However, behavioral maturation is not rigid and is regulated by social and dietary cues. For example, colony food shortage can induce precocious foraging (Schulz et al., 1998), whilst experimental exposure to excess levels of queen mandibular pheromone delays foraging onset (Pankiw et al., 1998). Behavioral maturation also is associated with dietary changes and changes in nutritional physiology. Dietary changes include a switch from a proteinaceous pollen-based diet early in adulthood to a carbohydrate-rich diet primarily comprised of honey and nectar in older bees (Crailsheim et al., 1992). Changes in nutritional physiology include a substantial decrease in abdominal lipid levels (Toth and Robinson, 2005), and experimental depletion of abdominal lipids has been shown to induce precocious foraging (Toth et al., 2005).
Research has shown that environmental cues regulate division of labor in honey bees through changes in brain gene expression. Behavioral maturation involves changes in the expression of thousands of genes in the brain (Whitfield et al., 2003; Alaux et al., 2009a) and a subset of these is regulated by social and nutritional cues (Grozinger et al., 2003; Wheeler et al., 2013). These transcriptomic changes are thought to converge on endocrine systems including juvenile hormone (JH), a multi-function hormone produced by the corpora allata (Whitfield et al., 2006). In honey bees, JH levels increase during behavioral maturation (Rutz et al., 1976; Huang et al., 1991; Huang and Robinson, 1995) and several manipulative experiments, including removal of the corpora allata, have shown that an increase in JH titers leads to the onset of foraging (Robinson, 1985, 1987; Sullivan et al., 2000). JH has co-regulatory interactions with insulin signalling pathways and peripheral nutritional signals related to lipid stores (Guidugli et al., 2005; Corona et al., 2007; Nilsen et al., 2011), and is known to elicit brain gene expression changes that also occur naturally during behavioral maturation (Whitfield et al., 2006). Moreover, recent bioinformatic analyses of brain transcriptomic data implicate transcription factors related to JH signalling as key regulators of behavioral maturation (Ament et al., 2012a).
These results indicate that environmental and physiological factors regulate behavioral maturation through JH signalling, but it is not known how JH interacts with neurosecretory factors in this process. Our goal was to develop a more complete understanding of the regulation of division labor by testing whether the PI integrates diet and hormonal signals to produce transcriptional changes that eventually regulate behavior.
We first compared PI transcriptional profiles for bees performing in-hive tasks and foragers. Using these PI transcriptional profiles as a baseline, we then performed manipulative experiments to test whether the PI is responsive to dietary changes and/or to treatment with a JH analog (JHA). In this study we tested a key prediction of our hypothesis, that diet and JH manipulations cause changes in PI gene expression consistent with changes that occur during behavioral maturation. The results of this study implicate, for the first time, the PI in the regulation of honey bee division of labor.
MATERIALS AND METHODS
Behavioral collections, juvenile hormone analog treatments and diet manipulations
Honey bees (a mixture of European subspecies, mostly Apis mellifera ligustica) were collected from colonies maintained according to standard beekeeping practices at the University of Illinois Bee Research Facility, Urbana, IL, USA. To partially control for genetic variation, we used colonies derived from queens that were instrumentally inseminated with semen from a single male. We collected nurses and foragers using standard behavioral assays. Briefly, we opened each hive and collected bees inserting their head into honeycomb cells containing larvae as nurse bees.
For foragers, we chose to focus on bees collecting pollen because their brightly colored loads of pollen attached to corbiculae on their hindlegs are easily visible, making foraging status unequivocal. In wild-type honey bees, pollen foragers either forage exclusively for pollen or collect both nectar and pollen; thus, limiting our analyses to these bees makes for a more homogeneous group than also including bees that forage for nectar and no pollen. As nectar and pollen foragers differ in a number of ways at the physiological and molecular levels (Page et al., 2000; Amdam et al., 2004; Schulz et al., 2004), the possible implications of this focus are considered in the Discussion. For brevity we call the bees sampled here ‘foragers’. Field collections were performed in the summer of 2010.
JHA and diet manipulation experiments were performed with bees that were 7–9 days old. To collect this focal group, we removed frames of honeycomb containing dark-eyed pupae from colonies and placed them in a 34°C incubator at 80% relative humidity (RH). Bees that emerged from these frames were marked with a dot of colored paint (Testor's Paint, Rockford, IL, USA) and returned to their natal colony. We repeated this process for three consecutive days to obtain a large population of marked bees (>1000 bees) in each colony. We collected marked bees from colonies when they were 7–9 days old, specifically from outer honeycomb frames (not brood frames) and placed them in Plexiglas cages (25–30 bees per cage) for JHA treatment and diet manipulations.
JHA and diet manipulations were set up as a 2×2 experiment and consisted of bees of uniform age that were: (1) fed a diet that was either nurse-like and protein rich, or forager-like and carbohydrate rich; and (2) treated with the JH analog methoprene (JHA+) or treated with a solvent control (Ctrl). We topically treated individuals on the abdomen with 200 μg methoprene in 5 μl acetone, a dose that reliably induces precocious foraging behavior (Robinson, 1985, 1987). After treatment, both JHA-treated and control bees were placed separately in Plexiglas cages supplied with either a high protein diet (45% pollen, 45% honey, 10% water; designated P+) or a diet with no protein (50% sucrose w/v; designated P−) ad libitum (Ament et al., 2011a, 2008). Sucrose was used in place of honey because honey contains low levels of pollen and we wanted to contrast a protein-rich diet with a no-protein diet, as in Ament et al. (2008). Cages were kept in a 34°C incubator at approximately 30% RH for 4 days. Consumption and mortality were monitored daily. We performed two independent trials of this experiment with unrelated bee colonies. These colonies were also unrelated to the colonies used in the nurse–forager experiment described above. The JHA and diet manipulation experiments were performed in the summer of 2011.
Laser capture microdissection
To collect cells from the PI, we first anesthetized bees using CO2 and then immobilized them further on wet ice. We then dissected brains from each individual using cold bee saline (Bicker, 1996) and embedded each brain into small tissue molds containing optimal cutting temperature (OCT) medium (Tissue-Tek). Each brain was embedded dorsal side down and frozen immediately using dry ice. Samples were placed at −80°C for long-term storage.
We performed laser capture microdissection (LCM) by sectioning frozen brains into 35 μm sections using a cryostat (Leica Biosystems). A total of 7–10 dorsal sections were collected onto polyethylene naphthalate (PEN) membrane glass slides (Applied Biosystems) and stained using the nuclear stain kit, Histogene LCM Frozen Section Staining Kit (Applied Biosystems). Cells from the PI were captured using an ArturusXT LCM instrument.
The PI is histologically recognized as the dorsal medial domain of the insect brain and is located between the calyces of the mushroom bodies and dorsal to the central complex (Strausfeld, 1976; Ludwig et al., 2002). We identified neurons in the PI from stained tissue sections based on its characteristic shape and on the color contrast between the cell bodies in the PI (nuclei were stained purple) and adjacent mushroom body nerve fibers (stained light pink). We captured PI neurons from all sections anterior to the central body (see Fig. 1) (Eichmüller et al., 1991). Cells from these sections were collected onto 2–3 Capsure HS LCM caps (Applied Biosystems). Extraction buffer (15 μl; PicoPure RNA Isolation Kit, Applied Biosystems) was applied directly onto each cap and incubated for 30 min at 42°C. After incubation, we collected buffer from each cap using a centrifuge and stored each aliquot at −80°C for subsequent RNA isolation.
Fig. 1.
