Abstract
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-associated death worldwide. Indeed, despite the benefit of sorafenib in the treatment of some patients with HCC, the majority of these patients have a poor response to or intolerance of sorafenib, resulting in further tumor progression. Exploring the mechanisms underlying sorafenib resistance is essential to the treatment of HCC. Long noncoding RNAs (lncRNAs) are known as participants in tumorigenesis. In this study, we identified that long intergenic non-protein coding RNA, regulator of reprogramming (LINC-ROR), was upregulated in HCC cell lines, which was transcriptionally activated by FOXM1. Furthermore, the sponging of miR-876-5p by LINC-ROR released FOXM1, thereby forming a positive-feedback loop. Additionally, we demonstrated that upregulation of both FOXM1 and LINC-ROR impaired the sensitivity to sorafenib in HCC cells. The role of this feedback loop was demonstrated by rescue assays. These results revealed a novel molecular feedback loop between LINC-ROR and FOXM1 and elucidated their functions in sorafenib sensitivity of HCC cell lines.
Keywords: LINC-ROR, FOXM1, miR-876-5p, HCC, sorafenib, sensitivity
Introduction
Hepatocellular carcinoma (HCC) is a prevalent malignancy worldwide with a high recurrence rate and poor prognosis.1, 2 Unfortunately, more than 500,000 new patients are diagnosed with HCC every year, and the situation is more serious in developing countries, especially in China.3 Because of late-stage detection and lack of effective therapies, the majority of patients lose the opportunity for surgery. Moreover, non-surgical treatments, including transarterial chemoembolization and ablation and systemic therapy, have no significant effect. As a result, the overall survival rate of HCC patients is still poor.4 Sorafenib, a multiple kinase inhibitor, has been approved by the U.S. Food and Drug Administration for advanced HCC treatment.5 Although sorafenib has been shown to improve HCC patients’ prognosis to some extent, inherent resistance and acquired resistance have limited its long-term value.6, 7, 8, 9 Hence, it is urgent to explore the molecular mechanisms underlying intrinsic sorafenib resistance in HCC patients.
Long noncoding RNAs (lncRNAs) are defined as a class of transcripts with at least 200 nt and limited protein-coding ability.10 Recent studies have uncovered that lncRNAs can exert various functions in different manners, including chromatin modification, transcription, and post-transcriptional regulation.11, 12, 13 In addition, aberrant expression of lncRNAs has been verified in multiple malignancies, including HCC.14, 15 More recently, a novel mechanism of post-transcriptional regulation has been proposed, in which lncRNAs interact with microRNAs (miRNAs) as competing endogenous RNAs (ceRNAs).16, 17
The long intergenic non-protein coding RNA, regulator of reprogramming (LINC-ROR) was first identified as a lincRNA involved in the reprogramming of differentiated cells into induced pluripotent stem cells.18 Previous studies indicated that LINC-ROR may function as a key ceRNA to regulate the expression of some core transcription factors, such as Oct4, Sox2, and Nanog.19, 20 Furthermore, LINC-ROR has been implicated in promoting proliferation and inducing metastasis, invasion, or chemoresistance.21, 22, 23 Our previous studies demonstrated that LINC-ROR promotes invasion and metastasis and radio resistance of HCC cells.24, 25 Actually, a previous study in 2014 implied that LINC-ROR is enriched in extracellular vesicles during chemotherapeutic stress and can functionally modulate chemotherapy-induced apoptosis and cell survival.26 However, LINC-ROR-related molecular mechanisms underlying primary sorafenib resistance of HCC cells has rarely been explored.
Forkhead Box M1b (FOXM1b or FOXM1) is a potent transcription factor that exerts oncogenic functions by activating many downstream targets in human malignant tumors. Accumulating evidence has indicated that FOXM1 plays significant roles in cancer initiation, progression, metastasis, and chemoresistance.27 Moreover, FOXM1 can cooperate with yes-associated protein 1 to induce chromosome instability in liver cancer.28 Based on a gene expression signature, high FOXM1 is significantly associated with poor prognosis of HCC patients.29, 30 A recent investigation suggested that lncRNAs may be involved in the functions of FOXM1.31 However, the interaction between LINC-ROR and FOXM1 and their effects on chemosensitivity in HCC have not been elucidated yet. Furthermore, previous studies have also focused on the role of FOXM1 in regulating the chemosensitivity or chemoresistance of various tumor cells. It has been verified that FOXM1 inhibition could enhance sensitivity of docetaxel-resistant A549 cells to docetaxel,32 but reverse docetaxel resistance in gastric cancer33 and contribute to curcumin-induced chemosensitivity in acute myeloid leukemia.34 Also, targeting FOXM1 with thiostrepton led to chemosensitizing effects in T-ALL Jurkat cells.35 However, the role of FOXM1 in the primary sorafenib resistance of HCC cells is poorly understood.
In the current study, we verified the high expression of LINC-ROR and FOXM1 in HCC cells and uncovered the positive regulatory correlation between LINC-ROR and FOXM1. Moreover, FOXM1 attenuates the chemosensitivity of HCC cells, partially through modulation of LINC-ROR. Furthermore, we validated that LINC-ROR functions as a molecular sponge of miR-876-5p to upregulate FOXM1. In general, FOXM1-mediated LINC-ROR activation contributes to poor sensitivity of HCC cells to sorafenib via partially regulating the miR-876-5p/FOXM1 axis, which forms a positive-feedback loop and provides new insight for exploring a potential therapeutic strategy for HCC.
Results
LINC-ROR Was Upregulated in HCC Cells and Activated by FOXM1
Our previous studies have demonstrated that high expression of LINC-ROR was related to advanced tumor node metastasis stage, high recurrence, poor prognosis of HCC patients and LINC-ROR promoted HCC metastasis and radioresistance.24, 25 However, the mechanisms of high LINC-ROR level and low chemosensitivity to sorafenib are still unclear.
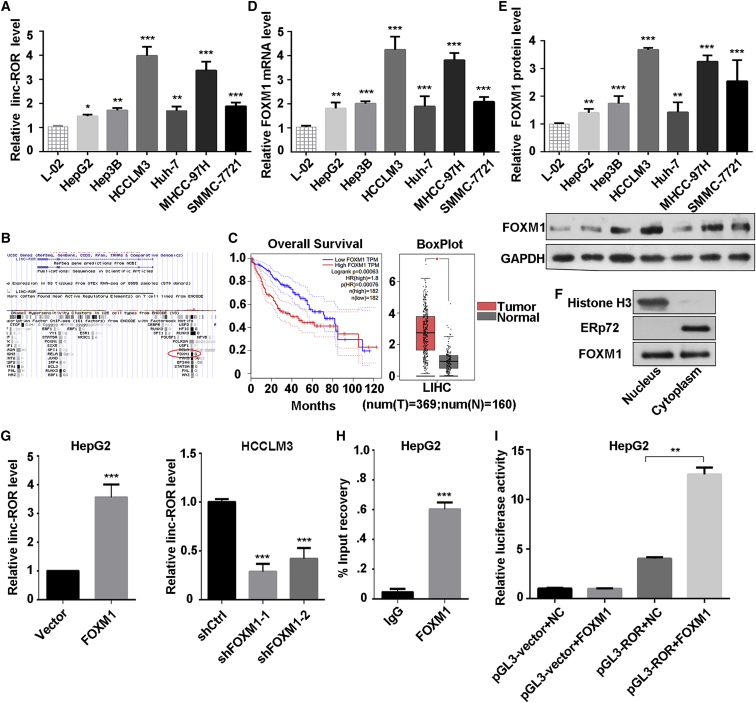
We performed quantitative real-time PCR to detect the expression of LINC-ROR in HCC cell lines. As shown in Figure 1A, the expression of LINC-ROR was upregulated in these six HCC cell lines, and the normal liver cell line L-02 was used as the control. Then, we searched for the mechanism by which LINC-ROR is upregulated in HCC cell lines. We searched and analyzed the ENCODE TFBS chromatin immunoprecipitation sequencing (ChIP-seq) data (https://genome-asia.ucsc.edu/index.html) and found that FOXM1 is one of the potential transcription factors of LINC-ROR (Figure 1B). Hence, we hypothesized that FOXM1 regulates the transcription of LINC-ROR. Based on The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) dataset (http://gepia.cancer-pku.cn/), FOXM1 expression was higher in tumor tissues compared to normal tissues. Also, the cohort analysis showed that overall survival of patients with high FOXM1 expression was significantly poorer than those with low FOXM1 expression (Figure 1C). Additionally, we investigated the FOXM1 expression in HCC cell lines. The results showed that both mRNA and protein levels of FOXM1 were upregulated in HCC cell lines (Figures 1D and 1E). Hence, we hypothesized that FOXM1 regulates the transcription of LINC-ROR. A nuclear and cytoplasmic protein extraction assay was performed to examine the subcellular localization of FOXM1 in HepG2 cells. It was found that FOXM1 was located in both the nucleus and the cytoplasm (Figure 1F). Subsequently, HepG2 cells were transfected with the FOXM1-overexpressing plasmid or control vector, and HCCLM3 cells were transfected with short hairpin FOXM1 (shFOXM1) or short hairpin of control (shCtrl) plasmid. Satisfactory transfection efficiency was obtained after 48 h (Figures S1A and S1B). Then, we observed that the LINC-ROR level was significantly decreased in FOXM1-knockdown HCCLM3 cells, whereas it was increased in FOXM1-overexpressing HepG2 cells (Figure 1G). These data indicated that FOXM1 may transcriptionally regulate LINC-ROR in HCC cells. Their binding sequences were listed in Table S1.
Figure 1.