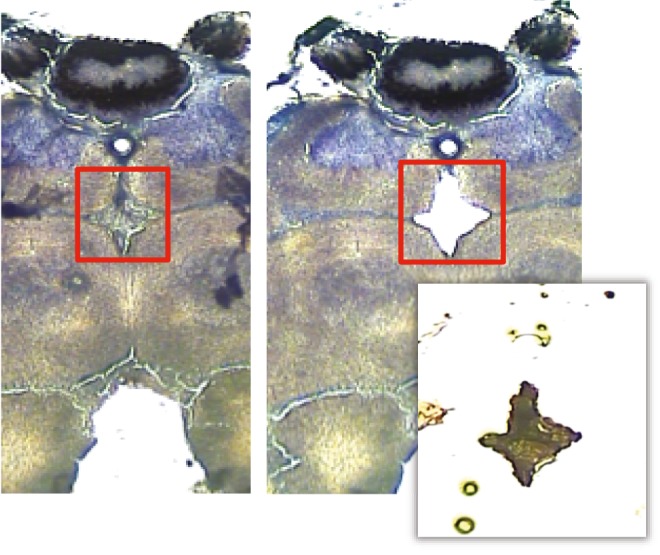
Stained section of the honey bee brain before and after laser capture microdissection. Frozen stained section of the honey bee brain depicting microdissection of cells in the pars intercerebralis. Cells were collected along the midline for sections anterior to the central body.
RNA extraction and amplification
RNA pertaining to the PI of individual bees (pooled extraction buffer aliquots from cells collected on 2–3 caps) was extracted using PicoPure RNA Isolation Kits (Applied Biosystems) following the manufacturer's guidelines for LCM tissue. After extraction, RNA from each sample was amplified using two rounds of linear antisense RNA (aRNA) amplification (Van Gelder et al., 1990). In vitro transcription was performed using the MEGAscript T7 Kit (Invitrogen) and carried out for 14 h for the first amplification round and 6 h for the second round of amplification. aRNA quantity was assessed on a spectrophotometer and aRNA quality was assessed using an Agilent 2100 Bioanalyzer.
Microarrays
PI samples from nurses and foragers were hybridized onto microarrays for gene expression profiling. The two-dye, two-sample microarray platform was characterized previously by Alaux et al. (2009a) and consists of 13,440 oligonucleotide probes based primarily on the A. mellifera genome sequence version 4.0 (Honeybee Genome Sequencing Consortium, 2006). Prior to hybridization, 2 μg of each sample were labelled with a Cy3 or a Cy5 dye using the universal linkage system (ULS) aRNA labelling kit (Kreatech Diagnostics). An aliquot of 60 pmol of each labelled sample was hybridized onto each microarray. Hybridization was carried out overnight using the MAUI Hybridization System (BioMicro Systems). Two samples labelled with different dyes were hybridized onto each microarray following a loop design that maximized the number of direct comparisons between contrasts of interest (Fig. S1). In total, we hybridized 32 samples, eight per behavioral group and colony. Microarray slides were scanned with an Axon 4000B Microarray Scanner (Molecular Devices) and analyzed with GENEPIX Pro 6.0 software (Molecular Devices).
Microarray data analysis
Expression intensities were normalized with a Lowess transformation with BEEHIVE (http://stagbeetle.animal.uiuc.edu/beehive.html). A linear mixed-effects model that accounted for the effects of dye, treatment, colony and individual bee was used to analyze the log2-transformed fluorescent intensities for each gene. The results were evaluated with an F-test statistic and false discovery rate (FDR) corrected P-values. Array probes were updated to the newest version of the A. mellifera genome, Assembly 4.5 (Elsik et al., 2014), by mapping array probes using Bowtie 2.
During the course of our analyses, we observed that seven major royal jelly protein genes were differentially expressed in PI samples of nurses and foragers, with substantial log-fold change differences. These genes are differentially expressed, and at high levels, in the hypopharyngeal glands (HPG) that surround the brain. To minimize the possibility that our results were affected by small amounts of HPG tissue, we filtered gene lists with a set of 113 genes expressed at very high levels in the HPG, two-fold higher than in the brain (Whitfield et al., 2003; Alaux et al., 2009a). A total of four ‘HPG genes’ were filtered from nurse–forager gene list (FDR<0.1), 22 ‘HPG genes’ were filtered from JHA (FDR<0.1), whilst none was filtered from the diet gene list (FDR<0.1). Expression of a few major royal jelly protein genes have been found in the mushroom bodies of the bee brain (Peixoto et al., 2009), so it is possible that some of these genes were expressed in the brain, but without further validation we cannot determine if these genes were expressed in the PI or the HPG. Because the number of genes is relatively small compared with the 13,774 total analyzed here, we chose this conservative approach.
cDNA libraries and RNA sequencing
Gene expression profiles for PI samples from JHA and diet-treated bees were generated using RNA sequencing. cDNA libraries from each PI sample were generated from 400 ng of aRNA with the TruSeq RNA Sample Preparation Kit (Illumina). Library concentrations were quantified using Qubit fluorometric quantitation and quantitative real-time PCR (qPCR) using Kapa Library Quant kits (KapaBiosystems). Average fragment size and overall quality were evaluated with the Agilent 2100 Bioanalyzer platform and an Agilent High Sensitivity DNA kit. For sequencing, all libraries were diluted to a 10 nmol l−1 concentration. We sequenced a total of 32 samples (eight samples per treatment group, four from each colony replicate). Eight samples were sequenced per lane, with two samples from each treatment group per lane. cDNA libraries were sequenced as single-end, 100 nt reads, using an Illumina HiSeq2000.
RNA sequencing data analysis
RNA sequencing generated an average of 23,833,961 reads per cDNA library. Reads from each library were aligned to the A. mellifera genome, Assembly 4.5 (Elsik et al., 2014) using Tophat (Trapnell et al., 2009). Approximately 80% of reads within each library mapped to the honey bee genome. Aligned read counts for each gene were counted using the program HTSeq version 0.5pv2 (http://www-huber.embl.de/users/anders/HTSeq) and the A. mellifera official gene set, version 3.2. Reads that did not map uniquely or that mapped to genomic locations outside genes were not included in subsequent analyses for differential expression. Differential expression was assessed using a negative binomial model with count data normalized by library size and library composition. Dispersion estimates for each individual gene were used to calculate differential expression. The model tested for the main effects of JHA treatment and diet, and an interaction between these two factors. Significance for these main effects was assessed using FDR corrected P-values (FDR<0.1). P-values were used to test significance (P<0.001) for pairwise contrasts to assess whether genes were significant due to JHA treatment, diet or the interaction between these two factors.
Gene ontology enrichment analyses
Functional inferences for microarray and RNA sequencing data sets were drawn using gene ontology (GO) enrichment analyses using DAVID Bioinformatic Resources Functional Annotation tool (Dennis et al., 2003). We tested for enrichment of terms in the functional categories: biological processes (BP), molecular function (MF) and cellular component (CC), and used the GO category GO FAT to filter out the broadest GO terms. We also examined GO terms annotated using InterPro and KEGG pathways. GO analyses were accomplished with the Drosophila melanogaster orthologs associated with each gene list. Significant enrichment was determined using a modified Fisher's exact test that used the number of honey bee genes with annotated Drosophila orthologs as a background.
Bioinformatic analysis of transcription factors and enrichment of transcription factor regulatory motifs
To identify transcription factors that were differentially expressed in our experiments, we drew upon a comprehensive list of transcription factors that was compiled previously (Chandrasekaran et al., 2011). Because few transcription factors have been experimentally validated in the honey bee, this transcription factor list (256 transcription factors, total) included robust orthologs of transcription factors experimentally characterized in D. melanogaster for which there is little or no critical residue changes between the honey bee gene and the D. melanogaster ortholog (Kim et al., 2010). We updated this list with information from the latest A. mellifera official gene set (version 3.2) (Elsik et al., 2014).
To determine whether our lists of differentially expressed genes (DEGs) suggest the involvement of specific transcription factors as regulators, we determined whether our lists were enriched for particular cis regulatory motifs, using the Motif Enrichment Tool (http://veda.cs.uiuc.edu/regSetFinder/interface_help.html). This bioinformatic tool predicts transcription factor regulators by finding significantly associated sets of genes that share a transcription factor motif in their promoter regions (Ament et al., 2012a). Genes thus targeted by a transcription factor motif were determined for the A. mellifera genome (version 4.5) using the official gene set (version 3.2) following methods similar to Alaux et al. (2009b). We analyzed 5 kbp of sequence upstream of each gene's transcriptional start site. We tested for significant associations between sets of significantly upregulated gene sets and specific cis regulatory motifs with a Fisher's exact test and an FDR correction, controlling for differences in guanine-cytosine (GC) content in the promoter regions (Sinha et al., 2006). To test whether cis motifs were significantly associated across multiple gene lists, we followed the methods of Alaux et al. (2009b). We calculated a combined P-value using the FDR-corrected P-values associated with motifs and upregulated gene sets using the following formula:
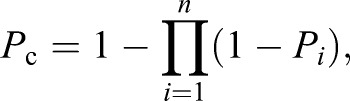 |
(1) |
where Pc is the combined P-value, Pi is the P-value for the ith gene list, i and n is the total number of gene lists. We looked for significant cis motifs (raw P<0.05) that occurred on different gene lists and calculated the combined P-value for each motif.
RESULTS
Gene expression differences associated with behavioral maturation in the PI
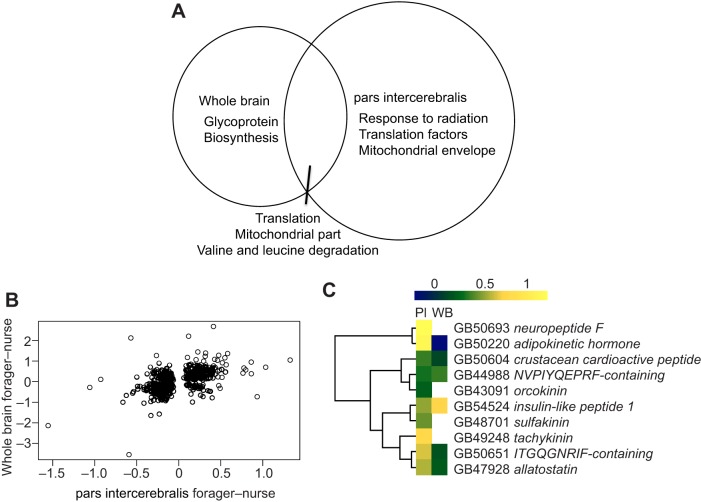
Microarray profiling of the PI revealed extensive differences in PI gene expression between nurses and foragers (2663 genes, ANOVA, FDR<0.1). The magnitude of the gene expression differences in the PI is in the same range as previously reported for whole brain microarray comparisons of nurses and foragers (Alaux et al., 2009a). Moreover, 22% (i.e. 589) of the genes differentially expressed between nurses and foragers in the PI were also differentially expressed in the whole brain (Fig. 2A). Correlation analysis of the log-fold changes of genes overlapping between the whole brain (Alaux et al., 2009a) and the present dataset revealed that the majority of genes were concordant in their direction of change (Fig. 2B).
Fig. 2.
Comparison of results from microarray analyses of nurse versus forager pars intercerebralis and previously published nurse versus forager whole brain (Alaux et al., 2009a). N=16 for each behavioral group. (A) Venn diagram showing significant gene ontology terms (Fisher's exact test, P≤0.001) enriched from the lists of overlapping (589 genes) and non-overlapping DEGs (PI: 2074 genes). (B) Pearson correlations of log-fold changes of differentially expressed genes that overlapped between whole brain and PI microarray results; r=0.532, P<0.0001. (C) Heat map for differentially expressed neuropeptide genes in whole brain (WB) and PI. Yellow indicates neuropeptide genes that are significantly upregulated in foragers, blue indicates upregulated genes in nurses.
GO enrichment analyses showed that the list of transcripts differentially expressed in the PI of nurses and foragers was enriched for terms related to energy metabolism and protein turnover (Table S1). Genes upregulated in foragers were related to detection of light stimulus and oxidative phosphorylation, while genes upregulated in nurses were related to ribosome and translation. Genes differentially expressed between nurses and foragers in both the PI and the whole brain were related to translation, mitochondrial processes and branched amino acid degradation (Fig. 2A, Table S2). Genes only differentially expressed in the PI were related to response to radiation, translation factor activity, mitochondrial envelope and amine transport whilst genes only differentially expressed in whole brain were related to glycoprotein biosynthesis (Fig. 2A, Table S2).
A finer-grained analysis, using genes differentially expressed in both the PI and the whole brain and with directionally concordant log-fold changes (323 genes), showed enrichment for terms related to protein turnover (ribosome, 28 genes, Benjamini=2.39×10–19). GO terms related to energy metabolism were not enriched in the concordant gene set because the relevant genes were regulated in the opposite direction in whole brain (downregulated in foragers) compared with the PI (upregulated in foragers).
As the PI contains neurosecretory cells, we expected to detect differentially expressed neuropeptide genes that had been undetected in prior whole brain microarray analyses. We found a total of 10 differentially expressed neuropeptide genes in the PI of nurse and forager bees (Fig. 2C); four of these genes were not detected in whole brain. Neuropeptide genes that were only differentially expressed in the PI encode neuropeptide F, orcokinin, sulfakinin and tachykinin. The neuropeptide genes that overlapped between both datasets showed similar upregulation in foragers in both the PI and the whole brain, with the exception of adipokinetic hormone, which was downregulated in forager whole brain and upregulated in forager PI.
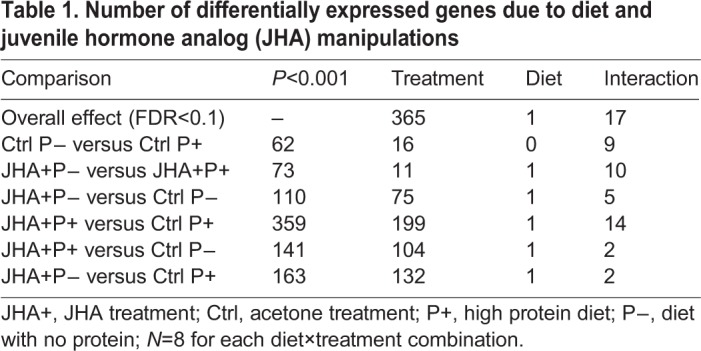
Effect of diet on gene expression in the PI
RNA sequencing results revealed that diet treatment did not influence gene expression in the PI (one differentially expressed gene, FDR<0.1, Table 1). Pairwise comparisons of diet-treated bees showed a total of 62 and 73 differentially expressed genes (P<0.001) for bees that received no pollen versus high pollen diets (Ctrl P− versus Ctrl P+) and for bees that received the same diet treatments and were also treated with JHA (JHA P− versus JHA P+) (Table 1). Due to the relatively low numbers of differentially expressed genes in each of these gene lists, GO functional enrichment analyses performed for diet pairwise comparisons only yielded significant enrichment for the JHA− P− versus JHA− P+ comparison; this pairwise contrast was enriched for GO terms related to cytochrome P450 expression (Table S3).
Table 1.
Number of differentially expressed genes due to diet and juvenile hormone analog (JHA) manipulations
Diet treatment influenced the expression of three neuropeptide genes, including corazonin (GB53951), LRNQLDIGDLQ-containing (GB43119) and NVPIYQEPRF-containing (GB44988). Corazonin and LRNQLDIGDLQ-containing were upregulated in response to high pollen diet in bees treated with JHA (JHA P− versus JHA P+), whilst NVPIYQEPRF-containing was upregulated by a high pollen diet in the Ctrl P− versus Ctrl P+ contrast.
Effect of JHA treatment on gene expression in the PI
In contrast to the diet treatment, JHA treatment caused changes in hundreds of genes in the PI (365 differentially expressed genes, FDR<0.1). Pairwise comparisons for hormone-treated bees showed 110–359 differentially expressed genes (P<0.001) in the PI, depending on the contrast (Table 1). Significantly enriched GO terms for hormonally induced differential gene expression were related to oxidation reduction and protein folding (heat shock proteins) for the JHA+ P− versus Ctrl P− contrast and translation (ribosome) and epidermal growth factor domain for the JHA+ P+ versus Ctrl P+ and JHA+ P+ versus Ctrl P− contrasts (Table S3).
JHA treatment influenced the expression of the same neuropeptide genes that were regulated by diet manipulations. Similar to diet comparisons, corazonin was upregulated in bees that received JHA treatment and a high-protein diet (JHA+ P+ versus Ctrl P+ and JHA+ P+ versus Ctrl P−), while LRNQLDIGDLQ-containing and NVPIYQEPRF-containing were upregulated in Ctrl P+ compared with JHA− P− bees. NVPIYQEPRF-containing gene was also differentially expressed in the PI of nurse and foragers but was upregulated in foragers relative to nurses (even though nurses have a diet richer in protein than foragers).