LINC-ROR Was Upregulated in HCC Cells and Activated by FOXM1
(A) The expression level of LINC-ROR in a normal liver cell line and six HCC cell lines were tested by quantitative real-time PCR. (B) The binding region of FOXM1 at the promoter of LINC-ROR in the UCSC Genome Browser (assembly >hg19, DNA range: chr18:54,745,025–54,745,392; strand, +). (C) The overall survival curve in HCC patients with high or low FOXM1 expression and a boxplot analysis of FOXM1 expression in normal and tumor tissues are shown, based on GEPIA data (http://gepia.cancer-pku.cn/). (D) The expression level of FOXM1 mRNA in a normal liver cell line and six HCC cell lines was tested by quantitative real-time PCR. (E) Western blot analysis of the protein level of FOXM1 in a normal liver cell line and six HCC cell lines (bottom) and the quantification of FOXM1 protein (top). (F) Relative enrichment of FOXM1 proteins in purified nuclear and cytoplasmic fraction by nuclear and cytoplasmic protein extraction assay. Histone 3 was used as the nuclear marker, and ERp72 was used as the cytoplasmic marker. (G) LINC-ROR expression was measured by quantitative real-time PCR in HepG2 or HCCLM3 cells transfected with FOXM1, shFOXM1-1, or shFOXM1-2, with relative control groups. (H) ChIP assays using anti-FOXM1 or anti-IgG antibody were applied to determine the affinity of FOXM1 on LINC-ROR promoter in HepG2 cells. Relative enrichment is normalized to IgG as the negative control. (I) Dual-luciferase reporter assays were performed to confirm the binding of FOXM1 and the LINC-ROR promoter in HepG2 cells. All data are presented as the mean ± SD of three independent experiments. The p values represent comparisons between groups (*p < 0.05, **p < 0.01, ***p < 0.001).
To further investigate the regulatory mechanism underlying the correlation between FOXM1 and LINC-ROR, ChIP was performed, and the result revealed that FOXM1 directly bound to the LINC-ROR promoter (Figure 1H; Figure S1C). Moreover, a dual-luciferase reporter assay indicated that overexpression of FOXM1 stimulated LINC-ROR promoter activity in HepG2 cells (Figure 1I). Taken together, the results showed that FOXM1 directly bound to the LINC-ROR promoter and promoted the transcription of LINC-ROR.
FOXM1 Conferred Poor Sorafenib Tolerance to HCC Cells, Partially through LINC-ROR
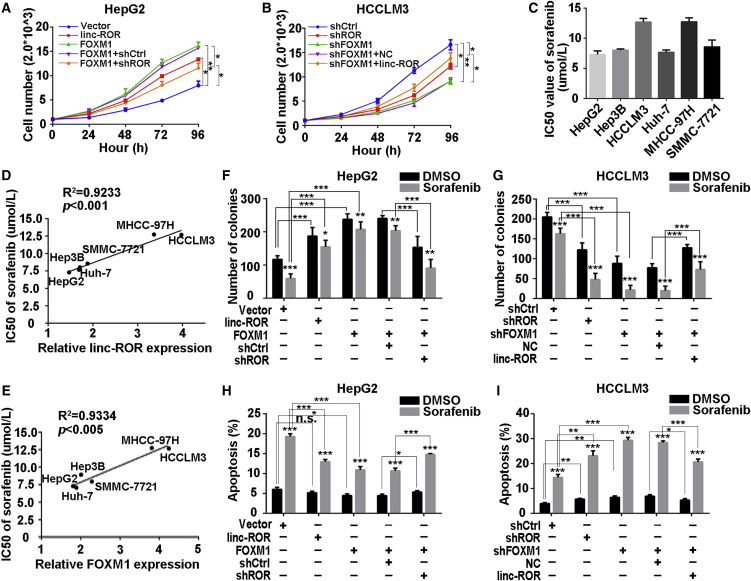
To explore the biological significance of FOXM1 and LINC-ROR in HCC, cells were constructed into stable FOXM1-overexpressing or FOXM1-silenced cell lines with puromycin. Similarly, we constructed LINC-ROR-specific-overexpressing HepG2 and LINC-ROR-silenced HCCLM3 cells. Satisfactory transfection efficiency was obtained after 48 h (Figure S1D). We chose the most efficient shRNA for the forthcoming experiments. Cell Counting Kit-8 (CCK-8) assays showed that FOXM1 facilitated HCC cell proliferation, whereas this effect was attenuated by LINC-ROR suppression in part, and inverse results were observed in HCCLM3 cells (Figures 2A and 2B). Additionally, we verified that LINC-ROR could promote proliferation capacity of HCC cells as well. To determine whether dysregulation of FOXM1 and LINC-ROR could regulate resistance of HCC cells to sorafenib, we examined the 50% inhibitory concentration (IC50) of the above six HCC cell lines. The results suggested that the IC50 in response to sorafenib correlated positively with relative LINC-ROR or FOXM1 levels in HCC cell lines (Figure 2C–2E; Figure S3A). Subsequently, we measured the sensitivity to sorafenib in different cell groups, with or without sorafenib. Colony-formation assays demonstrated that FOXM1 overexpression promoted cell colony-forming ability compared with the control group but attenuated sorafenib sensitivity, while the response was rescued in part by LINC-ROR knockdown (Figure 2F; Figure S2A). In contrast, suppression of FOXM1 expression impaired the colony-forming ability but enhanced the sensitivity to sorafenib, whereas the above effects could be restored partially by LINC-ROR overexpression (Figure 2G, Figure S2B). Moreover, flow cytometry assays indicated that forced FOXM1 expression decreased the cell apoptosis rate and enhanced the tolerance to sorafenib (Figure 2H; Figure S2C). Consistently, this effect could be reversed partially by suppression of LINC-ROR. On the contrary, silenced FOXM1 in HCCLM3 cells increased the apoptosis rate and promoted the sensitivity to sorafenib. Furthermore, this effect was reversed partially by forcing LINC-ROR (Figure 2I; Figure S2D). These findings suggested that FOXM1 conferred sorafenib tolerance to HCC cells via modulation of LINC-ROR.
Figure 2.
FOXM1 Conferred Poor Sorafenib Response on HCC Cells Partially through LINC-ROR
(A) CCK-8 assays were performed to detect the proliferation of HepG2 cells transfected with LINC-ROR or FOXM1 or co-transfected with FOXM1 and shROR, respectively. (B) CCK-8 assays were performed to detect the proliferation of HCCLM3 cells transfected with shROR or shFOXM1 or co-transfected with shFOXM1 and LINC-ROR. (C) IC50 values of sorafenib in HCC cell lines were measured by CCK-8. (D) The correlation plot of IC50 of sorafenib and relative LINC-ROR level. (E) A correlation plot of IC50 of sorafenib and relative FOXM1 level. (F) Quantification of colony-formation ability of HepG2 cells treated with LINC-ROR or FOXM1 or co-treated with FOXM1 and shROR individually, with sorafenib or DMSO added. (G) Quantification of colony-formation ability of HCCLM3 cells transfected with shROR or shFOXM1 or co-transfected with shFOXM1 and LINC-ROR individually, with sorafenib or DMSO treatment. (H) Quantification of total apoptosis rate of HepG2 cells after transfection of LINC-ROR or FOXM1 or co-transfection of FOXM1 and shROR, supplemented with sorafenib or DMSO. The percentage represents the sum of both the Annexin- and 7-AAD-positive populations. (I) Quantification of total apoptosis rate of HCCLM3 cells after transfection with shROR or shFOXM1 or co-transfection with shFOXM1 and LINC-ROR, supplemented with sorafenib or DMSO. The percentage represents a sum of both the Annexin- and 7-AAD-positive populations. All data are presented as the mean ± SD of three independent experiments. The p values represent comparisons between groups (*p < 0.05, **p < 0.01, ***p < 0.001).
LINC-ROR Served as a Competing Endogenous RNA for miR-876-5p to Regulate FOXM1 Expression
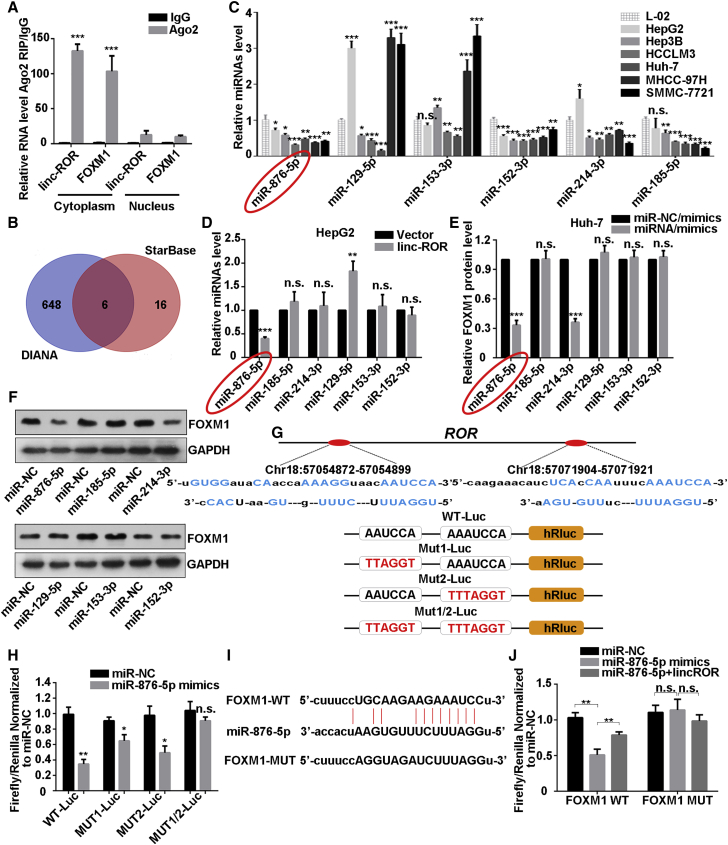
There has been much evidence that lncRNAs act as molecular sponges of specific miRNAs having complementary sequences with them, thus interfering in their functions. Also, increasing indications have revealed that LINC-ROR participated in the regulatory network of ceRNA. After LINC-ROR regulation, we also observed that both the mRNA and protein level of FOXM1 were positively regulated (Figures S3B and S3C). Hence, we hypothesized that endogenous LINC-ROR also acted as a ceRNA of FOXM1 in HCC. A previous study performed fluorescence in situ hybridization in breast cancer cells to examine the cellular localization of linc-ROR and confirmed that LINC-ROR was mainly located in cytoplasm.22 We demonstrated that LINC-ROR was mainly located in the cytoplasm in HepG2 cells by using the nuclear and cytoplasmic RNA extraction assay (Figure S3D). Studies have shown that cytoplasmic miRNAs are usually assembled into micro ribonucleoprotein (miRNP) complexes, to play roles in target mRNA recognition and translational repression. Ago2, which is a core component of the RNA-induced silencing complex (RISC), is also a component of miRNPs. A previous study observed that LINC-ROR was recruited to Ago2-related RISCs and interacted with miRNAs.36 Next, we carried out RNA immunoprecipitation (RIP) with anti-Ago2 protein and then performed real-time PCR assays to detect the enrichment of LINC-ROR and the mRNA of FOXM1. As shown in Figure 3A, LINC-ROR and FOXM1 mRNA were both enriched in Ago2 in the cytoplasm. What is interesting is that their enrichment was drastically reduced in Ago2 complexes purified from the nucleus. These results supported that LINC-ROR may serve as a ceRNA for FOXM1 mRNA in cytoplasm.