Regulatory changes associated with behavioral maturation and the PI
To determine whether there were diet and hormonally induced gene expression changes that were generally consistent with behavioral maturation in the PI, we compared the nurse versus forager DEG list with the list from each diet×treatment combination. The DEG list from each pairwise comparison showed 27–49% overlap with the nurse–forager list (Fig. 3A). For these genes, we tested whether the degree and direction of differential expression (using log-fold change) was significantly correlated with the results from the nurse–forager comparison. A significant positive correlation would suggest that the diet or JHA treatments led to gene expression patterns that resemble those observed as a result of behavioral maturation. There were no significant correlations between the nurse–forager and diet comparisons (Fig. 3B). In contrast, there were significant correlations between the nurse–forager and JHA treatment comparisons, in the expected direction. For example, the ortholog of the cytochrome P450 gene, Cyp4g1, was upregulated in foragers compared with nurses and upregulated by JHA treatment in the JHA+ P− versus Ctrl P− contrast.
Fig. 3.
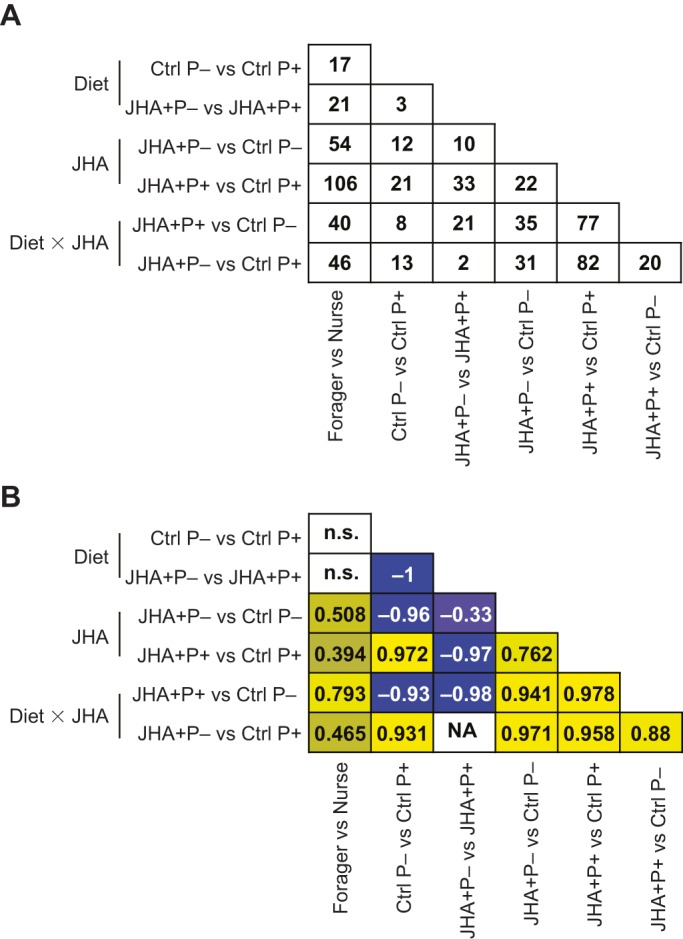
Overlap and correlation analyses for genes differentially expressed in the pars intercerebralis of nurses and foragers and as a function of diet and/or JH analog treatments. N=16 for each behavioral group and N=8 for each diet×treatment combination. (A) Number of genes overlapping in various pairwise comparisons. (B) Pearson's correlation coefficients for genes in A. Numbers represent the correlation coefficients that were significant (P<0.05). Yellow denotes positive correlation; blue denotes negative correlation; n.s., not significant, NA: not available; JHA+, JHA treatment; Ctrl, acetone treatment; P+, high protein diet; P−, diet with no protein.
Given the known causal relationship between JH and behavioral maturation in honey bees (Sullivan et al., 2000), these results suggested that differences in endogenous JH levels could be driving the nurse–forager differences we detected in the PI. If this is the case, we would expect that these shared gene expression changes were elicited by similar gene regulatory changes. To begin to explore this, we first looked for transcription factors (orthologs of transcription factors in D. melanogaster) that were differentially expressed in the PI of nurses versus foragers and as a function of JHA treatment. We found a total of 82 differentially expressed transcription factors in the PI of nurses and foragers (Table S4). These included ftz-f1, fruitless, Xpb1, Creb, Deaf1 and NFkβ, all of which have been proposed as major regulators of honey bee behavioral maturation (Chandrasekaran et al., 2011). Seven transcription factors were differentially expressed in JHA pairwise contrasts, and of these, two overlapped with the PI nurse versus forager data set (GB50553 and GB55635, Table S5). The JH-related transcription factor kr-h1 was differentially expressed in two of the JHA pairwise contrasts, but was not differentially expressed in the PI nurse versus forager dataset.
Next, we performed transcription factor motif analyses to indirectly identify transcription factors contributing to the observed differential gene expression. Genes upregulated in the PI of nurse bees were significantly enriched (FDR<0.1) for nine cis motifs (Table S6) including the cis motif for the JH-related transcription factor, usp. No transcription factor motifs were enriched for genes upregulated in the PI of forager bees. In diet×treatment comparisons, we found significant transcription factor motif enrichment for one gene list: genes upregulated by JHA treatment in the JHA+P− versus Ctrl P− comparison were enriched for four cis motifs, including the motif for NFkβ and the NFkβ-related heterodimers, dorsal-related immunity factor (DIF) and relish (Table S7).
To compare across nurse versus forager and diet×JHA experiments, we identified conserved transcription factor motifs (P<0.05) that overlapped between experiments. We found five cis motifs associated with genes upregulated in nurses and genes upregulated in bees that were not treated with JHA (Ctrl bees) (Table 2). These were indicative of changes in endocrine signaling and included the heterodimer, NR1H2-RXRA, which contains an isoform of the vertebrate homolog of usp, RXR (Oro et al., 1990). Overlapping cis motifs for genes upregulated in foragers and JHA-treated bees indicate that both of these gene lists were associated with NFkβ and the heat shock factor, HSF.
Table 2.
cis regulatory motifs and corresponding transcription factors common to pars intercerebralis DEG lists from nurse versus forager and control versus juvenile hormone analog experiments
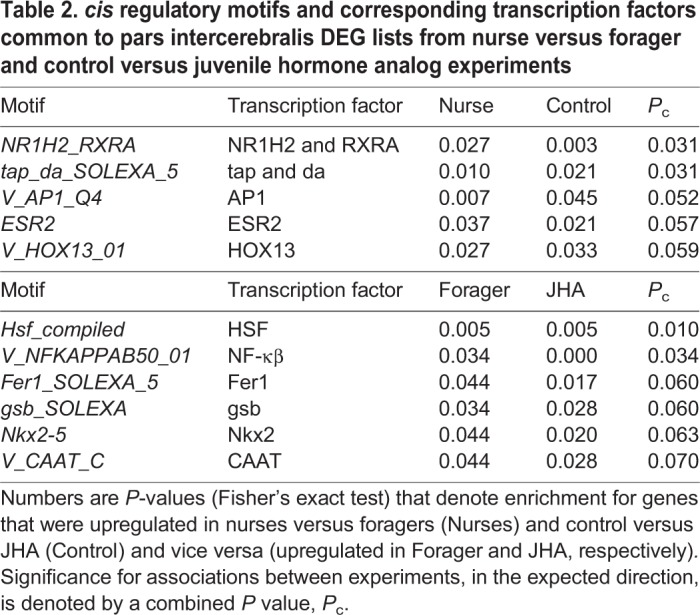
Several transcription factors were differentially expressed between nurses and foragers and also showed motif enrichment among differentially expressed genes. These included fruitless, Deaf1, HLH54F, ATF-2 and NFkβ, suggesting that these may be regulators of changes in the PI related to behavioral maturation (Table S8). We used the transcription factor list from the nurse versus forager experiment and looked for overlap with the JHA-based cis motif analyses. We found three transcription factors that met these criteria: Tkr, HLH54F and crc. All three were associated with genes upregulated by JHA treatment.
DISCUSSION
Our results reveal a robust molecular signature of honey bee behavioral maturation in the pars intercerebralis. Moreover, this signature appears to be strongly related to JH, a known regulator of honey bee behavioral maturation. These results support the hypothesis that interactions between the PI and JH signalling are involved in the regulation of behavioral maturation. This hypothesis should be tested in the future, by testing the effects of perturbations to PI gene expression on behavioral maturation. Such targeted manipulations are currently beyond the scope of what is available for honey bees, but the recent report of transgenic honey bees (Schulte et al., 2014), coupled with advances in genome editing (Doudna and Charpentier, 2014), promise new possibilities in the near future.
The molecular signature of behavioral maturation in the PI involves changes in protein turnover, heat shock protein genes, and GO terms related to energy metabolism. This suggests there is an increase in protein translation in the PI of nurses compared with foragers, something also inferred from whole brain analysis. However, many of the nurse–forager expression differences in the PI were not detected at the whole brain level, including differences in the neuropeptide gene neuropeptide F, whose expression has been shown to be restricted to cells in the PI in honey bees (Ament et al., 2011b) and involved in food searching and appetitive behaviors in vertebrates and invertebrates (Wu et al., 2003; Morton et al., 2006). Furthermore, gene expression differences related to energy metabolism were not directionally concordant between the PI and whole brain data sets. This result implicates regional differences in energy metabolism in the honey bee brain. It is possible that the smaller neuronal clusters, such as the PI, differ relative to the larger brain regions, which probably reflect the picture obtained with whole brain measures. Higher expression of energy metabolism genes in the PI of foragers fits with the previously reported involvement of insulin signalling in honey bee foraging (Ament et al., 2008; Wang et al., 2010; Ihle et al., 2014), and with our results showing that insulin-like peptide 1 is upregulated in the PI of foragers. These results demonstrate that the PI has a distinct transcriptional profile that reflects its neurosecretory function.
Both transcriptomic results and transcription factor motif analyses support the hypothesis that interactions between the PI and JH signalling are involved in the regulation of behavioral maturation in honey bees. Maturational and JH-related transcriptomic changes in the PI appear to be regulated by transcription factors already implicated in the regulation of behavioral maturation (Chandrasekaran et al., 2011; Ament et al., 2012b) including NFkβ and usp. If the PI is involved in the regulation of division of labor as suggested here, it may do so through the action of these transcription factors. With regard to NFkβ, our results suggest that this transcription factor may play a similar role in honey bees as it does in the hypothalamus of vertebrates where it is linked to changes in nutritional physiology and insulin signalling (Kaltschmidt and Kaltschmidt, 2009). In addition, neuronal circuits in the hypothalamus are central to sleep regulation in vertebrates and NFkβ-expressing cells in the hypothalamus are important to this process (Brandt et al., 2004). There is evidence that in the honey bee PI, protein synthesis follows a circadian rhythm (Vogel et al., 1977). Thus it would be interesting for future research to investigate whether NFkβ and other transcription factors expressed in the PI mediate the metabolic and behavioral changes related to sleep in honey bees (Eban-Rothschild and Bloch, 2008) as they do in other insects (Foltenyi et al., 2007).
The vertebrate homolog of usp, RXR, is also known as a regulator of metabolism and nutritional physiology (Keller et al., 1993). In insects, usp is linked to JH signalling (Jones and Sharp, 1997; Hiruma et al., 1999). In honey bees, usp has been shown to be expressed in the mushroom bodies of the honey bee brain (Velarde et al., 2006) and to influence maturation-related transcriptional networks that regulate behavior (Sinha et al., 2006; Ament et al., 2012b; Wang et al., 2012). Our results provide evidence that usp is involved in regulating transcriptional changes in a brain region that is structurally and functionally proximal to the corpora allata and JH production, a finding that is consistent with the idea that there is a feed-forward endocrine loop that regulates foraging behavior in honey bees (Amdam and Omholt, 2003).
There were thousands of genes differentially expressed in the PI of nurses and foragers, and hundreds in the PI of JHA-treated bees. One possible explanation for the difference in numbers is that JH regulates some, but not all, of the physiological and neurobiological changes that occur during honey bee behavioral maturation. Another possibility may be related to technology differences between the two experiments: the nurse–forager profiles were generated using microarrays, while JHA-treated bees were profiled using RNA sequencing. There are many differences between these two technologies, one of which is that RNA sequencing has greater dynamic range compared with microarrays (Ozsolak and Milos, 2011), making RNA sequencing a more sensitive technology, but possibly more susceptible to within-group variation. These technological differences may be related to our lack of an overlapping set of neuropeptide genes between the PI nurse–forager gene list and the gene lists associated with JHA treatment (with the exception of NVPIYQEPRF-containing). Given that insulin-like peptide 1 was differentially expressed in PI of nurses and foragers and that both neuropeptide F and insulin-like peptide 1 have been shown to be responsive to diet and JHA treatment in honey bees (respectively) (Corona et al., 2007; Ament et al., 2011a), we believe our RNA sequencing analysis may have lacked sufficient sensitivity or statistical power. Nevertheless, our RNA sequencing experiments do show that the neuropeptide genes corazonin, NVPIYQEPRF-containing and LRNQLDIGDLQ-containing were differentially expressed in the PI of JH-treated bees. These neuropeptides have been localized to the corpora cardiaca and corpora allata (Boerjan et al., 2010), indicating that these neuropeptides may act as neurohormones in honey bees. In addition, our analyses were able to detect a robust overlapping signal between the behavioral maturation and JHA experiments, for both differentially expressed genes in general and transcription factor motifs.
A surprising aspect of these results is that there is limited evidence to support our hypothesis that the PI is responsive to dietary changes related to behavioral maturation. Effects of JHA treatment were much stronger than dietary manipulation. We believe the JHA dosage produced results that were physiologically relevant, for two reasons. Firstly, as described above, this treatment caused differences in PI gene expression that were similar to the nurse–forager PI gene expression differences. Secondly, the same result was obtained for whole brain analyses (Whitfield et al., 2006). In contrast, dietary manipulations had little impact on PI gene expression and did not elicit changes consistent with behavioral maturation. One possible explanation is that age has strong effects on brain gene expression in honey bees (Whitfield et al., 2006) and the bees in the diet experiment were the same age. Nevertheless, this result is still surprising because the PI is known to integrate nutritional changes in other insects (Rajan and Perrimon, 2012) and in honey bees there is a switch from a high-protein diet to a low-protein diet prior to foraging onset (Crailsheim et al., 1992). There were also differences in gene expression measurement technology between these two experiments; however, this explanation is unlikely because, as noted above, there were substantial overlapping results between the nurse–forager and JHA experiments, across the same two analytical platforms. Another possibility is that there are differences in the endocrine responsiveness of the PI in pollen foragers compared with nectar foragers, given that there are known physiological and molecular differences between these two groups (Page et al., 2000; Amdam et al., 2004; Schulz et al., 2004). It would be interesting to compare the responsiveness of nectar and pollen foragers in the future, perhaps using the well-known selected lines of pollen and nectar foragers, where the specializations are probably more extreme than in our group of foragers. For the present, we present a verbal model that suggests that these results reflect an evolved attenuation of the relationship between diet and nutritional physiology, as follows.
We propose that adult worker honey bees have evolved an attenuation of the traditional relationship between diet and nutritional physiology, and instead have evolved a relationship between social factors and nutritional physiology (Fig. 4). The strongest effects linking nutrition to foraging onset in honey bees have come from starvation experiments (Schulz et al., 1998; Toth et al., 2005). These studies have shown that a depletion of a colony's food stores induces early foraging; however, depriving colonies of just pollen does not elicit these same effects (Toth et al., 2005). These results indicate that more moderate diet changes are not causal to foraging onset. Recent transcriptomic studies on the brain and peripheral tissues are consistent with our hypothesis; they show that diet did not elicit gene expression changes consistent with behavioral maturation (Ament et al., 2011a; Wheeler et al., 2013). According to our hypothesis, molecular and behavioral effects of experimental depletion of abdominal lipids and peripheral (not brain) RNAi knockdown of the lipid storage protein, vitellogenin (Nunes et al., 2013; Wheeler et al., 2013), relate to the fact that these physiological factors are modulated by social signals. The expression of vitellogenin, for example, is regulated by queen mandibular pheromone (QMP) (Fischer and Grozinger, 2008) and brood pheromone (BP) (Smedal et al., 2009). Moreover, recent transcriptomic studies show that unlike diet, both QMP and BP affect brain gene expression in a manner consistent with behavioral maturation (Grozinger et al., 2003; Alaux et al., 2009a). Together these findings suggest that pheromones are the primary extrinsic regulators of behavioral maturation.