Figure 3.
LINC-ROR Acts as a Competing Endogenous RNA for miR-876-5p to Regulate FOXM1 Expression
(A) RIP experiments revealed that LINC-ROR and FOXM1 mRNA coexisted in the anti-Ago2 complex in the cell cytoplasm. IgG was used as the negative control. (B) A Venn diagram shows the overlapping miRNAs that are predicted to bind with both LINC-ROR and the 3′ UTR of FOXM1 mRNA. (C) The expression levels of the miRNAs were detected with quantitative real-time PCR in a normal liver cell line and six HCC cell lines. (D) The expression levels of the six miRNAs were measured individually by quantitative real-time PCR methods in LINC-ROR-transfected HepG2 cells. (E) Quantitative real-time PCR and (F) western blot analysis of FOXM1 expression after all six miRNA mimics were introduced. (G) Binding sequences of LINC-ROR and miR-876-5p based on bioinformatics analysis and schematic constructions of WT-Luc (LINC-ROR 3′ partial region), Mut1-Luc, Mut2-Luc, and Mut1/2-Luc. (H) Dual-luciferase reporter assays were performed to examine the potential combination of LINC-ROR and miR-876-5p. (I) WT and mutant binding sites of miR-876-5p and 3′ UTR of FOXM1 mRNA from StarBase (http://starbase.sysu.edu.cn/). (J) Dual-luciferase reporter assays were conducted to confirm the association of FOXM1 and LINC-ROR or miR-876-5p and FOXM1. All data are presented as the mean ± SD of three independent experiments. The p values represent comparisons between groups (*p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant).
To search for miRNAs targeting the full-length transcripts of LINC-ROR and FOXM1, we applied the online bioinformatics tool DIANA (http://diana.imis.athena-innovation.gr/)37 and StarBase (http://starbase.sysu.edu.cn/).38 Of the miRNAs that fit the criteria (Figure 3B), six miRNAs-miR-152-3p, miR-153-3p, miR-129-5p, miR-185-5p, miR-214-3p, and miR-876-5p emerged as candidates as their predicted binding sites were shared by LINC-ROR and FOXM1. We then determined the expression of these six miRNAs in L-02 cells and six HCC cell lines. The results showed that the expression levels of miR-876-5p and miR-152-3p were higher in the cell lines whose LINC-ROR and FOXM1 mRNA levels were relative lower (Figure 3C). We examined the expression of all six of the miRNAs after overexpressing LINC-ROR in HepG2 cells. As shown in Figure 3D, only miR-876-5p was downregulated. We also confirmed that the miR-876-5p expression was upregulated in HCCLM3 cells after LINC-ROR silencing (Figure S3E). In addition, we transfected miR-NC/mimics and all six of the miRNA/mimics into Huh-7 cells, in which miRNAs levels were relatively lower. The transfection efficiency is confirmed in Figure S3F. Then, we investigated the FOXM1 protein levels after transfection of miR-mimics. As shown in Figures 3E and 3F, miR-876-5p and miR-214-3p had a negative effect on FOXM1 protein. Integrated analysis of the results indicated that there is a great possibility that miR-876-5p is the focus of the ceRNA model.
For further confirmation of the interaction between LINC-ROR and miR-876-5p, we subcloned LINC-ROR (wild type [WT] and the mutants Mut1, Mut2, and Mut1/2) downstream of the firefly luciferase gene into the pmirGLO vector, based on the online bioinformatics analysis DIANA (Figure 3G) and performed a luciferase reporter assay. As illustrated in Figure 3H, miR-876-5p greatly reduced the luciferase activity of WT and mutant Mut1 and Mut2 LINC-ROR reporters instead of the mutant Mut1/2 reporter. Referring to the putative binding sites on the online bioinformatics analysis (StarBase), another dual-luciferase reporter assay was applied. The results suggest that miR-876-5p mimic co-expression inhibited firefly luciferase activity of the WT but not the mutant type of FOXM1 3′ UTR, and this effect was abolished by the introduction of LINC-ROR (Figures 3I and 3J). Collectively, these data revealed that LINC-ROR serves as a competing endogenous RNA for miR-876-5p to regulate FOXM1 expression in HCC cells.
miR-876-5p Attenuated Proliferation, Promoted Apoptosis and Chemosensitivity of HCC Cells
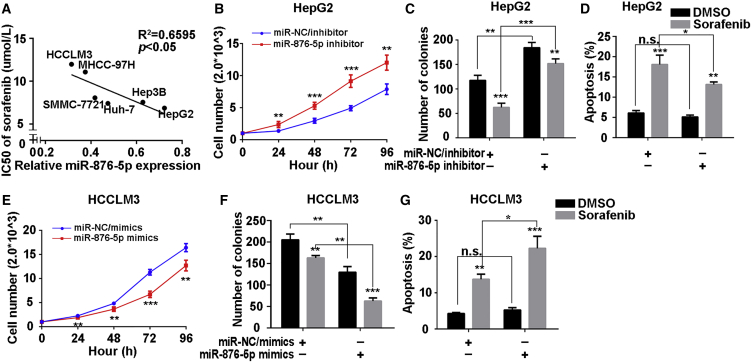
Since miR-876-5p had been shown to be sponged by LINC-ROR, we explored the effects of miR-876-5p on the proliferation, apoptosis, and sorafenib sensitivity of HCC cells. We first analyzed the correlation between the IC50 in response to sorafenib and relative miR-876-5p expression in six HCC cells and found that the IC50 values of the six HCC cell lines correlated negatively with the miR-876-5p levels (Figure 4A).
Figure 4.
miR-876-5p Attenuated Proliferation and Promoted Apoptosis and Chemosensitivity of HCC Cells
(A) A correlation plot of IC50 of sorafenib and relative miR-876-5p levels in six HCC cell lines. (B) CCK-8 assays were performed to detect the proliferation of HepG2 cells transfected with miR-NC/inhibitor or miR-876-5p/inhibitor. (C) Quantification of colony-formation ability of HepG2 cells treated with miR-NC/inhibitor or miR-876-5p/inhibitor, with sorafenib or DMSO added. (D) Quantification of the total apoptosis rate of HepG2 cells after transfection of miR-NC/inhibitor or miR-876-5p/inhibitor, supplemented with sorafenib or DMSO. The percentage represents the sum of both the Annexin- and 7-AAD-positive populations. (E) CCK-8 assays were performed to detect the proliferation of HCCLM3 cells after transfection with miR-NC/mimics or miR-876-5p/mimics. (F) Quantification of colony-formation ability of HCCLM3 cells treated with miR-NC/mimics or miR-876-5p/mimics, with sorafenib or DMSO added. (G) Quantification of total apoptosis rate of HCCLM3 cells after transfection with miR-NC/mimics or miR-876-5p/mimics, supplemented with sorafenib or DMSO. The percentage represents a sum of the Annexin- and 7-AAD-positive populations. All data are presented as the mean ± SD of three independent experiments. The p values represent comparisons between groups (*p < 0.05, **p < 0.01, ***p < 0.001).
Subsequently, we transfected miR-NC/inhibitor or miR-876-5p/inhibitor and miR-NC/mimics or miR-876-5p/mimics into HepG2 and HCCLM3 cells, respectively. The transfection efficiency was confirmed (Figure S3G). Results of the CCK-8, colony-formation, and apoptosis assays showed that miR-876-5p-inhibition enhanced proliferation, inhibited apoptosis and attenuated the sensitivity to sorafenib in HepG2 cells (Figures 4B–4D; Figures S2E and S2F). Furthermore, opposite results were observed in HCCLM3 cells after overexpression of miR-876-5p (Figures 4E–4G; Figures S2G and S2H).
Taken together, these results revealed that miR-876-5p correlated negatively with proliferation and positively with apoptosis and chemosensitivity of HCC cells.
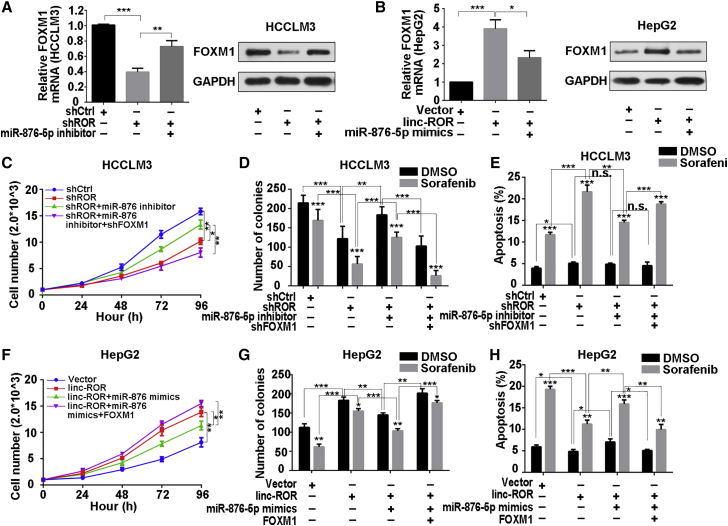
miR-876-5p-Mediated FOXM1 Suppression Was Involved with LINC-ROR-Mediated Poor Sorafenib Sensitivity
To assess the role of miR-876-5p in the LINC-ROR-mediated regulatory loop, we further employed the miR-876-5p inhibitor to abolish miR-876-5p elevation in LINC-ROR-silenced HCCLM3 cells. The results showed that after miR-876-5p inhibitor transfection, FOXM1 mRNA and protein level were rescued, and opposite results were acquired in HepG2 cells (Figures 5A and 5B). Also, the proliferation-inhibiting ability of LINC-ROR-suppression was enhanced partially by miR-876-5p inhibitor. Unsurprisingly, proliferative ability was attenuated again by suppressing FOXM1 (Figure 5C).
Figure 5.
FOXM1 Suppression, Mediated by miR-876-5p, Was Involved with LINC-ROR-Mediated Sorafenib Tolerance
(A) Quantitative real-time PCR and (B) western blotting were utilized to examine FOXM1 mRNA and protein levels in HCCLM3 cells, transfected with shROR or co-transfected with shROR and miR-876-5p inhibitor, or in HepG2 cells, transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics. (C) CCK-8 assays were carried out to detect the proliferation of HCCLM3 cells transfected with shROR or co-transfected with shROR and miR-876-5p inhibitor or co-transfected with shROR, miR-876-5p inhibitor, and shFOXM1. (D) Quantification of colony-formation ability of HCCLM3 cells transfected with shROR or co-transfected with shROR and miR-876-5p inhibitor or co-transfected with shROR, miR-876-5p inhibitor, and shFOXM1 under sorafenib treatment. (E) Quantification of total apoptosis rate of HCCLM3 cells treated with shROR or co-transfected with shROR and miR-876-5p inhibitor or co-transfected with shROR, miR-876-5p inhibitor, and shFOXM1, with the addition of DMSO or sorafenib. The percentage represents a sum of the Annexin- and 7-AAD-positive populations. (F) CCK-8 assays were carried out to detect the proliferation of HepG2 cells transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics or co-transfected with LINC-ROR, miR-876-5p mimics, and FOXM1. (G) Quantification of colony-formation ability of HepG2 cells transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics or co-transfected with LINC-ROR, miR-876-5p mimics, and FOXM1 after sorafenib treatment. (H) Quantification of total apoptosis rate of HepG2 cells transfected with LINC-ROR or co-transfected with LINC-ROR and miR-876-5p mimics or co-transfected with LINC-ROR, miR-876-5p mimics, and FOXM1, with the addition of DMSO or sorafenib. The percentage represents the sum of the Annexin- and 7-AAD-positive populations. All data are presented as the mean ± SD of three independent experiments. The p values represent comparisons between groups (*p < 0.05, **p < 0.01, ***p < 0.001).
Having confirmed that LINC-ROR could function as a ceRNA of FOXM1 through sponging miR-876-5p in HCC cells, we investigated whether LINC-ROR could affect the chemosensitivity of HCC cells, with or without sorafenib, via regulation of miR-876-5p. Colony-formation and apoptosis assays suggested that, despite sorafenib treatment, the colony-formation capacity decreased, and the apoptosis rate increased after LINC-ROR silencing. Furthermore, upon sorafenib supplementation, the colony-formation capacity decayed more obviously, and the apoptosis rate was enhanced more obviously as well, indicating that the sensitivity of cells to sorafenib was forced. Conversely, the introduction of miR-876-5p inhibitor increased proliferation, decreased apoptosis, and attenuated sorafenib sensitivity, thus resisting the effects mediated by suppressing LINC-ROR. Furthermore, suppressing FOXM1 reversed the effects of miR-876-5p inhibitor-weakened colony-forming ability, increased the apoptosis rate, and restored the response to sorafenib (Figures 5D and 5E; Figures S2I and S2J). On the contrary, overexpressed LINC-ROR induced an increase in proliferation and a decrease in apoptosis, and forced chemoresistance was compensated for by miR-876-5p mimics. Furthermore, overexpressing FOXM1 restored the effects of miR-876-5p mimics (Figures 5F–5H; Figures S2K and S2L). These results suggested that miR-876-5p-mediated FOXM1 suppression was involved with LINC-ROR-induced poor chemosensitivity.
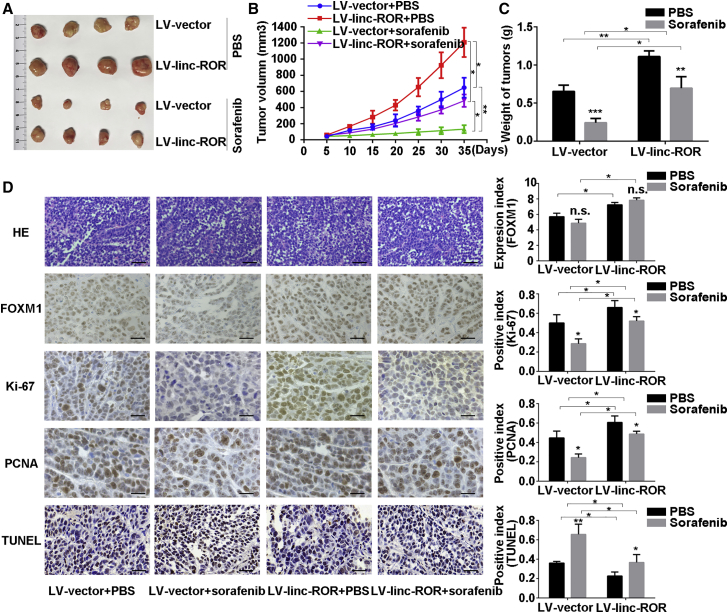
LINC-ROR or FOXM1 Overexpression Both Attenuated the Sensitivity to Sorafenib In Vivo
To verify the effects of LINC-ROR and FOXM1 on sensitivity to sorafenib in vivo, we employed the xenograft model with HepG2 cells with stable LINC-ROR and FOXM1 overexpression and used cells transfected with lv-vector or vector plasmid as negative controls, respectively. Consistent with in vitro observations, tumors derived from cells stably transfected with LINC-ROR- or FOXM1-overexpressing vectors grew faster than those derived from control cells, while this phenomenon was weakened compared with the control groups after continuous treatment with sorafenib (Figures 6A–6C; Figures S4A–S4C). Tumors derived from stably LINC-ROR- or FOXM1-transfected HepG2 cells exhibited increased Ki-67 and proliferating cell nuclear antigen (PCNA) expression compared with the control groups (Figure 6D; Figure S4D). We also observed that the FOXM1 protein level was upregulated in tumors derived from LINC-ROR-overexpressing cells, whereas no significant alternations were observed after sorafenib treatment. In addition, we applied the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining assay to detect the apoptosis rate in the tumors. As illustrated in Figure 6D, tumors derived from stably LINC-ROR-transfected HepG2 cells presented with significantly reduced apoptosis compared with tumors derived from control cells, and this effect was more obvious with continuous sorafenib treatment. With regard to FOXM1 overexpression, similar conclusions were obtained (Figure S4D). Therefore, the in vivo experiments supplemented the results of the in vitro experiments.
Figure 6.
Overexpression of LINC-ROR Attenuated Sorafenib Sensitivity In Vivo
(A) HepG2-lv-vector or HepG2-lv-LINC-ROR cells were implanted into nude mice. When the average tumor volume reached 50 mm3, the two groups of mice were subdivided into four groups randomly and were given 60 mg/kg sorafenib or PBS. Tumor sizes were calculated every 5 days. Images of subcutaneous xenograft tumors were taken. (B) Growth curves of the subcutaneous xenograft tumors were calculated; the bars indicate the SD. (C) The final tumor weights were measured and calculated. (D) Representative images of H&E staining, immunohistochemical staining (for FOXM1, Ki-67 and PCNA), and TUNEL staining are shown. H&E staining and TUNEL staining: scale bar, 50 μm; immunohistochemical staining for FOXM1, Ki-67, and PCNA: scale bar, 100 μm (*p < 0.05, **p < 0.01, ***p < 0.001; ns, not significant).