Fig. 4.
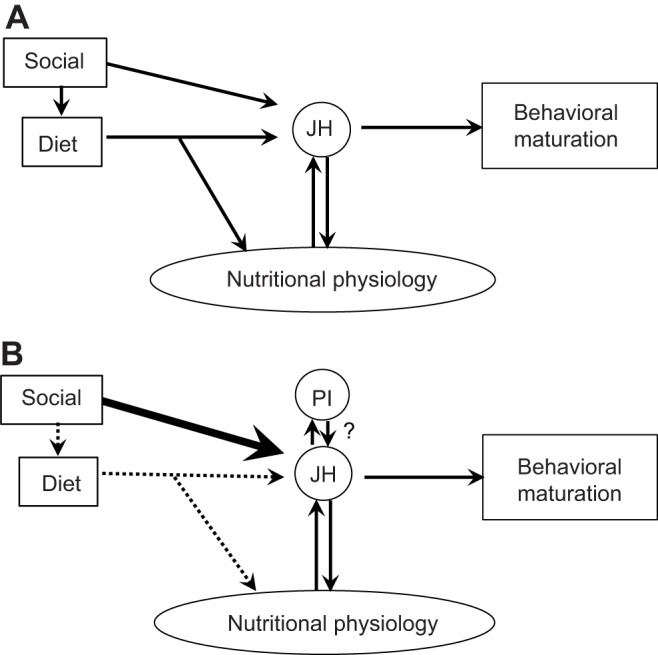
Verbal model for effects of diet, social factors and juvenile hormone on nutritional physiology and behavioral maturation in honey bees. (A) Previous understanding of the relationships between diet and nutritional physiology and between social factors and nutritional physiology (modified from Ament et al., 2011a). (B) New model, which proposes weaker role for diet, stronger role for social factors and a possible interaction between PI and JH signalling in the corpora allata. Social, pheromonal cues; JH, juvenile hormone; PI, pars intercerebralis.
The connection between pheromones and nutritional physiology is likely to be JH-mediated as QMP has been shown to modulate JH levels (Pankiw et al., 1998) and as JH has co-regulatory interactions with vitellogenin (Pinto et al., 2000; Guidugli et al., 2005). One additional possibility is that social factors mediate nutritional physiology through interactions with the corpora allata, followed by the PI. Our results provide indirect evidence for this idea by demonstrating that maturation-related changes are present in the PI, but these maturational changes are not regulated by variation in diet, or at least the variations that we tested. Future work should test these connections directly, and if confirmed the results would further suggest that neuroendocrine systems in honey bees have evolved to regulate worker division of labor by becoming less sensitive to dietary inputs and more sensitive to social factors tied to colony-level needs.
Acknowledgements
We acknowledge Jeff Hinchman and Danielle Ponsi for their invaluable assistance in the field and with laser capture microdissection; Dr Tom Newman and Dr Amy Cash for assistance in the laboratory; Charley Nye for assistance with beekeeping; and members of the Robinson laboratory for helpful thoughts and comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.M.W., S.A.A. and G.E.R. designed the study. M.M.W. performed the experiments. Data analysis was performed by M.M.W., S.L.R.-Z. and B.S. M.M.W. and G.E.R. wrote the manuscript.
Funding
This study was funded by the National Institutes of Health (NIH): grant 1R01DK082605-01A1 (G.E.R., principal investigator); and a fellowship on the NIH-University of Illinois at Urbana-Champaign Cell & Molecular Biology Training Grant to M.M.W. (James H. Morrissey, principal investigator). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jeb.biologists.org/lookup/suppl/doi:10.1242/jeb.119420/-/DC1
References
- Alaux C., Le Conte Y., Adams H. A., Rodriguez-Zas S., Grozinger C. M., Sinha S. and Robinson G. E. (2009a). Regulation of brain gene expression in honey bees by brood pheromone. Genes Brain Behav. 8, 309-319. 10.1111/j.1601-183X.2009.00480.x [DOI] [PubMed] [Google Scholar]
- Alaux C., Sinha S., Hasadsri L., Hunt G. J., Guzmán-Novoa E., DeGrandi-Hoffman G., Uribe-Rubio J. L., Southey B. R., Rodriguez-Zas S. and Robinson G. E. (2009b). Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. USA 106, 15400-15405. 10.1073/pnas.0907043106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam G. V. and Omholt S. W. (2003). The hive bee to forager transition in honeybee colonies: the double repressor hypothesis. J. Theor. Biol. 223, 451-464. 10.1016/S0022-5193(03)00121-8 [DOI] [PubMed] [Google Scholar]
- Amdam G. V., Norberg K., Fondrk M. K. and Page R. E. (2004). Reproductive ground plan may mediate colony-level selection effects on individual foraging behavior in honey bees. Proc. Natl. Acad. Sci. USA 101, 11350-11355. 10.1073/pnas.0403073101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Corona M., Pollock H. S. and Robinson G. E. (2008). Insulin signaling is involved in the regulation of worker division of labor in honey bee colonies. Proc. Natl. Acad. Sci. USA 105, 4226-4231. 10.1073/pnas.0800630105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Chan Q. W., Wheeler M. M., Nixon S. E., Johnson S. P., Rodriguez-Zas S. L., Foster L. J. and Robinson G. E. (2011a). Mechanisms of stable lipid loss in a social insect. J. Exp. Biol. 214, 3808-3821. 10.1242/jeb.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Velarde R. A., Kolodkin M. H., Moyse D. and Robinson G. E. (2011b). Neuropeptide Y-like signalling and nutritionally mediated gene expression and behaviour in the honey bee. Insect Mol. Biol. 20, 335-345. 10.1111/j.1365-2583.2011.01068.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Blatti C. A., Alaux C., Wheeler M. M., Toth A. L., Le Conte Y., Hunt G. J., Guzmán-Novoa E., DeGrandi-Hoffman G., Uribe-Rubio J. L. et al. (2012a). New meta-analysis tools reveal common transcriptional regulatory basis for multiple determinants of behavior. Proc. Natl. Acad. Sci. USA 109, E1801-E1810. 10.1073/pnas.1205283109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament S. A., Wang Y., Chen C.-C., Blatti C. A., Hong F., Liang Z. S., Negre N., White K. P., Rodriguez-Zas S. L., Mizzen C. A. et al. (2012b). The transcription factor ultraspiracle influences honey bee social behavior and behavior-related gene expression. PLoS Genet. 8, e1002596 10.1371/journal.pgen.1002596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgacem Y. H. and Martin J.-R. (2002). Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc. Natl. Acad. Sci. USA 99, 15154-15158. 10.1073/pnas.232244199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker G. (1996). Transmitter-induced calcium signalling in cultured neurons of the insect brain. J. Neurosci. Methods 69, 33-41. 10.1016/S0165-0270(96)00018-0 [DOI] [PubMed] [Google Scholar]
- Boerjan B., Cardoen D., Bogaerts A., Landuyt B., Schoofs L. and Verleyen P. (2010). Mass spectrometric profiling of (neuro)-peptides in the worker honeybee, Apis mellifera. Neuropharmacology 58, 248-258. 10.1016/j.neuropharm.2009.06.026 [DOI] [PubMed] [Google Scholar]
- Brandt J. A., Churchill L., Rehman A., Ellis G., Mémet S., Israël A. and Krueger J. M. (2004). Sleep deprivation increases the activation of nuclear factor κB in lateral hypothalamic cells. Brain Res. 1004, 91-97. 10.1016/j.brainres.2003.11.079 [DOI] [PubMed] [Google Scholar]
- Carrow G. M., Calabrese R. L. and Williams C. M. (1984). Architecture and physiology of insect cerebral neurosecretory cells. J. Neurosci. 4, 1034-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S., Ament S. A., Eddy J. A., Rodriguez-Zas S. L., Schatz B. R., Price N. D. and Robinson G. E. (2011). Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc. Natl. Acad. Sci. USA 108, 18020-18025. 10.1073/pnas.1114093108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona M., Velarde R. A., Remolina S., Moran-Lauter A., Wang Y., Hughes K. A. and Robinson G. E. (2007). Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc. Natl. Acad. Sci. USA 104, 7128-7133. 10.1073/pnas.0701909104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crailsheim K., Schneider L. H. W., Hrassnigg N., Bühlmann G., Brosch U., Gmeinbauer R. and Schöffmann B. (1992). Pollen consumption and utilization in worker honeybees (Apis mellifera carnica): dependence on individual age and function. J. Insect Physiol. 38, 409-419. 10.1016/0022-1910(92)90117-V [DOI] [Google Scholar]