Discussion
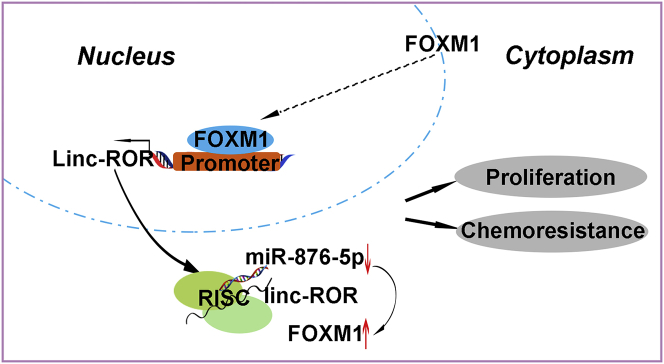
Dysregulation of lncRNAs has been demonstrated to contribute to malignant pathogenesis in many cancers.10, 12, 15 Although LINC-ROR was first identified as a key regulator of reprogramming, accumulating evidence has found aberrant regulation of LINC-ROR in different cancers.18, 23, 39, 40, 41 Furthermore, LINC-ROR has been proved to be an oncogene by facilitating the interaction of hnRNP I and AUF1 to enhance c-MYC mRNA stability.42 However, the roles of LINC-ROR in HCC have not been well understood. Our previous studies demonstrated that LINC-ROR promotes metastasis and radioresistance in HCC.24 In addition, high LINC-ROR correlates with advanced tumor node metastasis stage, high recurrence, and poor prognosis of HCC patients.25 Herein, we verified that LINC-ROR was upregulated in HCC cell lines, but not in a normal liver cell line. Mechanistically, we identified a positive-feedback loop, in which FOXM1 stimulated the transcription of LINC-ROR and LINC-ROR-regulated FOXM1 expression, then promoted HCC proliferation and hindered chemosensitivity to sorafenib via a ceRNA mechanism in HCC (Figure 7). Moreover, we performed immunofluorescence assays in L-02 cells and HCCLM3 cells, respectively. We found that FOXM1 localized in both the nucleus and the cytoplasm, whereas the nuclear FOXM1 was significantly enriched in the HCCLM3 cells (Figure S1E). Aside from this, after overexpressing FOXM1 in HepG2 cells, we found more nuclear gatherings of FOXM1 as well (Figure S1F). These results indicate that FOXM1 has important functions in the nucleus.
Figure 7.
Schematic Overview of FOXM1 and LINC-ROR Regulatory Signaling
LINC-ROR, which was activated by FOXM1 through promoter binding and transferred to the cytoplasm, then upregulated FOXM1 expression by competitively sponging miR-876-5p, forming a positive regulatory signaling pathway to promote proliferation and induce chemoresistance in HCC cells.
As a potent transcription factor, FOXM1 exerts functions by binding to the promoter of specific effector genes, such as CCNB1, AURKA, and c-MYC, and activating their transcription.27, 43, 44 It has been reported that lncRNA-PVT1 is actually a downstream lncRNA of FOXM1. In addition, PVT1 stabilizes FOXM1 protein.31 Moreover, c-MYC binds to the FOXM1 promoter in a TCPOBOP-dependent manner.45 Interestingly, Huang and colleagues42 reported that LINC-ROR interacts with hnRNP I and AUF1 and eventually enhances c-MYC mRNA stability, and our study suggested that FOXM1 stimulates the transcription of LINC-ROR. We propose that FOXM1 may stimulate the LINC-ROR promoter to elevate c-MYC expression and then elevate FOXM1 expression, thereby forming a complex feedback loop in the three genes.
A novel regulatory mechanism of lncRNAs has been described that they can interfere with miRNA pathways by competing for shared miRNA response elements and then affect post-transcriptional regulation. In this way, lncRNAs de-repress miRNA’s target gene expression and modulate downstream functions.16, 46 For example, lncRNA HOTAIR regulates HER2 expression by functioning as a sponge for miR-331-3p in gastric cancer.47 Similarly, LINC-ROR has been found to act as a miRNA sponge and was involved in lncRNA-miRNA-mRNA interaction in cancers, including HCC.20, 22, 25 In the present study, we found that overexpression of LINC-ROR resulted in decreased miR-876-5p expression in HepG2 cells and vice versa in HCCLM3 cells. As a tumor suppressor, miR-876-5p has been reported to exert suppressive functions in HCC progression by targeting several downstream genes.48, 49 Furthermore, a circular RNA, ciRS-7 has been reported to act as a miR-876-5p sponge to facilitate esophageal squamous cell carcinoma progression.50 In our study, we demonstrated that LINC-ROR could act as a sponge of miR-876-5p to release the expression of FOXM1.
Although the application of sorafenib as a treatment for HCC has achieved a good outcome, it is still challenging in treating those patients who are inherently nonresponsive. Therefore, it is of great significance to explore the mechanisms involved in sorafenib resistance and search for new powerful therapeutic targets. A previous study suggested that there are differences in the sensitivity of HCC cells to sorafenib and that some HCC cells may be spontaneously resistant to sorafenib.51 Our present investigation revealed that FOXM1 may attenuate the responsiveness to sorafenib partly through activating LINC-ROR, and in turn, LINC-ROR can impair sensitivity to sorafenib partially via the miR-876-5p-FOXM1 pathway. Considering that inhibiting FOXM1 with thiostrepton has been reported to inhibit tumor proliferation and angiogenesis and to induce apoptosis in different tumors,52, 53, 54 it may be that a potential combination of sorafenib and thiostrepton would synergistically inhibit HCC progression. This possibility needs to be explored further. Interestingly, in 2014, Takahashi et al.26 validated that LINC-ROR, which was enriched in the extracellular vesicles derived from HepG2 cells, transferred into recipient cells and thus participated in resistance to therapeutic agents in HCC cells, including sorafenib, partly depending on suppression of p53. This mechanism revealed the importance of tumor microenvironment in the functions of chemoresistance through signal transferring between tumor cells and recipient cells. Their research and our findings provide different mechanistic insights into resistance to sorafenib in HCC, which reveal that the formation of chemoresistance in human cancers involves different mechanisms including tumor cells themselves and their signal transmission in the tumor microenvironment.
In summary, our study identified the potential reciprocal link between LINC-ROR and FOXM1 and demonstrated that this correlation was involved in intrinsic resistance to sorafenib. These findings may help to improve the efficacy of sorafenib in HCC patients.
Materials and Methods
Cell Lines and Cell Culture
The cell lines L-02, HepG2, SMMC-7721, Huh-7, MHCC-97H, Hep3B, and HCCLM3 were cultured in DMEM (Gibco-BRL) supplemented with 10% FBS, streptomycin (100 μg/mL), and penicillin (100 U/mL). All cells were cultured at 37°C in an atmosphere containing 5% CO2. All the cell lines were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) where they were characterized by mycoplasma detection, DNA fingerprinting, isozyme detection, and cell vitality detection.
Quantitative Real-Time PCR
Total RNA from cells was isolated by using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Real-time PCR was performed with SYBR Prime Script RT-PCR Kits (Takara, Japan) based on the manufacturer’s instructions. The LINC-ROR, FOXM1, miR-129-5p, miR-153-3p, miR-152-3p, miR-876-5p, miR-214-3p, and miR-185-5p levels were detected with the 2−ΔΔCt method. GAPDH mRNA was employed as an endogenous control for mRNA and lncRNA, and U6 RNA was used as a miRNA internal control. For exact quantification of gene copies per cell, LINC-ROR and reverse-transcribed miR-145 cDNA were used as standard templates to formulate standard curves with limit dilution approaches. PCR primers are listed in Table S2.
Western Blot
Cells were lysed in RIPA lysis buffer and then lysate concentrations were detected with a bicinchoninic acid (BCA) kit (Thermo Scientific, Waltham, MA, USA). The protein lysates (50 μg/lane) were separated by 10% SDS-PAGE and were transferred to polyvinylidene difluoride membranes (Roche). Antibody dilutions of 1:1,000 were used for rabbit anti-FOXM1 (D3F2B) and mouse anti-GAPDH (D4C6R). The proteins were detected with an enhanced chemiluminescence system and exposed to X-ray film. All antibodies were purchased from Cell Signaling Technology (USA).
Cell Transfection
The FOXM1-overexpressing and shFOXM1-expressing plasmids were purchased from Ribobio (Guangzhou, China). HepG2 cells, transfected with FOXM1 plasmid into constructed cell lines stably expressing FOXM1, and HCCLM3 cells, transfected with shFOXM1 plasmid into constructed cell lines with stably silenced FOXM1, were screened with puromycin (2.0 μg/mL) for 4 weeks. The LINC-ROR-overexpressing plasmid was a gift from Dr. Y. Wang (College of Basic Medicine, Second Military Medical University, China). The short hairpin ROR (shROR)-expressing plasmid was a gift from Dr. P. Hou (The Institute of Genetics and Cytology, Northeast Normal University, Changchun, China). HepG2 cells transfected with LINC-ROR plasmid to constructed cell lines stably expressing LINC-ROR and HCCLM3 cells transfected with shROR plasmid to construct cell lines stably silencing LINC-ROR were screened with puromycin (2.0 μg/mL) for 4 weeks, as well. Has-miR-876-5p mimics and miR-NC/mimics and has-miR-876-5p inhibitor and miR-NC/inhibitor were purchased from Ribobio (Guangzhou, China). Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. All shRNA sequences are listed in Table S3.
Colony-Formation Assay
HCC cells were plated in 6-well culture plates at various cell densities (300–8,000 cells per well) for 6–8 h and were then were treated with sorafenib at different doses. After 10–14 days, cell colonies were stained by 0.1% crystal violet and scored by counting the number of colonies with an inverted microscope.
Flow Cytometry
Apoptosis was measured by an AnnexinV-7-AAD apoptosis detection kit (KeyGen BioTech, Nanjing, Jiangsu Province, China), according to the manufacturer’s instructions. The results were analyzed by fluorescence-activated cell sorting (FACS) cytometry (BD Biosciences, San Jose, CA, USA) to quantitatively measure the percentage of early apoptosis and late apoptosis cells. All of the analysis were performed in triplicate.
Cell Growth Assay
Cell viability was measured with CCK-8 (Dojindo, Japan), in accordance with the manufacturer’s protocol.
Preparation of Nuclear Extracts
The nuclear fraction of cells was extracted with a PARIS kit (AM1921; Ambion). Cells were washed three times with PBS on ice followed by centrifugation at 500 × g for 5 min. Subsequently, the cells were re-suspended in cell fraction buffer, incubated on ice for 10 min, and centrifuged at 500 × g for 5 min at 4°C. The supernatant was used for cytoplasmic RNA or protein purification. Nuclear fractions were washed with cell fraction buffer and finally homogenized with the cell disruption buffer from the kit.
ChIP Assay
We performed a ChIP assay, using the EZ ChIP Kit for cell line samples (Millipore) according to the manufacturer’s instruction. The primer sequences were (forward) 5′-GACCTTAACAGGCCCCATG-3′ and (reverse) 5′-ACGCCTTCCTCTTTGGGAC-3′.
RIP Assay
A RIP assay was performed with a kit (Thermo Fisher, USA), according to the manufacturer’s instructions. Ago2 antibody was purchased from Abcam (USA). Normal mouse IgG (Abcam, USA) was used as the negative control. Purified RNA was subjected to quantitative real-time PCR analysis.
Luciferase Reporter Assays
The pmirGLO, pmirGLO-WT, or pmirGLO-MUT for LINC-ROR and FOXM1 were co-transfected with miR-876-5p mimics or miRNA NC into HCCLM3 cells by using Lipofectamine-mediated gene transfer. Relative luciferase activity was normalized to Renilla luciferase activity at 48 h after transfection. The data are relative to the fold change of pair-matched control groups, which was defined as 1.0.
Immunofluorescence
L-02, HepG2, and HCCLM3 cells were seeded in chamber slides at 2.0 × 104 cells/well in DMEM with 10% FBS. After 24 h, the cells were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, blocked with 5% BSA, and stained with primary anti-FOXM1 antibody (D12D5; Cell Signaling Technology, USA) at 4°C overnight, followed by incubation with a fluorescent-dye-conjugated secondary antibody (Invitrogen) for 1 h, and then staining with DAPI. Images were taken with a Zeiss photomicroscope (Carl Zeiss, Oberkochen, Germany).
Mouse Xenograft Models
BALB/c nude mice (4–6 weeks old) were provided by the Animal Core Facility of Nanjing Medical University and housed in laminar flow cabinets under specific pathogen-free conditions. Approximately 5 × 106 HepG2-lv-vector cells and HepG2-lv-LINC-ROR cells were subcutaneously implanted into the right flank of nude mice (12 mice per group). Xenograft size was measured every 5 days and calculated by using the equation V (mm3) = (length × width2)/2. When the tumors grew to 50 mm3, the mice were randomly subdivided into four groups and received oral administration of sorafenib at a concentration of 0 or 60 mg/kg per day. All mice were sacrificed 30 days after the start of the sorafenib treatment, and the tumor tissues were weighed and used for subsequent experiments. Similarly, mice in xenograft models with stably FOXM1-overexpressing HepG2 cells were constructed and treated with sorafenib and then were sacrificed after 30 days of implantation. All animal experiments were performed with the approval of the Institutional Committee for Animal Research. The study protocol was also approved by the Committee on the Use of Live Animals in Research, Nanjing University (Nanjing, China).
Immunohistochemistry
Primary tumors were excised, paraffin embedded, and formalin fixed. H&E staining and Ki-67 immunostaining analyses were conducted, and FOXM1 expression and PCNA protein expression were evaluated according to the manufacturer’s instructions.
TUNEL Assay
In transplanted tumor tissues, apoptosis was measured by means of the TUNEL assay, which was carried out according to the kit (KeyGen BioTech, Nanjing, China) guidelines.
Statistical Analysis
SPSS version 21.0 software was applied for statistical analyses. The experimental results were expressed as means ± SD. Student’s t test or one-way ANOVA was used to detect the differences among groups. p values < 0.05 were considered statistically significant.
Author Contributions
Y.Z. and R.W. designed the research. Y.Z. and M.A. conducted the experiments and Y.Z. wrote the paper. H.Z., T.W., and B.F. supported the experiments and contributed in reagents/materials. R.W. provided the financial support, performed the data analysis, and helped to draft the manuscript. R.W. and X.C. supervised the laboratory work. All authors approved the final version and agreed to publish the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This project was supported by grants from the National Natural Science Foundation of China (81472266 and 81772196) and the Excellent Youth Foundation of Jiangsu Province, China (BK20140032).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.04.008.
Supplemental Information
References
- 1.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]; El-Serag, H.B., and Rudolph, K.L. (2007). Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557-2576. [DOI] [PubMed]
- 2.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]; Torre, L.A., Bray, F., Siegel, R.L., Ferlay, J., Lortet-Tieulent, J., and Jemal, A. (2015). Global cancer statistics, 2012. CA Cancer J. Clin. 65, 87-108. [DOI] [PubMed]
- 3.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]; Forner, A., Llovet, J.M., and Bruix, J. (2012). Hepatocellular carcinoma. Lancet 379, 1245-1255. [DOI] [PubMed]
- 4.Tejeda-Maldonado J., García-Juárez I., Aguirre-Valadez J., González-Aguirre A., Vilatobá-Chapa M., Armengol-Alonso A., Escobar-Penagos F., Torre A., Sánchez-Ávila J.F., Carrillo-Pérez D.L. Diagnosis and treatment of hepatocellular carcinoma: An update. World J. Hepatol. 2015;7:362–376. doi: 10.4254/wjh.v7.i3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tejeda-Maldonado, J., Garcia-Juarez, I., Aguirre-Valadez, J., Gonzalez-Aguirre, A., Vilatoba-Chapa, M., Armengol-Alonso, A., Escobar-Penagos, F., Torre, A., Sanchez-Avila, J.F., and Carrillo-Perez, D.L. (2015). Diagnosis and treatment of hepatocellular carcinoma: An update. World J. Hepatol. 7, 362-376. [DOI] [PMC free article] [PubMed]
- 5.SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]; SHARP Investigators Study Group (2008). Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378-390. [DOI] [PubMed]
- 6.Chen K.F., Chen H.L., Tai W.T., Feng W.C., Hsu C.H., Chen P.J., Cheng A.L. Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J. Pharmacol. Exp. Ther. 2011;337:155–161. doi: 10.1124/jpet.110.175786. [DOI] [PubMed] [Google Scholar]; Chen, K.F., Chen, H.L., Tai, W.T., Feng, W.C., Hsu, C.H., Chen, P.J., and Cheng, A.L. (2011). Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J. Pharmacol. Exp. Ther. 337, 155-161. [DOI] [PubMed]
- 7.Cheng A.L., Kang Y.K., Chen Z., Tsao C.J., Qin S., Kim J.S., Luo R., Feng J., Ye S., Yang T.S. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]; Cheng, A.L., Kang, Y.K., Chen, Z., Tsao, C.J., Qin, S., Kim, J.S., Luo, R., Feng, J., Ye, S., Yang, T.S., et al. (2009). Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 10, 25-34. [DOI] [PubMed]
- 8.Keating G.M. Sorafenib: A Review in Hepatocellular Carcinoma. Target. Oncol. 2017;12:243–253. doi: 10.1007/s11523-017-0484-7. [DOI] [PubMed] [Google Scholar]; Keating, G.M. (2017). Sorafenib: A Review in Hepatocellular Carcinoma. Target. Oncol. 12, 243-253. [DOI] [PubMed]
- 9.Villanueva A., Llovet J.M. Targeted therapies for hepatocellular carcinoma. Gastroenterology. 2011;140:1410–1426. doi: 10.1053/j.gastro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; Villanueva, A., and Llovet, J.M. (2011). Targeted therapies for hepatocellular carcinoma. Gastroenterology 140, 1410-1426. [DOI] [PMC free article] [PubMed]
- 10.Wilusz J.E., Sunwoo H., Spector D.L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wilusz, J.E., Sunwoo, H., and Spector, D.L. (2009). Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23, 1494-1504. [DOI] [PMC free article] [PubMed]
- 11.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]; Geisler, S., and Coller, J. (2013). RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699-712. [DOI] [PMC free article] [PubMed]
- 12.Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]; Mercer, T.R., Dinger, M.E., and Mattick, J.S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155-159. [DOI] [PubMed]
- 13.Adams B.D., Parsons C., Walker L., Zhang W.C., Slack F.J. Targeting noncoding RNAs in disease. J. Clin. Invest. 2017;127:761–771. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]; Adams, B.D., Parsons, C., Walker, L., Zhang, W.C., and Slack, F.J. (2017). Targeting noncoding RNAs in disease. J. Clin. Invest. 127, 761-771. [DOI] [PMC free article] [PubMed]
- 14.Kopp F., Mendell J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kopp, F., and Mendell, J.T. (2018). Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 172, 393-407. [DOI] [PMC free article] [PubMed]
- 15.Huo X., Han S., Wu G., Latchoumanin O., Zhou G., Hebbard L., George J., Qiao L. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol. Cancer. 2017;16:165. doi: 10.1186/s12943-017-0734-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huo, X., Han, S., Wu, G., Latchoumanin, O., Zhou, G., Hebbard, L., George, J., and Qiao, L. (2017). Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol. Cancer 16, 165. [DOI] [PMC free article] [PubMed]
- 16.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P.P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353-358. [DOI] [PMC free article] [PubMed]