- Crocker A. and Sehgal A. (2010). Genetic analysis of sleep. Genes Dev. 24, 1220-1235. 10.1101/gad.1913110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. M., Thomas A. L., Nomie K. J., Huang L. and Dierick H. A. (2014). Tailless and Atrophin control Drosophila aggression by regulating neuropeptide signalling in the pars intercerebralis. Nat. Commun. 5, 3177 10.1038/ncomms4177 [DOI] [PubMed] [Google Scholar]
- de Velasco B., Erclik T., Shy D., Sclafani J., Lipshitz H., McInnes R. and Hartenstein V. (2007). Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev. Biol. 302, 309-323. 10.1016/j.ydbio.2006.09.035 [DOI] [PubMed] [Google Scholar]
- Dennis G., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C. and Lempicki R. A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Doudna J. A. and Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Dulac C. and Torello A. T. (2003). Molecular detection of pheromone signals in mammals: from genes to behaviour. Nat. Rev. Neurosci. 4, 551-562. 10.1038/nrn1140 [DOI] [PubMed] [Google Scholar]
- Eban-Rothschild A. D. and Bloch G. (2008). Differences in the sleep architecture of forager and young honeybees (Apis mellifera). J. Exp. Biol. 211, 2408-2416. 10.1242/jeb.016915 [DOI] [PubMed] [Google Scholar]
- Eichmüller S., Hammer M. and Schäfer S. (1991). Neurosecretory cells in the honeybee brain and suboesophageal ganglion show FMRFamide-like immunoreactivity. J. Comp. Neurol. 312, 164-174. 10.1002/cne.903120112 [DOI] [PubMed] [Google Scholar]
- Elsik C. G., Worley K. C., Bennett A. K., Beye M., Camara F., Childers C. P., de Graaf D. C., Debyser G., Deng J., Devreese B. et al. (2014). Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics 15, 86 10.1186/1471-2164-15-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer P. and Grozinger C. M. (2008). Pheromonal regulation of starvation resistance in honey bee workers (Apis mellifera). Naturwissenschaften 95, 723-729. 10.1007/s00114-008-0378-8 [DOI] [PubMed] [Google Scholar]
- Flatt T., Tu M.-P. and Tatar M. (2005). Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays 27, 999-1010. 10.1002/bies.20290 [DOI] [PubMed] [Google Scholar]
- Foltenyi K., Greenspan R. J. and Newport J. W. (2007). Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10, 1160-1167. 10.1038/nn1957 [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., Sharabash N. M., Whitfield C. W. and Robinson G. E. (2003). Pheromone-mediated gene expression in the honey bee brain. Proc. Natl. Acad. Sci. USA 100 Suppl. 2, 14519-14525. 10.1073/pnas.2335884100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidugli K. R., Nascimento A. M., Amdam G. V., Barchuk A. R., Omholt S., Simões Z. L. P. and Hartfelder K. (2005). Vitellogenin regulates hormonal dynamics in the worker caste of a eusocial insect. FEBS Lett. 579, 4961-4965. 10.1016/j.febslet.2005.07.085 [DOI] [PubMed] [Google Scholar]
- Hartenstein V. (2006). The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J. Endocrinol. 190, 555-570. 10.1677/joe.1.06964 [DOI] [PubMed] [Google Scholar]
- Hiruma K., Shinoda T., Malone F. and Riddiford L. M. (1999). Juvenile hormone modulates 20-hydroxyecdysone-inducible ecdysone receptor and ultraspiracle gene expression in the tobacco hornworm, Manduca sexta. Dev. Genes Evol. 209, 18-30. 10.1007/s004270050223 [DOI] [PubMed] [Google Scholar]
- Hodková M. (1976). Nervous inhibition of corpora allata by photoperiod in Pyrrhocoris apterus. Nature 263, 521-523. 10.1038/263521a0 [DOI] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931-949. 10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.-Y. and Robinson G. E. (1995). Seasonal changes in juvenile hormone titers and rates of biosynthesis in honey bees. J. Comp. Physiol. B Biochem. Syst. Environ. Physiol. 165, 18-28. 10.1007/BF00264682 [DOI] [PubMed] [Google Scholar]
- Huang Z.-Y., Robinson G. E., Tobe S. S., Yagi K. J., Strambi C., Strambi A. and Stay B. (1991). Hormonal regulation of behavioural development in the honey bee is based on changes in the rate of juvenile hormone biosynthesis. J. Insect Physiol. 37, 733-741. 10.1016/0022-1910(91)90107-B [DOI] [Google Scholar]
- Ihle K. E., Baker N. A. and Amdam G. V. (2014). Insulin-like peptide response to nutritional input in honey bee workers. J. Insect Physiol. 69, 49-55. 10.1016/j.jinsphys.2014.05.026 [DOI] [PubMed] [Google Scholar]
- Insel T. R. and Fernald R. D. (2004). How the brain processes social information: searching for the social brain. Annu. Rev. Neurosci. 27, 697-722. 10.1146/annurev.neuro.27.070203.144148 [DOI] [PubMed] [Google Scholar]
- Jones G. and Sharp P. A. (1997). Ultraspiracle: an invertebrate nuclear receptor for juvenile hormones. Proc. Natl. Acad. Sci. USA 94, 13499-13503. 10.1073/pnas.94.25.13499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt B. and Kaltschmidt C. (2009). NF-κB in the nervous system. Cold Spring Harb. Perspect. Biol. 1, a001271 10.1101/cshperspect.a001271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller H., Dreyer C., Medin J., Mahfoudi A., Ozato K. and Wahli W. (1993). Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc. Natl. Acad. Sci. USA 90, 2160-2164. 10.1073/pnas.90.6.2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Cunningham R., James B., Wyder S., Gibson J. D., Niehuis O., Zdobnov E. M., Robertson H. M., Robinson G. E., Werren J. H. et al. (2010). Functional characterization of transcription factor motifs using cross-species comparison across large evolutionary distances. PLoS Comput. Biol. 6, e1000652 10.1371/journal.pcbi.1000652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig P., Williams L. and Boyan G. (2002). The pars intercerebralis of the locust brain: a developmental and comparative study. Microsc. Res. Tech. 56, 174-188. 10.1002/jemt.10023 [DOI] [PubMed] [Google Scholar]
- Morton G. J., Cummings D. E., Baskin D. G. and Barsh G. S. (2006). Central nervous system control of food intake and body weight. Nature 443, 289-295. 10.1038/nature05026 [DOI] [PubMed] [Google Scholar]
- Nilsen K.-A., Ihle K. E., Frederick K., Fondrk M. K., Smedal B., Hartfelder K. and Amdam G. V. (2011). Insulin-like peptide genes in honey bee fat body respond differently to manipulation of social behavioral physiology. J. Exp. Biol. 214, 1488-1497. 10.1242/jeb.050393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes F. M. F., Ihle K. E., Mutti N. S., Simões Z. L. P. and Amdam G. V. (2013). The gene vitellogenin affects microRNA regulation in honey bee (Apis mellifera) fat body and brain. J. Exp. Biol. 216, 3724-3732. 10.1242/jeb.089243 [DOI] [PubMed] [Google Scholar]
- Oro A. E., McKeown M. and Evans R. M. (1990). Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature 347, 298-301. 10.1038/347298a0 [DOI] [PubMed] [Google Scholar]