- 17.Juan L., Wang G., Radovich M., Schneider B.P., Clare S.E., Wang Y., Liu Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med. Genomics. 2013;6(Suppl 1):S7. doi: 10.1186/1755-8794-6-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Juan, L., Wang, G., Radovich, M., Schneider, B.P., Clare, S.E., Wang, Y., and Liu, Y. (2013). Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC Med. Genomics 6 (Suppl 1), S7. [DOI] [PMC free article] [PubMed]
- 18.Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H., Garber M., Curran M., Onder T., Agarwal S. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]; Loewer, S., Cabili, M.N., Guttman, M., Loh, Y.H., Thomas, K., Park, I.H., Garber, M., Curran, M., Onder, T., Agarwal, S., et al. (2010). Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113-1117. [DOI] [PMC free article] [PubMed]
- 19.Feng L., Shi L., Lu Y.F., Wang B., Tang T., Fu W.M., He W., Li G., Zhang J.F. Linc-ROR Promotes Osteogenic Differentiation of Mesenchymal Stem Cells by Functioning as a Competing Endogenous RNA for miR-138 and miR-145. Mol. Ther. Nucleic Acids. 2018;11:345–353. doi: 10.1016/j.omtn.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feng, L., Shi, L., Lu, Y.F., Wang, B., Tang, T., Fu, W.M., He, W., Li, G., and Zhang, J.F. (2018). Linc-ROR Promotes Osteogenic Differentiation of Mesenchymal Stem Cells by Functioning as a Competing Endogenous RNA for miR-138 and miR-145. Mol. Ther. Nucleic Acids 11, 345-353. [DOI] [PMC free article] [PubMed]
- 20.Wang L., Yu X., Zhang Z., Pang L., Xu J., Jiang J., Liang W., Chai Y., Hou J., Li F. Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J. Exp. Clin. Cancer Res. 2017;36:182. doi: 10.1186/s13046-017-0658-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang, L., Yu, X., Zhang, Z., Pang, L., Xu, J., Jiang, J., Liang, W., Chai, Y., Hou, J., and Li, F. (2017). Linc-ROR promotes esophageal squamous cell carcinoma progression through the derepression of SOX9. J. Exp. Clin. Cancer Res. 36, 182. [DOI] [PMC free article] [PubMed]
- 21.Zhan H.X., Wang Y., Li C., Xu J.W., Zhou B., Zhu J.K., Han H.F., Wang L., Wang Y.S., Hu S.Y. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016;374:261–271. doi: 10.1016/j.canlet.2016.02.018. [DOI] [PubMed] [Google Scholar]; Zhan, H.X., Wang, Y., Li, C., Xu, J.W., Zhou, B., Zhu, J.K., Han, H.F., Wang, L., Wang, Y.S., and Hu, S.Y. (2016). LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 374, 261-271. [DOI] [PubMed]
- 22.Eades G., Wolfson B., Zhang Y., Li Q., Yao Y., Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol. Cancer Res. 2015;13:330–338. doi: 10.1158/1541-7786.MCR-14-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]; Eades, G., Wolfson, B., Zhang, Y., Li, Q., Yao, Y., and Zhou, Q. (2015). lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol. Cancer Res. 13, 330-338. [DOI] [PMC free article] [PubMed]
- 23.Peng W.X., Huang J.G., Yang L., Gong A.H., Mo Y.-Y. Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer. 2017;16:161. doi: 10.1186/s12943-017-0727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; Peng, W.X., Huang, J.G., Yang, L., Gong, A.H., and Mo, Y.-Y. (2017). Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol. Cancer 16, 161. [DOI] [PMC free article] [PubMed]
- 24.Chen Y., Shen Z., Zhi Y., Zhou H., Zhang K., Wang T., Feng B., Chen Y., Song H., Wang R., Chu X. Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch. Biochem. Biophys. 2018;645:117–125. doi: 10.1016/j.abb.2018.03.018. [DOI] [PubMed] [Google Scholar]; Chen, Y., Shen, Z., Zhi, Y., Zhou, H., Zhang, K., Wang, T., Feng, B., Chen, Y., Song, H., Wang, R., and Chu, X. (2018). Long non-coding RNA ROR promotes radioresistance in hepatocelluar carcinoma cells by acting as a ceRNA for microRNA-145 to regulate RAD18 expression. Arch. Biochem. Biophys. 645, 117-125. [DOI] [PubMed]
- 25.Li C., Lu L., Feng B., Zhang K., Han S., Hou D., Chen L., Chu X., Wang R. The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci. Rep. 2017;7:4637. doi: 10.1038/s41598-017-04113-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; Li, C., Lu, L., Feng, B., Zhang, K., Han, S., Hou, D., Chen, L., Chu, X., and Wang, R. (2017). The lincRNA-ROR/miR-145 axis promotes invasion and metastasis in hepatocellular carcinoma via induction of epithelial-mesenchymal transition by targeting ZEB2. Sci. Rep. 7, 4637. [DOI] [PMC free article] [PubMed] [Retracted]
- 26.Takahashi K., Yan I.K., Kogure T., Haga H., Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; Takahashi, K., Yan, I.K., Kogure, T., Haga, H., and Patel, T. (2014). Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 4, 458-467. [DOI] [PMC free article] [PubMed]
- 27.Raychaudhuri P., Park H.J. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]; Raychaudhuri, P., and Park, H.J. (2011). FoxM1: a master regulator of tumor metastasis. Cancer Res. 71, 4329-4333. [DOI] [PMC free article] [PubMed]
- 28.Weiler S.M.E., Pinna F., Wolf T., Lutz T., Geldiyev A., Sticht C., Knaub M., Thomann S., Bissinger M., Wan S. Induction of Chromosome Instability by Activation of Yes-Associated Protein and Forkhead Box M1 in Liver Cancer. Gastroenterology. 2017;152:2037–2051.e22. doi: 10.1053/j.gastro.2017.02.018. [DOI] [PubMed] [Google Scholar]; Weiler, S.M.E., Pinna, F., Wolf, T., Lutz, T., Geldiyev, A., Sticht, C., Knaub, M., Thomann, S., Bissinger, M., Wan, S., et al. (2017). Induction of Chromosome Instability by Activation of Yes-Associated Protein and Forkhead Box M1 in Liver Cancer. Gastroenterology 152, 2037-2051.e22. [DOI] [PubMed]
- 29.Song B.-N., Chu I.-S. A gene expression signature of FOXM1 predicts the prognosis of hepatocellular carcinoma. Exp. Mol. Med. 2018;50:e418. doi: 10.1038/emm.2017.159. [DOI] [PMC free article] [PubMed] [Google Scholar]; Song, B.-N., and Chu, I.-S. (2018). A gene expression signature of FOXM1 predicts the prognosis of hepatocellular carcinoma. Exp. Mol. Med. 50, e418. [DOI] [PMC free article] [PubMed]
- 30.Xia L., Huang W., Tian D., Zhu H., Zhang Y., Hu H., Fan D., Nie Y., Wu K. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J. Hepatol. 2012;57:600–612. doi: 10.1016/j.jhep.2012.04.020. [DOI] [PubMed] [Google Scholar]; Xia, L., Huang, W., Tian, D., Zhu, H., Zhang, Y., Hu, H., Fan, D., Nie, Y., and Wu, K. (2012). Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J. Hepatol. 57, 600-612. [DOI] [PubMed]
- 31.Xu M.D., Wang Y., Weng W., Wei P., Qi P., Zhang Q., Tan C., Ni S.J., Dong L., Yang Y. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin. Cancer Res. 2017;23:2071–2080. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]; Xu, M.D., Wang, Y., Weng, W., Wei, P., Qi, P., Zhang, Q., Tan, C., Ni, S.J., Dong, L., Yang, Y., et al. (2017). A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin. Cancer Res. 23, 2071-2080. [DOI] [PubMed]
- 32.Wang K., Zhu X., Zhang K., Zhu L., Zhou F. FoxM1 inhibition enhances chemosensitivity of docetaxel-resistant A549 cells to docetaxel via activation of JNK/mitochondrial pathway. Acta Biochim. Biophys. Sin. (Shanghai) 2016;48:804–809. doi: 10.1093/abbs/gmw072. [DOI] [PubMed] [Google Scholar]; Wang, K., Zhu, X., Zhang, K., Zhu, L., and Zhou, F. (2016). FoxM1 inhibition enhances chemosensitivity of docetaxel-resistant A549 cells to docetaxel via activation of JNK/mitochondrial pathway. Acta Biochim. Biophys. Sin. (Shanghai) 48, 804-809. [DOI] [PubMed]
- 33.Li X., Yao R., Yue L., Qiu W., Qi W., Liu S., Yao Y., Liang J. FOXM1 mediates resistance to docetaxel in gastric cancer via up-regulating Stathmin. J. Cell. Mol. Med. 2014;18:811–823. doi: 10.1111/jcmm.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, X., Yao, R., Yue, L., Qiu, W., Qi, W., Liu, S., Yao, Y., and Liang, J. (2014). FOXM1 mediates resistance to docetaxel in gastric cancer via up-regulating Stathmin. J. Cell. Mol. Med. 18, 811-823. [DOI] [PMC free article] [PubMed]
- 34.Zhang J.R., Lu F., Lu T., Dong W.H., Li P., Liu N., Ma D.X., Ji C.Y. Inactivation of FoxM1 transcription factor contributes to curcumin-induced inhibition of survival, angiogenesis, and chemosensitivity in acute myeloid leukemia cells. J. Mol. Med. (Berl.) 2014;92:1319–1330. doi: 10.1007/s00109-014-1198-2. [DOI] [PubMed] [Google Scholar]; Zhang, J.R., Lu, F., Lu, T., Dong, W.H., Li, P., Liu, N., Ma, D.X., and Ji, C.Y. (2014). Inactivation of FoxM1 transcription factor contributes to curcumin-induced inhibition of survival, angiogenesis, and chemosensitivity in acute myeloid leukemia cells. J. Mol. Med. (Berl.) 92, 1319-1330. [DOI] [PubMed]
- 35.Wang J.Y., Jia X.H., Xing H.Y., Li Y.J., Fan W.W., Li N., Xie S.Y. Inhibition of Forkhead box protein M1 by thiostrepton increases chemosensitivity to doxorubicin in T-cell acute lymphoblastic leukemia. Mol. Med. Rep. 2015;12:1457–1464. doi: 10.3892/mmr.2015.3469. [DOI] [PubMed] [Google Scholar]; Wang, J.Y., Jia, X.H., Xing, H.Y., Li, Y.J., Fan, W.W., Li, N., and Xie, S.Y. (2015). Inhibition of Forkhead box protein M1 by thiostrepton increases chemosensitivity to doxorubicin in T-cell acute lymphoblastic leukemia. Mol. Med. Rep. 12, 1457-1464. [DOI] [PubMed]
- 36.Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L., Wu M., Xiong J., Guo X., Liu H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]; Wang, Y., Xu, Z., Jiang, J., Xu, C., Kang, J., Xiao, L., Wu, M., Xiong, J., Guo, X., and Liu, H. (2013). Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 25, 69-80. [DOI] [PubMed]
- 37.Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Reczko M., Maragkakis M., Dalamagas T.M., Hatzigeorgiou A.G. DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41:D239–D245. doi: 10.1093/nar/gks1246. [DOI] [PMC free article] [PubMed] [Google Scholar]; Paraskevopoulou, M.D., Georgakilas, G., Kostoulas, N., Reczko, M., Maragkakis, M., Dalamagas, T.M., and Hatzigeorgiou, A.G. (2013). DIANA-LncBase: experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 41, D239-D245. [DOI] [PMC free article] [PubMed]
- 38.Li J.-H., Liu S., Zhou H., Qu L.-H., Yang J.-H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, J.-H., Liu, S., Zhou, H., Qu, L.-H., and Yang, J.-H. (2014). starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92-D97. [DOI] [PMC free article] [PubMed]
- 39.Hou P., Zhao Y., Li Z., Yao R., Ma M., Gao Y., Zhao L., Zhang Y., Huang B., Lu J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014;5:e1287. doi: 10.1038/cddis.2014.249. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hou, P., Zhao, Y., Li, Z., Yao, R., Ma, M., Gao, Y., Zhao, L., Zhang, Y., Huang, B., and Lu, J. (2014). LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 5, e1287. [DOI] [PMC free article] [PubMed]
- 40.Zhang R., Hardin H., Huang W., Buehler D., Lloyd R.V. Long Non-coding RNA Linc-ROR Is Upregulated in Papillary Thyroid Carcinoma. Endocr. Pathol. 2018;29:1–8. doi: 10.1007/s12022-017-9507-2. [DOI] [PubMed] [Google Scholar]; Zhang, R., Hardin, H., Huang, W., Buehler, D., and Lloyd, R.V. (2018). Long Non-coding RNA Linc-ROR Is Upregulated in Papillary Thyroid Carcinoma. Endocr. Pathol. 29, 1-8. [DOI] [PubMed]
- 41.Li L., Gu M., You B., Shi S., Shan Y., Bao L., You Y. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016;107:1215–1222. doi: 10.1111/cas.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li, L., Gu, M., You, B., Shi, S., Shan, Y., Bao, L., and You, Y. (2016). Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 107, 1215-1222. [DOI] [PMC free article] [PubMed]
- 42.Huang J., Zhang A., Ho T.T., Zhang Z., Zhou N., Ding X., Zhang X., Xu M., Mo Y.Y. Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 2016;44:3059–3069. doi: 10.1093/nar/gkv1353. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang, J., Zhang, A., Ho, T.T., Zhang, Z., Zhou, N., Ding, X., Zhang, X., Xu, M., and Mo, Y.Y. (2016). Linc-RoR promotes c-Myc expression through hnRNP I and AUF1. Nucleic Acids Res. 44, 3059-3069. [DOI] [PMC free article] [PubMed]
- 43.Zeng J., Wang L., Li Q., Li W., Björkholm M., Jia J., Xu D. FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27 kip1. J. Pathol. 2009;218:419–427. doi: 10.1002/path.2530. [DOI] [PubMed] [Google Scholar]; Zeng, J., Wang, L., Li, Q., Li, W., Bjorkholm, M., Jia, J., and Xu, D. (2009). FoxM1 is up-regulated in gastric cancer and its inhibition leads to cellular senescence, partially dependent on p27 kip1. J. Pathol. 218, 419-427. [DOI] [PubMed]
- 44.Yang N., Wang C., Wang Z., Zona S., Lin S.X., Wang X., Yan M., Zheng F.M., Li S.S., Xu B. FOXM1 recruits nuclear Aurora kinase A to participate in a positive feedback loop essential for the self-renewal of breast cancer stem cells. Oncogene. 2017;36:3428–3440. doi: 10.1038/onc.2016.490. [DOI] [PMC free article] [PubMed] [Google Scholar]; Yang, N., Wang, C., Wang, Z., Zona, S., Lin, S.X., Wang, X., Yan, M., Zheng, F.M., Li, S.S., Xu, B., et al. (2017). FOXM1 recruits nuclear Aurora kinase A to participate in a positive feedback loop essential for the self-renewal of breast cancer stem cells. Oncogene 36, 3428-3440. [DOI] [PMC free article] [PubMed]
- 45.Blanco-Bose W.E., Murphy M.J., Ehninger A., Offner S., Dubey C., Huang W., Moore D.D., Trumpp A. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008;48:1302–1311. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]; Blanco-Bose, W.E., Murphy, M.J., Ehninger, A., Offner, S., Dubey, C., Huang, W., Moore, D.D., and Trumpp, A. (2008). C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology 48, 1302-1311. [DOI] [PubMed]
- 46.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tay, Y., Rinn, J., and Pandolfi, P.P. (2014). The multilayered complexity of ceRNA crosstalk and competition. Nature 505, 344-352. [DOI] [PMC free article] [PubMed]
- 47.Liu X.H., Sun M., Nie F.Q., Ge Y.B., Zhang E.B., Yin D.D., Kong R., Xia R., Lu K.H., Li J.H. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu, X.H., Sun, M., Nie, F.Q., Ge, Y.B., Zhang, E.B., Yin, D.D., Kong, R., Xia, R., Lu, K.H., Li, J.H., et al. (2014). Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 13, 92. [DOI] [PMC free article] [PubMed]
- 48.Xu Q., Zhu Q., Zhou Z., Wang Y., Liu X., Yin G., Tong X., Tu K. MicroRNA-876-5p inhibits epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by targeting BCL6 corepressor like 1. Biomed. Pharmacother. 2018;103:645–652. doi: 10.1016/j.biopha.2018.04.037. [DOI] [PubMed] [Google Scholar]; Xu, Q., Zhu, Q., Zhou, Z., Wang, Y., Liu, X., Yin, G., Tong, X., and Tu, K. (2018). MicroRNA-876-5p inhibits epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by targeting BCL6 corepressor like 1. Biomed. Pharmacother. 103, 645-652. [DOI] [PubMed]
- 49.Wang Y., Xie Y., Li X., Lin J., Zhang S., Li Z., Huo L., Gong R. MiR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol. Res. Pract. 2018;214:1024–1030. doi: 10.1016/j.prp.2018.04.012. [DOI] [PubMed] [Google Scholar]; Wang, Y., Xie, Y., Li, X., Lin, J., Zhang, S., Li, Z., Huo, L., and Gong, R. (2018). MiR-876-5p acts as an inhibitor in hepatocellular carcinoma progression by targeting DNMT3A. Pathol. Res. Pract. 214, 1024-1030. [DOI] [PubMed]
- 50.Sang M., Meng L., Sang Y., Liu S., Ding P., Ju Y., Liu F., Gu L., Lian Y., Li J. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 2018;426:37–46. doi: 10.1016/j.canlet.2018.03.049. [DOI] [PubMed] [Google Scholar]; Sang, M., Meng, L., Sang, Y., Liu, S., Ding, P., Ju, Y., Liu, F., Gu, L., Lian, Y., Li, J., et al. (2018). Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett. 426, 37-46. [DOI] [PubMed]
- 51.Ezzoukhry Z., Louandre C., Trécherel E., Godin C., Chauffert B., Dupont S., Diouf M., Barbare J.C., Mazière J.C., Galmiche A. EGFR activation is a potential determinant of primary resistance of hepatocellular carcinoma cells to sorafenib. Int. J. Cancer. 2012;131:2961–2969. doi: 10.1002/ijc.27604. [DOI] [PubMed] [Google Scholar]; Ezzoukhry, Z., Louandre, C., Trecherel, E., Godin, C., Chauffert, B., Dupont, S., Diouf, M., Barbare, J.C., Maziere, J.C., and Galmiche, A. (2012). EGFR activation is a potential determinant of primary resistance of hepatocellular carcinoma cells to sorafenib. Int. J. Cancer 131, 2961-2969. [DOI] [PubMed]
- 52.Ju S.Y., Huang C.Y., Huang W.C., Su Y. Identification of thiostrepton as a novel therapeutic agent that targets human colon cancer stem cells. Cell Death Dis. 2015;6:e1801. doi: 10.1038/cddis.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ju, S.Y., Huang, C.Y., Huang, W.C., and Su, Y. (2015). Identification of thiostrepton as a novel therapeutic agent that targets human colon cancer stem cells. Cell Death Dis. 6, e1801. [DOI] [PMC free article] [PubMed]
- 53.Jiang L., Wu X., Wang P., Wen T., Yu C., Wei L., Chen H. Targeting FoxM1 by thiostrepton inhibits growth and induces apoptosis of laryngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2015;141:971–981. doi: 10.1007/s00432-014-1872-3. [DOI] [PubMed] [Google Scholar]; Jiang, L., Wu, X., Wang, P., Wen, T., Yu, C., Wei, L., and Chen, H. (2015). Targeting FoxM1 by thiostrepton inhibits growth and induces apoptosis of laryngeal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 141, 971-981. [DOI] [PubMed]
- 54.Jiang L., Wang P., Chen L., Chen H. Down-regulation of FoxM1 by thiostrepton or small interfering RNA inhibits proliferation, transformation ability and angiogenesis, and induces apoptosis of nasopharyngeal carcinoma cells. Int. J. Clin. Exp. Pathol. 2014;7:5450–5460. [PMC free article] [PubMed] [Google Scholar]; Jiang, L., Wang, P., Chen, L., and Chen, H. (2014). Down-regulation of FoxM1 by thiostrepton or small interfering RNA inhibits proliferation, transformation ability and angiogenesis, and induces apoptosis of nasopharyngeal carcinoma cells. Int. J. Clin. Exp. Pathol. 7, 5450-5460. [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.