- Ozsolak F. and Milos P. M. (2011). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87-98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R. E. Jr, Fondrk M. K., Hunt G. J., Guzmán-Novoa E., Humphries M. A., Nguyen K. and Greene A. S. (2000). Genetic dissection of honeybee (Apis mellifera L.) foraging behavior. J. Hered. 91, 474-479. 10.1093/jhered/91.6.474 [DOI] [PubMed] [Google Scholar]
- Pankiw T., Huang Z.-Y., Winston M. L. and Robinson G. E. (1998). Queen mandibular gland pheromone influences worker honey bee (Apis mellifera L.) foraging ontogeny and juvenile hormone titers. J. Insect Physiol. 44, 685-692. 10.1016/S0022-1910(98)00040-7 [DOI] [PubMed] [Google Scholar]
- Peixoto L. G., Calábria L. K., Garcia L., Capparelli F. E., Goulart L. R., de Sousa M. V. and Espindola F. S. (2009). Identification of major royal jelly proteins in the brain of the honeybee Apis mellifera. J. Insect Physiol. 55, 671-677. 10.1016/j.jinsphys.2009.05.005 [DOI] [PubMed] [Google Scholar]
- Pinto L. Z., Bitondi M. M. G. and Simões Z. L. P. (2000). Inhibition of vitellogenin synthesis in Apis mellifera workers by a juvenile hormone analogue, pyriproxyfen. J. Insect Physiol. 46, 153-160. 10.1016/S0022-1910(99)00111-0 [DOI] [PubMed] [Google Scholar]
- Rajan A. and Perrimon N. (2012). Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151, 123-137. 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. E. (1985). Effects of a juvenile hormone analogue on honey bee foraging behaviour and alarm pheromone production. J. Insect Physiol. 31, 277-282. 10.1016/0022-1910(85)90003-4 [DOI] [Google Scholar]
- Robinson G. E. (1987). Regulation of honey bee age polyethism by juvenile hormone. Behav. Ecol. Sociobiol. 20, 329-338. 10.1007/BF00300679 [DOI] [Google Scholar]
- Robinson G. E., Fernald R. D. and Clayton D. F. (2008). Genes and social behavior. Science 322, 896-900. 10.1126/science.1159277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K. and Nusse R. (2002). Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296, 1118-1120. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- Rutz W., Gerig L., Wille H. and Lüscher M. (1976). The function of juvenile hormone in adult worker honeybees, Apis mellifera. J. Insect Physiol. 22, 1485-1491. 10.1016/0022-1910(76)90214-6 [DOI] [Google Scholar]
- Schulte C., Theilenberg E., Müller-Borg M., Gempe T. and Beye M. (2014). Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. USA 111, 9003-9008. 10.1073/pnas.1402341111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D. J., Huang Z.-Y. and Robinson G. E. (1998). Effects of colony food shortage on behavioral development in honey bees. Behav. Ecol. Sociobiol. 42, 295-303. 10.1007/s002650050442 [DOI] [Google Scholar]
- Schulz D. J., Pankiw T., Fondrk M. K., Robinson G. E. and Page R. E. Jr (2004). Comparisons of juvenile hormone hemolymph and octopamine brain titers in honey bees (Hymenoptera: Apidae) selected for high and low pollen hoarding. Ann. Entomol. Soc. Am. 97, 1313-1319. 10.1603/0013-8746(2004)097[1313:COJHHA]2.0.CO;2 [DOI] [Google Scholar]
- Shiga S. and Numata H. (2000). The role of neurosecretory neurons in the pars intercerebralis and pars lateralis in reproductive diapause of the blowfly, Protophormia terraenovae. Naturwissenschaften 87, 125-128. 10.1007/s001140050689 [DOI] [PubMed] [Google Scholar]
- Sinha S., Ling X., Whitfield C. W., Zhai C. and Robinson G. E. (2006). Genome scan for cis-regulatory DNA motifs associated with social behavior in honey bees. Proc. Natl. Acad. Sci. USA 103, 16352-16357. 10.1073/pnas.0607448103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedal B., Brynem M., Kreibich C. D. and Amdam G. V. (2009). Brood pheromone suppresses physiology of extreme longevity in honeybees (Apis mellifera). J. Exp. Biol. 212, 3795-3801. 10.1242/jeb.035063 [DOI] [PubMed] [Google Scholar]
- Strausfeld N. J. (1976). Atlas of an Insect Brain. Berlin: Springer. [Google Scholar]
- Sullivan J. P., Jassim O., Fahrbach S. E. and Robinson G. E. (2000). Juvenile hormone paces behavioral development in the adult worker honey bee. Horm. Behav. 37, 1-14. 10.1006/hbeh.1999.1552 [DOI] [PubMed] [Google Scholar]
- Toth A. L. and Robinson G. E. (2005). Worker nutrition and division of labour in honeybees. Anim. Behav. 69, 427-435. 10.1016/j.anbehav.2004.03.017 [DOI] [Google Scholar]
- Toth A. L., Kantarovich S., Meisel A. F. and Robinson G. E. (2005). Nutritional status influences socially regulated foraging ontogeny in honey bees. J. Exp. Biol. 208, 4641-4649. 10.1242/jeb.01956 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. and Salzberg S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105-1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder R. N., von Zastrow M. E., Yool A., Dement W. C., Barchas J. D. and Eberwine J. H. (1990). Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA 87, 1663-1667. 10.1073/pnas.87.5.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velarde R. A., Robinson G. E. and Fahrbach S. E. (2006). Nuclear receptors of the honey bee: annotation and expression in the adult brain. Insect Mol. Biol. 15, 583-595. 10.1111/j.1365-2583.2006.00679.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H., Heinzeller T. and Renner M. (1977). Daily protein synthesis in the pars intercerebralis and corpora allata of Apis mellifica L. with and without training to a foraging schedule. J. Comp. Physiol. A 118, 51-60. 10.1007/BF00612336 [DOI] [Google Scholar]
- Wang Y., Mutti N. S., Ihle K. E., Siegel A., Dolezal A., Kaftanoglu O. and Amdam G. V. (2010). Down-regulation of honey bee IRS gene biases behavior toward food rich in protein. PLoS Genet. 6, e1000896 10.1371/journal.pgen.1000896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Brent C. S., Fennern E. and Amdam G. V. (2012). Gustatory perception and fat body energy metabolism are jointly affected by vitellogenin and juvenile hormone in honey bees. PLoS Genet. 8, e1002779 10.1371/journal.pgen.1002779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard M. J. (2003). Developmental Plasticity and Evolution. New York, NY: The Oxford University Press. [Google Scholar]
- Wheeler M. M., Ament S. A., Rodriguez-Zas S. L. and Robinson G. E. (2013). Brain gene expression changes elicited by peripheral vitellogenin knockdown in the honey bee. Insect Mol. Biol. 22, 562-573. 10.1111/imb.12043 [DOI] [PubMed] [Google Scholar]
- Whitfield C. W., Cziko A.-M. and Robinson G. E. (2003). Gene expression profiles in the brain predict behavior in individual honey bees. Science 302, 296-299. 10.1126/science.1086807 [DOI] [PubMed] [Google Scholar]
- Whitfield C. W., Ben-Shahar Y., Brillet C., Leoncini I., Crauser D., LeConte Y., Rodriguez-Zas S. and Robinson G. E. (2006). Genomic dissection of behavioral maturation in the honey bee. Proc. Natl. Acad. Sci. USA 103, 16068-16075. 10.1073/pnas.0606909103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston M. L. (1987). The Biology of the Honey Bee. Cambridge, MA: Harvard University Press. [Google Scholar]
- Wu Q., Wen T., Lee G., Park J. H., Cai H. N. and Shen P. (2003). Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 39, 147-161. 10.1016/S0896-6273(03)00396-9 [DOI] [PubMed] [Google Scholar]